Unravelling the Diagnostic Dilemma: A MicroRNA Panel of Circulating MiR-16 and MiR-877 as A Diagnostic Classifier for Distal Bile Duct Tumors
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
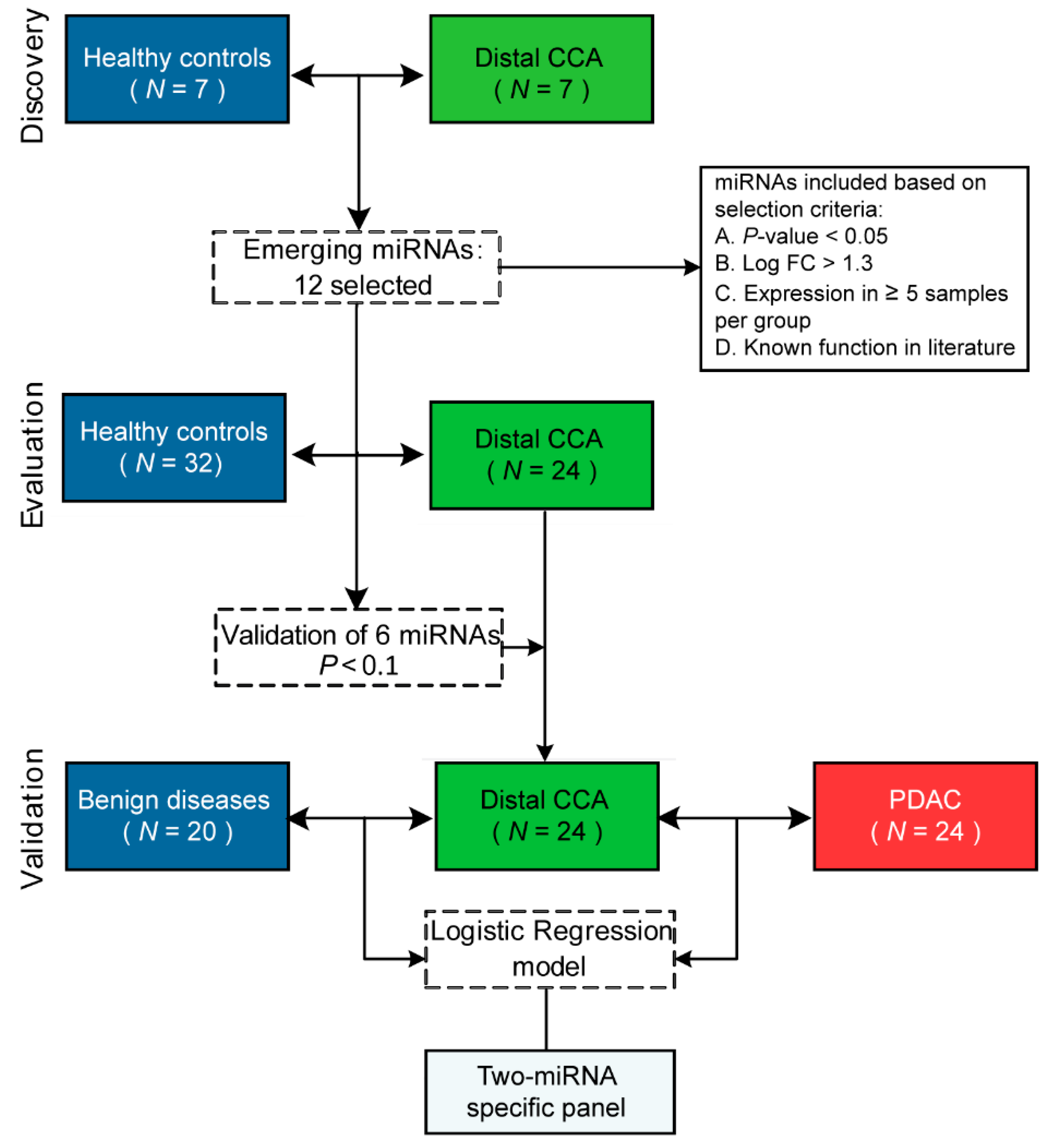
2.2. PCR Panel Profiling of MiRNAs in the Discovery Phase
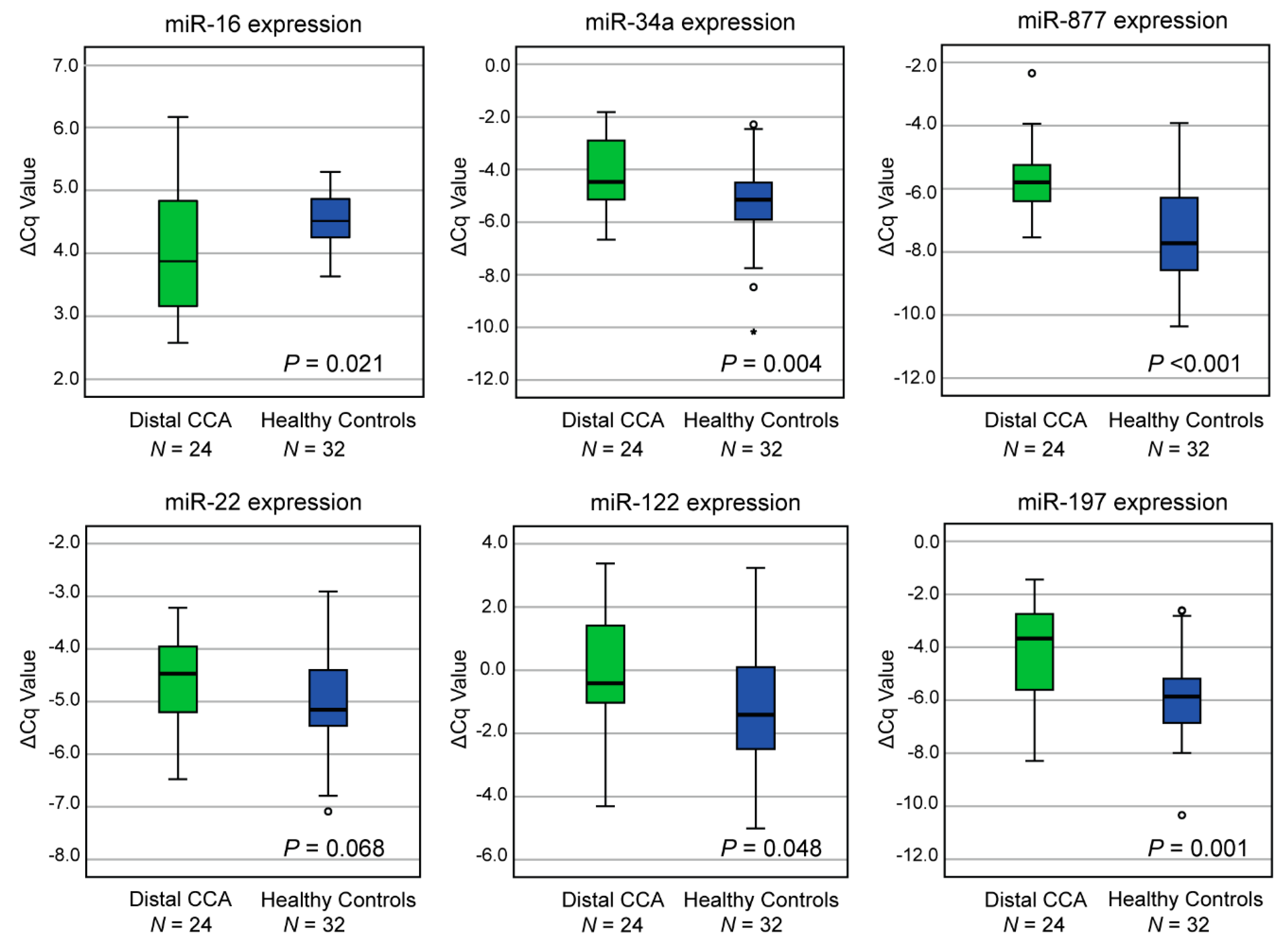
2.3. Evaluation of MiRNA Expression in an Independent Cohort Of Distal CCA and Healthy Individuals Reveals Six Dysregulated MiRNAs
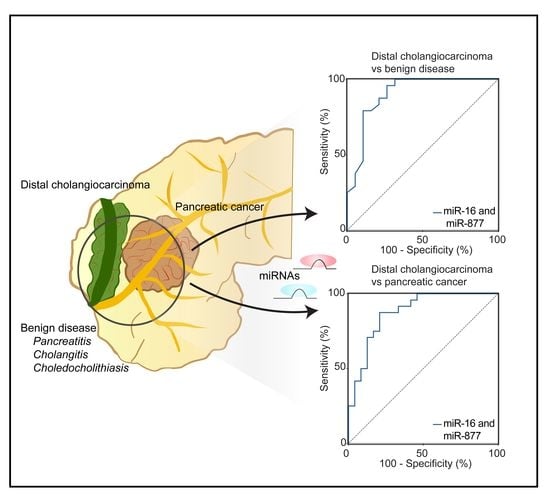
2.4. Diagnostic Performance of the Optimal MiRNA Panel in the Validation Phase of Distal CCA and BD
2.5. Validation of MiRNA-Based Differentiation between Distal CCA and PDAC
2.6. Prediction of Target Genes of MiR-16 and MiR-877
2.7. Influence of Bilirubin on the Diagnostic Accuracy of the Two-MiRNA Panel
2.8. Expression of MiR-877 and MiR-16 in Plasma Samples of Patients with Distal CCA Compared to Perihilar CCA and Intrahepatic CCA
3. Discussion
4. Materials and Methods
4.1. Study Design and Patients
4.2. Sample Collection and MiRNA Expression
4.3. Detection of CA19–9 and Bilirubin Levels
4.4. Statistical Analysis
4.5. Prediction of Target Genes of Emerging MiRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bledsoe, J.R.; Shinagare, S.A.; Deshpande, V. Difficult Diagnostic Problems in Pancreatobiliary Neoplasia. Arch. Pathol. Lab. Med. 2014, 139, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Blechacz, B.; Komuta, M.; Roskams, T.; Gores, G.J. Clinical diagnosis and staging of cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 512–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ethun, C.G.; Lopez-Aguiar, A.G.; Pawlik, T.M.; Poultsides, G.; Idrees, K.; Fields, R.C.; Weber, S.M.; Cho, C.; Martin, R.C.; Scoggins, C.R.; et al. Distal Cholangiocarcinoma and Pancreas Adenocarcinoma: Are They Really the Same Disease? A 13-Institution Study from the US Extrahepatic Biliary Malignancy Consortium and the Central Pancreas Consortium. J. Am. Coll. Surg. 2017, 224, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Caparello, C.; Meijer, L.L.; Garajova, I.; Falcone, A.; Le Large, T.Y.; Funel, N.; Kazemier, G.; Peters, G.J.; Vasile, E.; Giovannetti, E. Folfirinox and translational studies: Towards personalized therapy in pancreatic cancer. World J. Gastroenterol. 