Novel Approaches to Target Mutant FLT3 Leukaemia
Abstract
Simple Summary
Abstract
1. Introduction
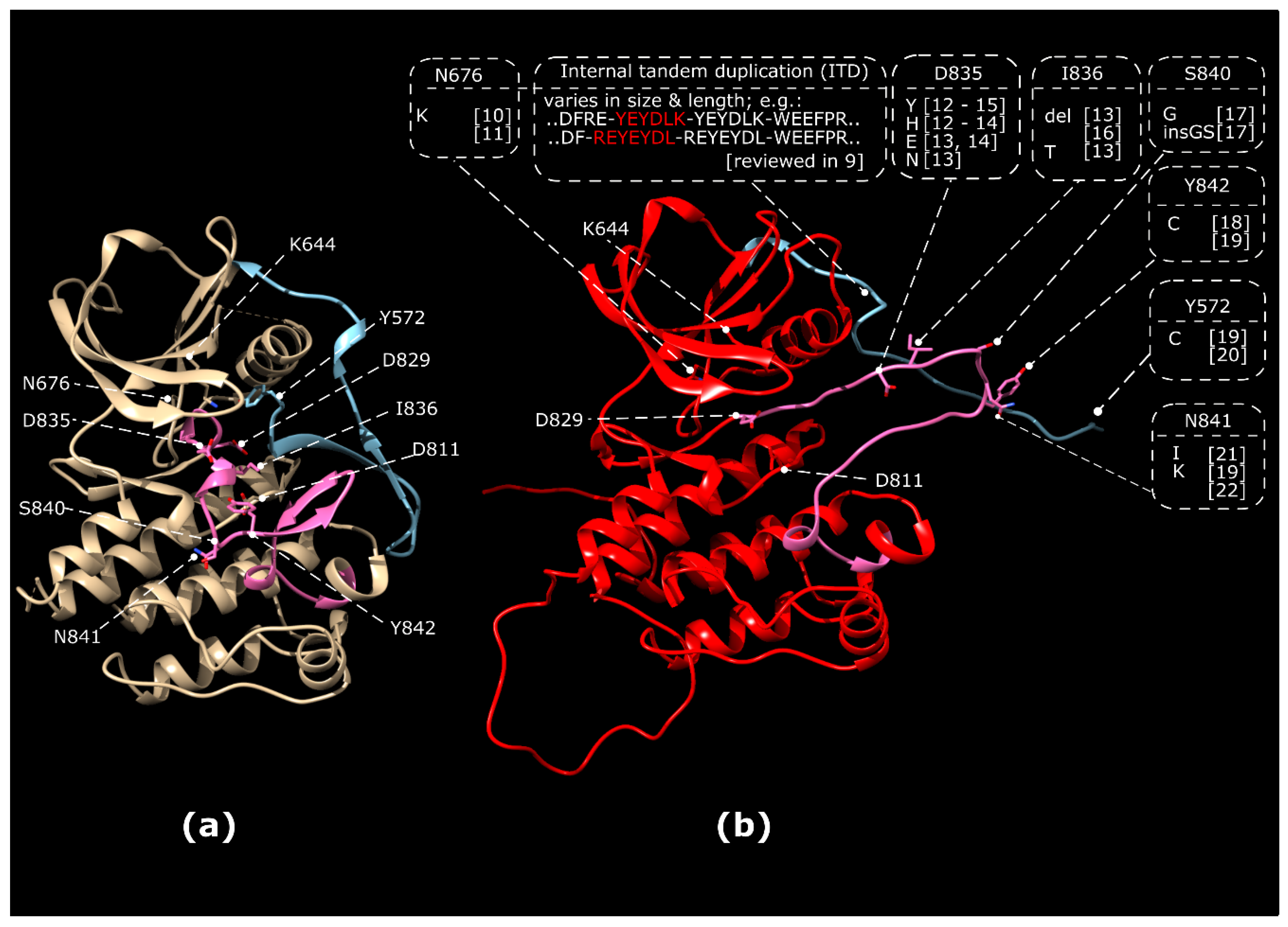
1.1. Classes of Activating FLT3 Mutations
1.2. Biogenesis, Signalling and Regulation of the FLT3 Receptor Tyrosine Kinase
1.3. FLT3 Regulates HSC Self-Renewal and Aging
2. FLT3 in Leukaemia
2.1. Initiating Events in Leukemogenesis
2.2. Involvement of FLT3 in Other Haematological Diseases
2.3. Cell-Intrinsic Oncogenes Cooperating With Mutant FLT3
2.4. Interaction of FLT3 Mutant Blasts With the Haematopoietic Niche
3. FLT3 as Therapeutic Target in Leukaemia
3.1. Small Molecule Inhibition of FLT3
3.2. Novel Alternative Approaches to Affect Oncogenic FLT3 Activity
3.3. Exploiting Novel Vulnerabilities in FLT3 Mutant Blasts
3.4. Preventing Protective Signals from the Bone Marrow Niche
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Maroc, N.; Rottapel, R.; Rosnet, O.; Marchetto, S.; Lavezzi, C.; Mannoni, P.; Birnbaum, D.; Dubreuil, P. Biochemical characterization and analysis of the transforming potential of the FLT3/FLK2 receptor tyrosine kinase. Oncogene 1993, 8, 909–918. [Google Scholar]
- Kikushige, Y.; Yoshimoto, G.; Miyamoto, T.; Iino, T.; Mori, Y.; Iwasaki, H.; Niiro, H.; Takenaka, K.; Nagafuji, K.; Harada, M.; et al. Human Flt3 is expressed at the hematopoietic stem cell and the granulocyte/macrophage progenitor stages to maintain cell survival. J. Immunol. 2008, 180, 7358–7367. [Google Scholar] [CrossRef]
- Toffalini, F.; Demoulin, J.B. New insights into the mechanisms of hematopoietic cell transformation by activated receptor tyrosine kinases. Blood 2010, 116, 2429–2437. [Google Scholar] [CrossRef]
- Stirewalt, D.L.; Radich, J.P. The role of FLT3 in haematopoietic malignancies. Nat. Rev. Cancer 2003, 3, 650–665. [Google Scholar] [CrossRef]
- Sanz, M.; Burnett, A.; Lo-Coco, F.; Lowenberg, B. FLT3 inhibition as a targeted therapy for acute myeloid leukemia. Curr. Opin. Oncol. 2009, 21, 594–600. [Google Scholar] [CrossRef]
- Dehlin, M.; Bokarewa, M.; Rottapel, R.; Foster, S.J.; Magnusson, M.; Dahlberg, L.E.; Tarkowski, A. Intra-articular fms-like tyrosine kinase 3 ligand expression is a driving force in induction and progression of arthritis. PLoS ONE 2008, 3, e3633. [Google Scholar] [CrossRef]
- Hogan, F.L.; Williams, V.; Knapper, S. FLT3 Inhibition in Acute Myeloid Leukaemia—Current Knowledge and Future Prospects. Curr. Cancer Drug Targets 2020, 20, 513–531. [Google Scholar] [CrossRef]
- Levis, M.; Perl, A.E. Gilteritinib: Potent targeting of FLT3 mutations in AML. Blood Adv. 2020, 4, 1178–1191. [Google Scholar] [CrossRef]
