Expression of Oncogenic Drivers in 3D Cell Culture Depends on Nuclear ATP Synthesis by NUDT5
Abstract
:1. Introduction
2. Results and Discussion
2.1. NUDT5 in Cancer
2.1.1. NUDT5 in Expression in Human Cancers Correlates with an Aggressive Phenotype
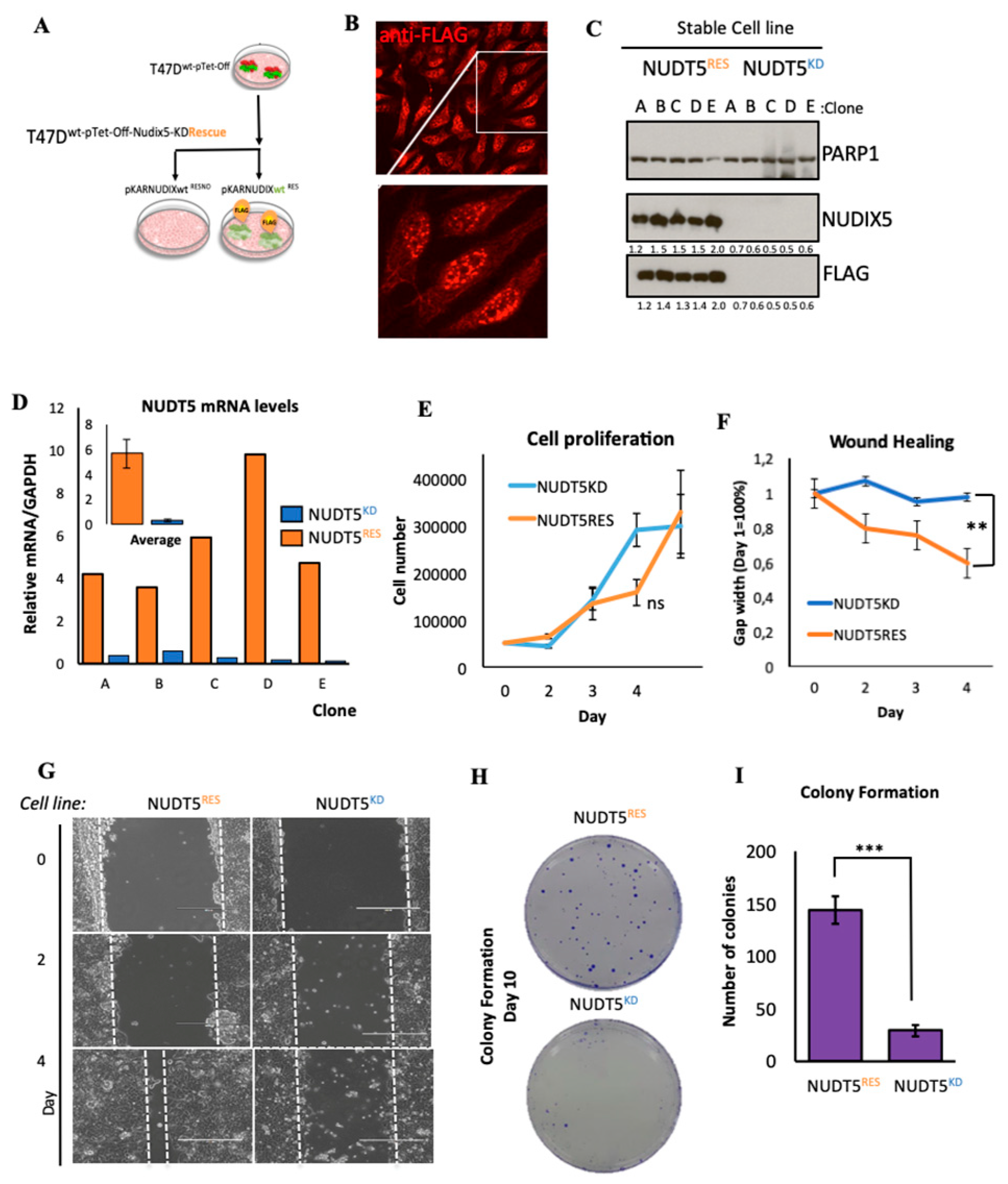
2.1.2. Effect of Nudt5 Knockdown on Cell Growth in 2D Cultures, Cell Migration and Colony Formation Assays
2.1.3. Characterisation of the Expression Profile Changes in T47D Oncosphere Model
2.1.4. NUDT5 Knock-Down Prevents Oncosphere Formation
2.1.5. NUDT5 Promotes the Expression of Genes Involved in EMT and in the Aggressive Tumour Phenotype
2.1.6. NUDT5-Catalysed Synthesis of Nuclear ATP is Required for Oncosphere Formation
2.1.7. NUDT5-Dependent Genes are Required for Oncosphere Growth and Correlate with Poor Cancer Prognosis
3. Materials and Methods
3.1. 2D and 3D Cell Culture
3.1.1. Stable Cell Line Generation
3.1.2. Secondary Oncosphere Assays
3.1.3. RNA Extraction and RT-PCR
3.1.4. Protein Extract Preparation, Western Blotting
3.1.5. Immunofluorescence, Image Acquisition and Analysis
3.1.6. BrdU Cell Proliferation
3.1.7. Scratch Wound Healing Assays
3.1.8. Colony Formation Assays
3.2. Bioinformatic Procedures
3.2.1. RNA-Seq Data Processing
3.2.2. Gene Expression Analysis
3.2.3. Gene Ontology Analysis
3.2.4. Kaplan Meyer Survival Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Y.D.; Wang, L.; Kabatas, S.; Ulrich, H.; Zafonte, R.D. Cancer Stem Cells or Tumor Survival Cells? Stem Cells Dev. 2018, 27, 1466–1478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunes, T.; Hamdan, D.; Leboeuf, C.; El Bouchtaoui, M.; Gapihan, G.; Nguyen, T.T.; Meles, S.; Angeli, E.; Ratajczak, P.; Lu, H.; et al. Targeting Cancer Stem Cells to Overcome Chemoresistance. Int. J. Mol. Sci. 2018, 19, 4036. [Google Scholar] [CrossRef] [PubMed]
- Palomeras, S.; Ruiz-Martinez, S.; Puig, T. Targeting Breast Cancer Stem Cells to Overcome Treatment Resistance. Molecules 2018, 23, 2193. [Google Scholar] [CrossRef] [PubMed]
- Doherty, M.R.; Smigiel, J.M.; Junk, D.J.; Jackson, M.W. Cancer Stem Cell Plasticity Drives Therapeutic Resistance. Cancers 2016, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Wright, R.H.; Castellano, G.; Bonet, J.; Le Dily, F.; Font-Mateu, J.; Ballare, C.; Nacht, A.S.; Soronellas, D.; Oliva, B.; Beato, M. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012, 26, 1972–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, R.H.; Beato, M. PARty promoters: Hormone-dependent gene regulation requires CDK2 activation of PARP1. Cell Cycle 2012, 11, 4291–4293. [Google Scholar] [CrossRef] [PubMed]
- Fatokun, A.A.; Dawson, V.L.; Dawson, T.M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014, 171, 2000–2016. [Google Scholar] [CrossRef]