2016, 22, 6987–7005. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.W.; Furuse, J.; Jitlal, M.; Beare, S.; Mizuno, N.; Wasan, H.; Bridgewater, J.; Okusaka, T. Cisplatin and gemcitabine for advanced biliary tract cancer: A meta-analysis of two randomised trials. Ann. Oncol. 2014, 25, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.; Gores, G.J. Emerging molecular therapeutic targets for cholangiocarcinoma. J. Hepatol. 2017, 67, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Wood, L.D.; Suzuki, M.; Takai, E.; Totoki, Y.; Kato, M.; Luchini, C.; Arai, Y.; Nakamura, H.; Hama, N.; et al. Genomic Sequencing Identifies ELF3 as a Driver of Ampullary Carcinoma. Cancer Cell 2016, 29, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; De La Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Korc, P.; Sherman, S. ERCP tissue sampling. Gastrointest Endosc 2016, 84, 557–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weynand, B.; Deprez, P. Endoscopic ultrasound guided fine needle aspiration in biliary and pacreatic diseases: Pitfalls and performances. Acta Gastro Enterol. Belg. 2004, 67, 294–300. [Google Scholar]
- Khan, S.A.; Davidson, B.R.; Goldin, R.D.; Heaton, N.; Karani, J.; Pereira, S.P.; Rosenberg, W.M.; Tait, P.; Taylor-Robinson, S.D.; Thillainayagam, A.V.; et al. British Society of Gastroenterology, Guidelines for the diagnosis and treatment of cholangiocarcinoma: An update. Gut 2012, 61, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.R.; Gurusamy, K. Is preoperative histological diagnosis necessary for cholangiocarcinoma? HPB 2008, 10, 94–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soer, E.; Brosens, L.; Van De Vijver, M.; Dijk, F.; Van Velthuysen, M.L.; Farina-Sarasqueta, A.; Morreau, H.; Offerhaus, J.; Koens, L.; Verheij, J. Dilemmas for the pathologist in the oncologic assessment of pancreatoduodenectomy specimens: An overview of different grossing approaches and the relevance of the histopathological characteristics in the oncologic assessment of pancreatoduodenectomy specimens. Virchows Arch. 2018, 472, 533–543. [Google Scholar] [PubMed]