- Kayser, S.; Schlenk, R.F.; Londono, M.C.; Breitenbuecher, F.; Wittke, K.; Du, J.; Groner, S.; Spath, D.; Krauter, J.; Ganser, A.; et al. Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood 2009, 114, 2386–2392. [Google Scholar] [CrossRef]
- Huang, K.; Yang, M.; Pan, Z.; Heidel, F.H.; Scherr, M.; Eder, M.; Fischer, T.; Busche, G.; Welte, K.; von Neuhoff, N.; et al. Leukemogenic potency of the novel FLT3-N676K mutant. Ann. Hematol. 2016, 95, 783–791. [Google Scholar] [CrossRef]
- Heidel, F.; Solem, F.K.; Breitenbuecher, F.; Lipka, D.B.; Kasper, S.; Thiede, M.H.; Brandts, C.; Serve, H.; Roesel, J.; Giles, F.; et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood 2006, 107, 293–300. [Google Scholar] [CrossRef]
- Abu-Duhier, F.M.; Goodeve, A.C.; Wilson, G.A.; Care, R.S.; Peake, I.R.; Reilly, J.T. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br. J. Haematol. 2001, 113, 983–988. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kiyoi, H.; Nakano, Y.; Suzuki, R.; Kodera, Y.; Miyawaki, S.; Asou, N.; Kuriyama, K.; Yagasaki, F.; Shimazaki, C.; et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 2001, 97, 2434–2439. [Google Scholar] [CrossRef]
- Taketani, T.; Taki, T.; Sugita, K.; Furuichi, Y.; Ishii, E.; Hanada, R.; Tsuchida, M.; Sugita, K.; Ida, K.; Hayashi, Y. FLT3 mutations in the activation loop of tyrosine kinase domain are frequently found in infant ALL with MLL rearrangements and pediatric ALL with hyperdiploidy. Blood 2004, 103, 1085–1088. [Google Scholar] [CrossRef]
- Frohling, S.; Schlenk, R.F.; Breitruck, J.; Benner, A.; Kreitmeier, S.; Tobis, K.; Dohner, H.; Dohner, K.; leukemia AMLSGUAm. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: A study of the AML Study Group Ulm. Blood 2002, 100, 4372–4380. [Google Scholar] [CrossRef]
- Thiede, C.; Steudel, C.; Mohr, B.; Schaich, M.; Schakel, U.; Platzbecker, U.; Wermke, M.; Bornhauser, M.; Ritter, M.; Neubauer, A.; et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002, 99, 4326–4335. [Google Scholar] [CrossRef]
- Ghosh, K.; Swaminathan, S.; Madkaikar, M.; Gupta, M.; Kerketta, L.; Vundinti, B. FLT3 and NPM1 mutations in a cohort of AML patients and detection of a novel mutation in tyrosine kinase domain of FLT3 gene from Western India. Ann. Hematol. 2012, 91, 1703–1712. [Google Scholar] [CrossRef]
- Kindler, T.; Breitenbuecher, F.; Kasper, S.; Estey, E.; Giles, F.; Feldman, E.; Ehninger, G.; Schiller, G.; Klimek, V.; Nimer, S.D.; et al. Identification of a novel activating mutation (Y842C) within the activation loop of FLT3 in patients with acute myeloid leukemia (AML). Blood 2005, 105, 335–340. [Google Scholar] [CrossRef]