- Pascal, J.M. The comings and goings of PARP-1 in response to DNA damage. DNA Repair (Amst) 2018, 71, 177–182. [Google Scholar] [CrossRef]
- Hanzlikova, H.; Kalasova, I.; Demin, A.A.; Pennicott, L.E.; Cihlarova, Z.; Caldecott, K.W. The Importance of Poly(ADP-Ribose) Polymerase as a Sensor of Unligated Okazaki Fragments during DNA Replication. Mol. Cell 2018, 71, 319–331.e313. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Zha, M.; Zhong, C.; Peng, Y.; Hu, H.; Ding, J. Crystal structures of human NUDT5 reveal insights into the structural basis of the substrate specificity. J. Mol. Biol. 2006, 364, 1021–1033. [Google Scholar] [CrossRef] [PubMed]
- Wright, R.H.; Lioutas, A.; Le Dily, F.; Soronellas, D.; Pohl, A.; Bonet, J.; Nacht, A.S.; Samino, S.; Font-Mateu, J.; Vicent, G.P.; et al. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science 2016, 352, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, T.; Ohata, H.; Sato, A.; Yamawaki, K.; Enomoto, T.; Okamoto, K. Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications. Cancer Sci. 2017, 108, 283–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Xiong, L.; Li, Q.; Lin, L.; Miao, X.; Yan, S.; Hong, Z.; Yang, L.; Wen, Y.; Deng, X. 3D modeling of cancer stem cell niche. Oncotarget 2018, 9, 1326–1345. [Google Scholar] [CrossRef] [PubMed]
- Forte, E.; Chimenti, I.; Rosa, P.; Angelini, F.; Pagano, F.; Calogero, A.; Giacomello, A.; Messina, E. EMT/MET at the Crossroad of Stemness, Regeneration and Oncogenesis: The Ying-Yang Equilibrium Recapitulated in Cell Spheroids. Cancers 2017, 9, 98. [Google Scholar] [CrossRef]
- Morrison, B.J.; Hastie, M.L.; Grewal, Y.S.; Bruce, Z.C.; Schmidt, C.; Reynolds, B.A.; Gorman, J.J.; Lopez, J.A. Proteomic comparison of mcf-7 tumoursphere and monolayer cultures. PLoS ONE 2012, 7, e52692. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Vechot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef]
- Ma, H.T.; On, K.F.; Tsang, Y.H.; Poon, R.Y. An inducible system for expression and validation of the specificity of short hairpin RNA in mammalian cells. Nucleic Acids Res. 2007, 35, e22. [Google Scholar] [CrossRef]
- Callari, M.; Guffanti, A.; Solda, G.; Merlino, G.; Fina, E.; Brini, E.; Moles, A.; Cappelletti, V.; Daidone, M.G. In-depth characterization of breast cancer tumor-promoting cell transcriptome by RNA sequencing and microarrays. Oncotarget 2016, 7, 976–994. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dontu, G.; Abdallah, W.M.; Foley, J.M.; Jackson, K.W.; Clarke, M.F.; Kawamura, M.J.; Wicha, M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003, 17, 1253–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballesta, A.M.; Molina, R.; Filella, X.; Jo, J.; Gimenez, N. Carcinoembryonic antigen in staging and follow-up of patients with solid tumors. Tumour. Biol. 1995, 16, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, F.; Wicklein, D.; Horst, J.; Sundermann, P.; Maar, H.; Streichert, T.; Tachezy, M.; Izbicki, J.R.; Bockhorn, M.; Schumacher, U. Carcinoembryonic antigen-related cell adhesion molecules (CEACAM) 1, 5 and 6 as biomarkers in pancreatic cancer. PLoS ONE 2014, 9, e113023. [Google Scholar] [CrossRef]