- Pomianowska, E.; Grzyb, K.; Westgaard, A.; Clausen, O.P.; Gladhaug, I.P. Reclassification of tumour origin in resected periampullary adenocarcinomas reveals underestimation of distal bile duct cancer. Eur. J. Surg. Oncol. 2012, 38, 1043–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballehaninna, U.K.; Chamberlain, R.S. The clinical utility of serum CA 19–9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J. Gastrointest. Oncol. 2012, 3, 105–119. [Google Scholar]
- Marrelli, D.; Caruso, S.; Pedrazzani, C.; Neri, A.; Fernandes, E.; Marini, M.; Pinto, E.; Roviello, F. CA19–9 serum levels in obstructive jaundice: Clinical value in benign and malignant conditions. Am. J. Surg. 2009, 198, 333–339. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, C.J.; Farid, W.R.; De Jonge, J.; Metselaar, H.J.; Kazemier, G.; Van Der Laan, L.J. Biomarkers to assess graft quality during conventional and machine preservation in liver transplantation. J. Hepatol. 2014, 61, 672–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Liu, S.-M.; Huang, J.-T.; Zhang, X.; Yan, D.; Xia, Q.; Ji, C.; Chen, W.; Zhang, X.; Xu, J.; et al. Clinically relevant circulating microRNA profiling studies in pancreatic cancer using meta-analysis. Oncotarget 2017, 8, 22616–22624. [Google Scholar] [CrossRef] [Green Version]
- Correa-Gallego, C.; Maddalo, D.; Doussot, A.; Kemeny, N.; Kingham, T.P.; Allen, P.J.; D’Angelica, M.I.; DeMatteo, R.P.; Betel, D.; Klimstra, D.; et al. Circulating Plasma Levels of MicroRNA-21 and MicroRNA-221 Are Potential Diagnostic Markers for Primary Intrahepatic Cholangiocarcinoma. PLoS ONE 2016, 11, e0163699. [Google Scholar] [CrossRef] [PubMed]
- Letelier, P.; Riquelme, I.; Hernandez, A.H.; Guzman, N.; Farias, J.G.; Roa, J.C. Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers. Int. J. Mol. Sci. 2016, 17, 791. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Jeong, S.; Zhu, Y.; Chen, L.; Xia, Q. miRNA and lncRNA as biomarkers in cholangiocarcinoma(CCA). Oncotarget 2017, 8, 100819–100830. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H.; Da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Liao, B.Y.; Yu, L.; Gao, X.; Lu, S.; Wang, S.; Dai, Z.; Zhang, X.; Chen, Q.; et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int. J. Cancer 2014, 135, 1187–1194. [Google Scholar] [CrossRef]