- Zhang, H.; Savage, S.; Schultz, A.R.; Bottomly, D.; White, L.; Segerdell, E.; Wilmot, B.; McWeeney, S.K.; Eide, C.A.; Nechiporuk, T.; et al. Clinical resistance to crenolanib in acute myeloid leukemia due to diverse molecular mechanisms. Nat. Commun. 2019, 10, 244. [Google Scholar] [CrossRef]
- Frohling, S.; Scholl, C.; Levine, R.L.; Loriaux, M.; Boggon, T.J.; Bernard, O.A.; Berger, R.; Dohner, H.; Dohner, K.; Ebert, B.L.; et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell 2007, 12, 501–513. [Google Scholar] [CrossRef]
- Jiang, J.; Paez, J.G.; Lee, J.C.; Bo, R.; Stone, R.M.; DeAngelo, D.J.; Galinsky, I.; Wolpin, B.M.; Jonasova, A.; Herman, P.; et al. Identifying and characterizing a novel activating mutation of the FLT3 tyrosine kinase in AML. Blood 2004, 104, 1855–1858. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, N.; Nanri, T.; Kawakita, T.; Mitsuya, H.; Asou, N. A novel FLT3 activation loop mutation N841K in acute myeloblastic leukemia. Leukemia 2005, 19, 480–481. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.; Black, J.; Faerman, C.; Swenson, L.; Wynn, M.; Lu, F.; Lippke, J.; Saxena, K. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol. Cell. 2004, 13, 169–178. [Google Scholar] [CrossRef]
- Nguyen, B.; Williams, A.B.; Young, D.J.; Ma, H.; Li, L.; Levis, M.; Brown, P.; Small, D. FLT3 activating mutations display differential sensitivity to multiple tyrosine kinase inhibitors. Oncotarget 2017, 8, 10931–10944. [Google Scholar] [CrossRef] [PubMed]
- Opatz, S.; Polzer, H.; Herold, T.; Konstandin, N.P.; Ksienzyk, B.; Zellmeier, E.; Vosberg, S.; Graf, A.; Krebs, S.; Blum, H.; et al. Exome sequencing identifies recurring FLT3 N676K mutations in core-binding factor leukemia. Blood 2013, 122, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Hyrenius-Wittsten, A.; Pilheden, M.; Falques-Costa, A.; Eriksson, M.; Sturesson, H.; Schneider, P.; Wander, P.; Garcia-Ruiz, C.; Liu, J.; Agerstam, H.; et al. FLT3(N676K) drives acute myeloid leukemia in a xenograft model of KMT2A-MLLT3 leukemogenesis. Leukemia 2019, 33, 2310–2314. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Arras, D.; Bohmer, F.D. Mislocalisation of Activated Receptor Tyrosine Kinases—Challenges for Cancer Therapy. Trends Mol. Med. 2020, 26, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Arras, D.E.; Böhmer, A.; Markova, B.; Choudhary, C.; Serve, H.; Böhmer, F.D. Tyrosine phosphorylation regulates maturation of receptor tyrosine kinases. Mol. Cell. Biol. 2005, 25, 3690–3703. [Google Scholar] [CrossRef]
- Schmidt-Arras, D.; Böhmer, S.A.; Koch, S.; Müller, J.P.; Blei, L.; Cornils, H.; Bauer, R.; Korasikha, S.; Thiede, C.; Böhmer, F.D. Anchoring of FLT3 in the endoplasmic reticulum alters signaling quality. Blood 2009, 113, 3568–3576. [Google Scholar] [CrossRef]