- Beauchemin, N.; Arabzadeh, A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013, 32, 643–671. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.; Mahadevan, D. Emerging Role and Targeting of Carcinoembryonic Antigen-related Cell Adhesion Molecule 6 (CEACAM6) in Human Malignancies. Clin. Cancer Drugs 2015, 2, 100–111. [Google Scholar] [CrossRef]
- Meng, Q.; Shi, S.; Liang, C.; Liang, D.; Xu, W.; Ji, S.; Zhang, B.; Ni, Q.; Xu, J.; Yu, X. Diagnostic and prognostic value of carcinoembryonic antigen in pancreatic cancer: A systematic review and meta-analysis. Onco Targets Ther. 2017, 10, 4591–4598. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, S.W. The Roles of Carcinoembryonic Antigen in Liver Metastasis and Therapeutic Approaches. Gastroenterol. Res. Pract. 2017, 2017, 7521987. [Google Scholar] [CrossRef]
- Borras, G.; Molina, R.; Xercavins, J.; Ballesta, A.; Iglesias, J. Tumor antigens CA 19.9, CA 125, and CEA in carcinoma of the uterine cervix. Gynecol. Oncol. 1995, 57, 205–211. [Google Scholar] [CrossRef]
- Powell, E.; Shao, J.; Picon, H.M.; Bristow, C.; Ge, Z.; Peoples, M.; Robinson, F.; Jeter-Jones, S.L.; Schlosberg, C.; Grzeskowiak, C.L.; et al. A functional genomic screen in vivo identifies CEACAM5 as a clinically relevant driver of breast cancer metastasis. NPJ Breast Cancer 2018, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Liang, H.; Hao, C.; Yang, X.; Cui, X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol. Rep. 2019, 41, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.; Barraclough, R.; Vasieva, O. Gene Expression Meta-Analysis of Potential Metastatic Breast Cancer Markers. Curr. Mol. Med. 2017, 17, 200–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Masri, A.; Gendler, S.J. Muc1 affects c-Src signaling in PyV MT-induced mammary tumorigenesis. Oncogene 2005, 24, 5799–5808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizcaino, C.; Mansilla, S.; Portugal, J. Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol. Ther. 2015, 152, 111–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaboli, M.; Kotsi, P.A.; Gurrieri, C.; Cattoretti, G.; Ronchetti, S.; Cordon-Cardo, C.; Broxmeyer, H.E.; Hromas, R.; Pandolfi, P.P. Mzf1 controls cell proliferation and tumorigenesis. Genes Dev. 2001, 15, 1625–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nygaard, M.; Terkelsen, T.; Vidas Olsen, A.; Sora, V.; Salamanca Viloria, J.; Rizza, F.; Bergstrand-Poulsen, S.; Di Marco, M.; Vistesen, M.; Tiberti, M.; et al. The Mutational Landscape of the Oncogenic MZF1 SCAN Domain in Cancer. Front. Mol. Biosci. 2016, 3, 78. [Google Scholar] [CrossRef] [Green Version]