- Niu, Y.; Wu, Y.; Huang, J.; Li, W.; Kang, K.; Qu, J.; Gou, D. Identification of reference genes for circulating microRNA analysis in colorectal cancer. Sci. Rep. 2016, 6, 35611. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Bai, Z.; Han, W.; Zhang, J.; Meng, H.; Bi, J.; Ma, X.; Han, S.; Zhang, Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig. Dis. Sci. 2012, 57, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, P.; Dong, Y.; Cai, X.; Hou, D.; Guo, J.; Yin, Y.; Zhang, Y.; Li, J.; Liang, H.; et al. A microarray-based approach identifies ADP ribosylation factor-like protein 2 as a target of microRNA-16. J. Biol. Chem. 2011, 286, 9468–9476. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liang, H.; Deng, T.; Zhu, K.; Zhang, S.; Wang, N.; Jiang, X.; Wang, X.; Liu, R.; Zen, K.; et al. The identification of novel targets of miR-16 and characterization of their biological functions in cancer cells. Mol. Cancer 2013, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lu, X.; Liu, Z.; Chen, L.; Xu, Y.; Wang, Y.; Wei, G.; Chen, Y. FBXW7 suppresses epithelial-mesenchymal transition, stemness and metastatic potential of cholangiocarcinoma cells. Oncotarget 2015, 6, 6310–6325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, T.H.; Qiu, C.; Sun, J.; Li, W.H. MiR-877–5p suppresses cell growth, migration and invasion by targeting cyclin dependent kinase 14 and predicts prognosis in hepatocellular carcinoma. Eur. Rev. Med. Pharm. Sci. 2018, 22, 3038–3046. [Google Scholar]
- Schmuck, R.B.; De Carvalho-Fischer, C.V.; Neumann, C.; Pratschke, J.; Bahra, M. Distal bile duct carcinomas and pancreatic ductal adenocarcinomas: Postulating a common tumor entity. Cancer Med. 2016, 5, 88–99. [Google Scholar] [CrossRef]
- Ploeger, C.; Waldburger, N.; Fraas, A.; Goeppert, B.; Pusch, S.; Breuhahn, K.; Wang, X.W.; Schirmacher, P.; Roessler, S. Chromosome 8p tumor suppressor genes SH2D4A and SORBS3 cooperate to inhibit interleukin-6 signaling in hepatocellular carcinoma. Hepatology 2016, 64, 828–842. [Google Scholar] [CrossRef] [Green Version]
- Roessler, S.; Long, E.L.; Budhu, A.; Chen, Y.; Zhao, X.; Ji, J.; Walker, R.; Jia, H.L.; Ye, Q.H.; Qin, L.X.; et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 2012, 142, 957–966. [Google Scholar] [CrossRef]