- Choudhary, C.; Olsen, J.V.; Brandts, C.; Cox, J.; Reddy, P.N.; Böhmer, F.D.; Gerke, V.; Schmidt-Arras, D.E.; Berdel, W.E.; Müller-Tidow, C.; et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol. Cell. 2009, 36, 326–339. [Google Scholar] [CrossRef]
- Reiter, K.; Polzer, H.; Krupka, C.; Maiser, A.; Vick, B.; Rothenberg-Thurley, M.; Metzeler, K.H.; Dorfel, D.; Salih, H.R.; Jung, G.; et al. Tyrosine kinase inhibition increases the cell surface localization of FLT3-ITD and enhances FLT3-directed immunotherapy of acute myeloid leukemia. Leukemia 2018, 32, 313–322. [Google Scholar] [CrossRef]
- Chan, P.M. Differential signaling of Flt3 activating mutations in acute myeloid leukemia: A working model. Protein Cell 2011, 2, 108–115. [Google Scholar] [CrossRef]
- Hubbard, G.P.; Wolffram, S.; Lovegrove, J.A.; Gibbins, J.M. Ingestion of quercetin inhibits platelet aggregation and essential components of the collagen-stimulated platelet activation pathway in humans. J. Thromb. Haemost. 2004, 2, 2138–2145. [Google Scholar] [CrossRef]
- Schmidt-Arras, D.; Schwable, J.; Böhmer, F.D.; Serve, H. Flt3 receptor tyrosine kinase as a drug target in leukemia. Curr. Pharm. Des. 2004, 10, 1867–1883. [Google Scholar] [CrossRef]
- Kazi, J.U.; Ronnstrand, L. FMS-like Tyrosine Kinase 3/FLT3, From Basic Science to Clinical Implications. Physiol. Rev. 2019, 99, 1433–1466. [Google Scholar] [CrossRef]
- Lamaze, C.; Schmid, S.L. Recruitment of epidermal growth factor receptors into coated pits requires their activated tyrosine kinase. J. Cell Biol. 1995, 129, 47–54. [Google Scholar] [CrossRef]
- Jahn, T.; Seipel, P.; Coutinho, S.; Urschel, S.; Schwarz, K.; Miething, C.; Serve, H.; Peschel, C.; Duyster, J. Analysing c-kit internalization using a functional c-kit-EGFP chimera containing the fluorochrome within the extracellular domain. Oncogene 2002, 21, 4508–4520. [Google Scholar] [CrossRef][Green Version]
- Kellner, F.; Keil, A.; Schindler, K.; Tschongov, T.; Hunninger, K.; Loercher, H.; Rhein, P.; Bohmer, S.A.; Bohmer, F.D.; Muller, J.P. Wild-type FLT3 and FLT3 ITD exhibit similar ligand-induced internalization characteristics. J. Cell Mol. Med. 2020. [Google Scholar] [CrossRef]
- Frangioni, J.V.; Beahm, P.H.; Shifrin, V.; Jost, C.A.; Neel, B.G. The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell 1992, 68, 545–560. [Google Scholar] [CrossRef]
- Tenev, T.; Bohmer, S.A.; Kaufmann, R.; Frese, S.; Bittorf, T.; Beckers, T.; Bohmer, F.D. Perinuclear localization of the protein-tyrosine phosphatase SHP-1 and inhibition of epidermal growth factor-stimulated STAT1/3 activation in A431 cells. Eur. J. Cell Biol. 2000, 79, 261–271. [Google Scholar] [CrossRef]