- Mollen, E.W.J.; Ient, J.; Tjan-Heijnen, V.C.G.; Boersma, L.J.; Miele, L.; Smidt, M.L.; Vooijs, M. Moving Breast Cancer Therapy up a Notch. Front. Oncol. 2018, 8, 518. [Google Scholar] [CrossRef] [Green Version]
- Farnie, G.; Willan, P.M.; Clarke, R.B.; Bundred, N.J. Combined inhibition of ErbB1/2 and Notch receptors effectively targets breast ductal carcinoma in situ (DCIS) stem/progenitor cell activity regardless of ErbB2 status. PLoS ONE 2013, 8, e56840. [Google Scholar] [CrossRef]
- Cochrane, C.R.; Szczepny, A.; Watkins, D.N.; Cain, J.E. Hedgehog Signaling in the Maintenance of Cancer Stem Cells. Cancers 2015, 7, 1554–1585. [Google Scholar] [CrossRef]
- Monkkonen, T.; Lewis, M.T. New paradigms for the Hedgehog signaling network in mammary gland development and breast Cancer. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Grudzien, P.; Lo, S.; Albain, K.S.; Robinson, P.; Rajan, P.; Strack, P.R.; Golde, T.E.; Miele, L.; Foreman, K.E. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res. 2010, 30, 3853–3867. [Google Scholar] [PubMed]
- Liu, S.; Dontu, G.; Mantle, I.D.; Patel, S.; Ahn, N.S.; Jackson, K.W.; Suri, P.; Wicha, M.S. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006, 66, 6063–6071. [Google Scholar] [CrossRef] [PubMed]
- Wright, R.H.; Fernandez-Fuentes, N.; Oliva, B.; Beato, M. Insight into the machinery that oils chromatin dynamics. Nucleus 2016, 7, 532–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, B.D.G.; Valerie, N.C.K.; Wright, R.H.G.; Wallner, O.; Isaksson, R.; Carter, M.; Rudd, S.G.; Loseva, O.; Jemth, A.S.; Almlof, I.; et al. Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells. Nat. Commun. 2018, 9, 250. [Google Scholar] [CrossRef] [Green Version]
- Mehrotra, S.; Morimiya, A.; Agarwal, B.; Konger, R.; Badve, S. Microsomal prostaglandin E2 synthase-1 in breast cancer: A potential target for therapy. J. Pathol. 2006, 208, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Brockschmidt, A.; Trost, D.; Peterziel, H.; Zimmermann, K.; Ehrler, M.; Grassmann, H.; Pfenning, P.N.; Waha, A.; Wohlleber, D.; Brockschmidt, F.F.; et al. KIAA1797/FOCAD encodes a novel focal adhesion protein with tumour suppressor function in gliomas. Brain 2012, 135, 1027–1041. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, D.B.; Zoncu, R.; Root, D.E.; Sabatini, D.M.; Sawyers, C.L. Identification of an oncogenic RAB protein. Science 2015, 350, 211–217. [Google Scholar] [CrossRef] [Green Version]
- Villagomez, F.R.; Medina-Contreras, O.; Cerna-Cortes, J.F.; Patino-Lopez, G. The role of the oncogenic Rab35 in cancer invasion, metastasis, and immune evasion, especially in leukemia. Small GTPases 2018, 1–12. [Google Scholar] [CrossRef]