- Yamaura, Y.; Nakajima, M.; Takagi, S.; Fukami, T.; Tsuneyama, K.; Yokoi, T. Plasma microRNA profiles in rat models of hepatocellular injury, cholestasis, and steatosis. PLoS ONE 2012, 7, e30250. [Google Scholar] [CrossRef]
- Puik, J.R.; Meijer, L.L.; Le Large, T.Y.; Prado, M.M.; Frampton, A.E.; Kazemier, G.; Giovannetti, E. miRNA profiling for diagnosis, prognosis and stratification of cancer treatment in cholangiocarcinoma. Pharmacogenomics 2017, 18, 1343–1358. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Arai, Y.; Totoki, Y.; Shirota, T.; Elzawahry, A.; Kato, M.; Hama, N.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Genomic spectra of biliary tract cancer. Nat. Genet. 2015, 47, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Komuta, M.; Govaere, O.; Vandecaveye, V.; Akiba, J.; Van Steenbergen, W.; Verslype, C.; Laleman, W.; Pirenne, J.; Aerts, R.; Yano, H.; et al. Histological diversity in cholangiocellular carcinoma reflects the different cholangiocyte phenotypes. Hepatology 2012, 55, 1876–1888. [Google Scholar] [CrossRef] [PubMed]
- Jusakul, A.; Cutcutache, I.; Yong, C.H.; Lim, J.Q.; Huang, M.N.; Padmanabhan, N.; Nellore, V.; Kongpetch, S.; Ng, A.W.T.; Ng, L.M.; et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017, 7, 1116–1135. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, D.; Tang, G.; Yang, X.; Jiao, C.; Yang, R.; Zhang, Y.; Huo, L.; Shao, Z.; Lu, Z.; et al. Suppression of miR-16 promotes tumor growth and metastasis through reversely regulating YAP1 in human cholangiocarcinoma. Oncotarget 2017, 8, 56635–56650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, M.; Sudo, H.; Kawauchi, J.; Takizawa, S.; Kondou, S.; Nobumasa, H.; Ochiai, A. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS ONE 2015, 10, e0118220. [Google Scholar] [CrossRef] [PubMed]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16–1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Bonci, D.; Coppola, V.; Musumeci, M.; Addario, A.; Giuffrida, R.; Memeo, L.; D’Urso, L.; Pagliuca, A.; Biffoni, M.; Labbaye, C.; et al. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008, 14, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010, 251, 499–505. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Sandanayake, N.S.; Sinclair, J.; Andreola, F.; Chapman, M.H.; Xue, A.; Webster, G.J.; Clarkson, A.; Gill, A.; Norton, I.D.; Smith, R.C.; et al. A combination of serum leucine-rich alpha-2-glycoprotein 1, CA19–9 and interleukin-6 differentiate biliary tract cancer from benign biliary strictures. Br. J. Cancer 2011, 105, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Versteijne, E.; Van Eijck, C.H.; Punt, C.J.; Suker, M.; Zwinderman, A.H.; Dohmen, M.A.; Groothuis, K.B.; Busch, O.R.; Besselink, M.G.; De Hingh, I.H.; et al. Preoperative radiochemotherapy versus immediate surgery for resectable and borderline resectable pancreatic cancer (PREOPANC trial): Study protocol for a multicentre randomized controlled trial. Trials 2016, 17, 127. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.M.; Reitsma, J.B.; Bruns, D.E.; Gatstonis, C.A.; Glasziou, P.P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; De Vet, H.C.W.; et al. STARD 2015: An Updated List of Essential Items for Reporting Diagnostic Accuracy Studies. Clin. Chem. 2015, 61, 1446–1452. [Google Scholar] [CrossRef] [Green Version]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Hess, V.; Glimelius, B.; Grawe, P.; Dietrich, D.; Bodoky, G.; Ruhstaller, T.; Bajetta, E.; Saletti, P.; Figer, A.; Scheithauer, W.; et al. CA 19–9 tumour-marker response to chemotherapy in patients with advanced pancreatic cancer enrolled in a randomised controlled trial. Lancet Oncol. 2008, 9, 132–138. [Google Scholar] [CrossRef]



| Discovery Phase | Evaluation and Validation Phase | ||||||
|---|---|---|---|---|---|---|---|
| Healthy control (N = 7) | Distal CCA (N = 7) | Healthy control (N = 32) | Benign disease (N = 20) | Distal CCA (N = 24) | PDAC (N = 24) | P-value * | |
| Age—years | 0.989 | ||||||
| Mean (± SD) | 68 (± 9) | 68 (± 14) | 63 (± 9) | 60 (± 12) | 68 (± 11) | 68 (± 10) | |
| Sex—No. (%) | 1.000 | ||||||
| Male | 4 (57) | 4 (57) | 19 (59) | 10 (50) | 15 (63) | 15 (63) | |
| Female | 3 (43) | 3 (43) | 13 (41) | 10 (50) | 9 (37) | 9 (37) | |
| Stage+—No. (%) | 1.000 | ||||||
| I | - | 0 (0) | - | - | 2 (8) | 2 (8) | |
| II | - | 6 (86) | - | - | 17 (71) | 17 (71) | |
| III | - | 1 (14) | - | - | 2 (8) | 2 (8) | |
| IV | - | 0 (0) | - | - | 3 (13) | 3 (13) | |
| CA19–9—No. (%) | 0.056 | ||||||