- Müller, J.P.; Schönherr, C.; Markova, B.; Bauer, R.; Stocking, C.; Böhmer, F.D. Role of SHP2 for FLT3-dependent proliferation and transformation in 32D cells. Leukemia 2008, 22, 1945–1948. [Google Scholar] [CrossRef][Green Version]
- Zhang, S.; Broxmeyer, H.E. Flt3 ligand induces tyrosine phosphorylation of gab1 and gab2 and their association with shp-2, grb2, and PI3 kinase. Biochem. Biophys. Res. Commun. 2000, 277, 195–199. [Google Scholar] [CrossRef]
- Nabinger, S.C.; Li, X.J.; Ramdas, B.; He, Y.; Zhang, X.; Zeng, L.; Richine, B.; Bowling, J.D.; Fukuda, S.; Goenka, S.; et al. The protein tyrosine phosphatase, Shp2, positively contributes to FLT3-ITD-induced hematopoietic progenitor hyperproliferation and malignant disease in vivo. Leukemia 2013, 27, 398–408. [Google Scholar] [CrossRef][Green Version]
- Pandey, R.; Ramdas, B.; Wan, C.; Sandusky, G.; Mohseni, M.; Zhang, C.; Kapur, R. SHP2 inhibition reduces leukemogenesis in models of combined genetic and epigenetic mutations. J. Clin. Investig. 2019, 129, 5468–5473. [Google Scholar] [CrossRef]
- Arora, D.; Kothe, S.; van den Eijnden, M.; Hooft van Huijsduijnen, R.; Heidel, F.; Fischer, T.; Scholl, S.; Tolle, B.; Bohmer, S.A.; Lennartsson, J.; et al. Expression of protein-tyrosine phosphatases in Acute Myeloid Leukemia cells: FLT3 ITD sustains high levels of DUSP6 expression. Cell Commun. Signal. 2012, 10, 19. [Google Scholar] [CrossRef]
- Arora, D.; Stopp, S.; Bohmer, S.A.; Schons, J.; Godfrey, R.; Masson, K.; Razumovskaya, E.; Ronnstrand, L.; Tanzer, S.; Bauer, R.; et al. Protein-tyrosine phosphatase DEP-1 controls receptor tyrosine kinase FLT3 signaling. J. Biol. Chem. 2011, 286, 10918–10929. [Google Scholar] [CrossRef]
- Bohmer, S.A.; Weibrecht, I.; Soderberg, O.; Bohmer, F.D. Association of the protein-tyrosine phosphatase DEP-1 with its substrate FLT3 visualized by in situ proximity ligation assay. PLoS ONE 2013, 8, e62871. [Google Scholar] [CrossRef]
- Godfrey, R.; Arora, D.; Bauer, R.; Stopp, S.; Muller, J.P.; Heinrich, T.; Bohmer, S.A.; Dagnell, M.; Schnetzke, U.; Scholl, S.; et al. Cell transformation by FLT3 ITD in acute myeloid leukemia involves oxidative inactivation of the tumor suppressor protein-tyrosine phosphatase DEP-1/ PTPRJ. Blood 2012, 119, 4499–4511. [Google Scholar] [CrossRef]
- Lee, B.H.; Williams, I.R.; Anastasiadou, E.; Boulton, C.L.; Joseph, S.W.; Amaral, S.M.; Curley, D.P.; Duclos, N.; Huntly, B.J.; Fabbro, D.; et al. FLT3 internal tandem duplication mutations induce myeloproliferative or lymphoid disease in a transgenic mouse model. Oncogene 2005, 24, 7882–7892. [Google Scholar] [CrossRef]