- Glinsky, G.V. Death-from-cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle 2005, 4, 1171–1175. [Google Scholar] [CrossRef]
- Schrecengost, R.S.; Dean, J.L.; Goodwin, J.F.; Schiewer, M.J.; Urban, M.W.; Stanek, T.J.; Sussman, R.T.; Hicks, J.L.; Birbe, R.C.; Draganova-Tacheva, R.A.; et al. USP22 regulates oncogenic signaling pathways to drive lethal cancer progression. Cancer Res. 2014, 74, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Varthi, M.; Sykes, S.M.; Phillips, C.; Warzecha, C.; Zhu, W.; Wyce, A.; Thorne, A.W.; Berger, S.L.; McMahon, S.B. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 2008, 29, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Yun, X.; Zhang, K.; Wang, J.; Pangeni, R.P.; Yang, L.; Bonner, M.; Wu, J.; Wang, J.; Nardi, I.K.; Gao, M.; et al. Targeting USP22 Suppresses Tumorigenicity and Enhances Cisplatin Sensitivity Through ALDH1A3 Downregulation in Cancer-Initiating Cells from Lung Adenocarcinoma. Mol. Cancer Res. 2018, 16, 1161–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 1, 2114–2120. [Google Scholar] [CrossRef]
- Bray, N.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 2017, 14, 687–690. [Google Scholar] [CrossRef]
- Yi, L.; Pimentel, H.; Bray, N.L.; Pachter, L. Gene-level differential analysis at transcript-level resolution. Genome Biol. 2018, 19, 53. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Kong, L.; Liu, Y.; Qu, H. dbEMT: An epithelial-mesenchymal transition associated gene resource. Sci. Rep. 2015, 5, 11459. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nuc. Acid. Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 6352, eaan2507. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pickup, K.E.; Pardow, F.; Carbonell-Caballero, J.; Lioutas, A.; Villanueva-Cañas, J.L.; Wright, R.H.G.; Beato, M. Expression of Oncogenic Drivers in 3D Cell Culture Depends on Nuclear ATP Synthesis by NUDT5. Cancers 2019, 11, 1337. https://doi.org/10.3390/cancers11091337
Pickup KE, Pardow F, Carbonell-Caballero J, Lioutas A, Villanueva-Cañas JL, Wright RHG, Beato M. Expression of Oncogenic Drivers in 3D Cell Culture Depends on Nuclear ATP Synthesis by NUDT5. Cancers. 2019; 11(9):1337. https://doi.org/10.3390/cancers11091337
Chicago/Turabian StylePickup, Katherine E., Felicitas Pardow, José Carbonell-Caballero, Antonios Lioutas, José Luis Villanueva-Cañas, Roni H. G. Wright, and Miguel Beato. 2019. "Expression of Oncogenic Drivers in 3D Cell Culture Depends on Nuclear ATP Synthesis by NUDT5" Cancers 11, no. 9: 1337. https://doi.org/10.3390/cancers11091337
APA StylePickup, K. E., Pardow, F., Carbonell-Caballero, J., Lioutas, A., Villanueva-Cañas, J. L., Wright, R. H. G., & Beato, M. (2019). Expression of Oncogenic Drivers in 3D Cell Culture Depends on Nuclear ATP Synthesis by NUDT5. Cancers, 11(9), 1337. https://doi.org/10.3390/cancers11091337