| Normal § | - | 3 (42) | - | 15 (75) | 6 (25) | 1 (4) | |
| ULN to <59 × ULN | - | 2 (29) | - | 4 (20) | 15 (63) | 19 (79) | |
| High ≥59 × ULN | - | 0 (0) | - | 0 (0) | 1 (4) | 4 (17) | |
| Missing | 2 (29) | - | 1 (5) | 2 (8) | 0 (0) | ||
| CA19–9 (U/mL) | 0.119 | ||||||
| Median (± SD) | - | 31 (± 337) | - | 15 (± 123) | 86 (± 844) | 461 (± 3114) | |
| Bilirubin—No. (%) | 0.267 | ||||||
| High | - | 6 (86) | 0 (0) | 6 (30) | 21 (87) | 18 (75) | |
| Low | - | 1 (14) | 31 (97) | 13 (65) | 3 (13) | 6 (25) | |
| Missing | 0 (0) | 1 (3) | 1 (5) | 0 (0) | 0 (0) | ||
| Bilirubin (µmol/L) | 0.908 | ||||||
| Median (± SD) | - | 52 (± 196) | 4 (± 3) | 7 (± 26) | 104 (± 166) | 105 (± 190) | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meijer, L.L.; Puik, J.R.; Le Large, T.Y.S.; Heger, M.; Dijk, F.; Funel, N.; Wurdinger, T.; Garajová, I.; van Grieken, N.C.T.; van de Wiel, M.A.; et al. Unravelling the Diagnostic Dilemma: A MicroRNA Panel of Circulating MiR-16 and MiR-877 as A Diagnostic Classifier for Distal Bile Duct Tumors. Cancers 2019, 11, 1181. https://doi.org/10.3390/cancers11081181
Meijer LL, Puik JR, Le Large TYS, Heger M, Dijk F, Funel N, Wurdinger T, Garajová I, van Grieken NCT, van de Wiel MA, et al. Unravelling the Diagnostic Dilemma: A MicroRNA Panel of Circulating MiR-16 and MiR-877 as A Diagnostic Classifier for Distal Bile Duct Tumors. Cancers. 2019; 11(8):1181. https://doi.org/10.3390/cancers11081181
Chicago/Turabian StyleMeijer, Laura L., Jisce R. Puik, Tessa Y.S. Le Large, Michal Heger, Frederike Dijk, Niccola Funel, Thomas Wurdinger, Ingrid Garajová, Nicole C.T. van Grieken, Mark A. van de Wiel, and et al. 2019. "Unravelling the Diagnostic Dilemma: A MicroRNA Panel of Circulating MiR-16 and MiR-877 as A Diagnostic Classifier for Distal Bile Duct Tumors" Cancers 11, no. 8: 1181. https://doi.org/10.3390/cancers11081181
APA StyleMeijer, L. L., Puik, J. R., Le Large, T. Y. S., Heger, M., Dijk, F., Funel, N., Wurdinger, T., Garajová, I., van Grieken, N. C. T., van de Wiel, M. A., Giovannetti, E., & Kazemier, G. (2019). Unravelling the Diagnostic Dilemma: A MicroRNA Panel of Circulating MiR-16 and MiR-877 as A Diagnostic Classifier for Distal Bile Duct Tumors. Cancers, 11(8), 1181. https://doi.org/10.3390/cancers11081181