- Kresinsky, A.; Bauer, R.; Schnoder, T.M.; Berg, T.; Meyer, D.; Ast, V.; Konig, R.; Serve, H.; Heidel, F.H.; Bohmer, F.D.; et al. Loss of DEP-1 (Ptprj) promotes myeloproliferative disease in FLT3-ITD acute myeloid leukemia. Haematologica 2018, 103, e505–e509. [Google Scholar] [CrossRef]
- Kresinsky, A.; Schnoder, T.M.; Jacobsen, I.D.; Rauner, M.; Hofbauer, L.C.; Ast, V.; Konig, R.; Hoffmann, B.; Svensson, C.M.; Figge, M.T.; et al. Lack of CD45 in FLT3-ITD mice results in a myeloproliferative phenotype, cortical porosity, and ectopic bone formation. Oncogene 2019, 38, 4773–4787. [Google Scholar] [CrossRef]
- Sundaresan, M.; Yu, Z.X.; Ferrans, V.J.; Irani, K.; Finkel, T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 1995, 270, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.M.; Kalantar-Zadeh, K.; Streja, E.; Carrero, J.J.; Ma, J.Z.; Lu, J.L.; Kovesdy, C.P. The relationship between thyroid function and estimated glomerular filtration rate in patients with chronic kidney disease. Nephrol. Dial. Transpl. 2015, 30, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Bohmer, A.; Barz, S.; Schwab, K.; Kolbe, U.; Gabel, A.; Kirkpatrick, J.; Ohlenschlager, O.; Gorlach, M.; Bohmer, F.D. Modulation of FLT3 signal transduction through cytoplasmic cysteine residues indicates the potential for redox regulation. Redox Biol. 2020, 28, 101325. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, D.G.; Griffin, J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood 2002, 100, 1532–1542. [Google Scholar] [CrossRef]
- Parcells, B.W.; Ikeda, A.K.; Simms-Waldrip, T.; Moore, T.B.; Sakamoto, K.M. FMS-like tyrosine kinase 3 in normal hematopoiesis and acute myeloid leukemia. Stem Cells 2006, 24, 1174–1184. [Google Scholar] [CrossRef]
- Adolfsson, J.; Mansson, R.; Buza-Vidas, N.; Hultquist, A.; Liuba, K.; Jensen, C.T.; Bryder, D.; Yang, L.; Borge, O.J.; Thoren, L.A.; et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 2005, 121, 295–306. [Google Scholar] [CrossRef]
- Chu, S.H.; Heiser, D.; Li, L.; Kaplan, I.; Collector, M.; Huso, D.; Sharkis, S.J.; Civin, C.; Small, D. FLT3-ITD knockin impairs hematopoietic stem cell quiescence/homeostasis, leading to myeloproliferative neoplasm. Cell Stem Cell 2012, 11, 346–358. [Google Scholar] [CrossRef]
- Ocampo, A.; Izpisua Belmonte, J.C. Stem cells: Holding your breath for longevity. Science 2015, 347, 1319–1320. [Google Scholar] [CrossRef]
- Rossi, D.J.; Bryder, D.; Zahn, J.M.; Ahlenius, H.; Sonu, R.; Wagers, A.J.; Weissman, I.L. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA 2005, 102, 9194–9199. [Google Scholar] [CrossRef]
- Mohrin, M.; Shin, J.; Liu, Y.; Brown, K.; Luo, H.; Xi, Y.; Haynes, C.M.; Chen, D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 2015, 347, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, A.; Schmidt, M.; Huber, O.; Frietsch, J.J.; Scholl, S.; Heidel, F.H.; Hochhaus, A.; Muller, J.P.; Ernst, T. SIRT7, an influence factor in healthy aging and the development of age-dependent myeloid stem-cell disorders. Leukemia 2020, 34, 2206–2216. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Jing, Z.X.; Ke, Z.; Yi, P. Sirtuin 7 plays an oncogenic role in human osteosarcoma via downregulating CDC4 expression. Am. J. Cancer Res. 2017, 7, 1788–1803. [Google Scholar] [PubMed]
- Li, L.; Osdal, T.; Ho, Y.; Chun, S.; McDonald, T.; Agarwal, P.; Lin, A.; Chu, S.; Qi, J.; Li, L.; et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell 2014, 15, 431–446. [Google Scholar] [CrossRef]
- Ding, L.; Ley, T.J.; Larson, D.E.; Miller, C.A.; Koboldt, D.C.; Welch, J.S.; Ritchey, J.K.; Young, M.A.; Lamprecht, T.; McLellan, M.D.; et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012, 481, 506–510. [Google Scholar] [CrossRef]
- Dohner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. New Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef]
- Genovese, G.; Kahler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef]
- Ganguly, B.B.; Kadam, N.N. Mutations of myelodysplastic syndromes (MDS): An update. Mutat. Res. 2016, 769, 47–62. [Google Scholar] [CrossRef]
- Martin-Izquierdo, M.; Abaigar, M.; Hernandez-Sanchez, J.M.; Tamborero, D.; Lopez-Cadenas, F.; Ramos, F.; Lumbreras, E.; Madinaveitia-Ochoa, A.; Megido, M.; Labrador, J.; et al. Co-occurrence of cohesin complex and Ras signaling mutations during progression from myelodysplastic syndromes to secondary acute myeloid leukemia. Haematologica 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Easton, J.; Shao, Y.; Maciaszek, J.; Wang, Z.; Wilkinson, M.R.; McCastlain, K.; Edmonson, M.; Pounds, S.B.; Shi, L.; et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017, 49, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. New Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Tomizawa, D. Acute leukemia in adolescents and young adults. Rinsho Ketsueki 2017, 58, 2160–2167. [Google Scholar] [PubMed]
- Zhang, J.; Zhang, Y.; Zhang, M.; Liu, C.; Liu, X.; Yin, J.; Wu, P.; Chen, X.; Yang, W.; Zhang, L.; et al. FLT3 pathway is a potential therapeutic target for PRC2-mutated T-cell acute lymphoblastic leukemia. Blood 2018, 132, 2520–2524. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.I.; Park, J.; Ahn, K.S.; Won, N.H.; Kim, B.K.; Shin, W.G.; Yoon, S.S.; Oh, J.M. Molecular characterization and prognostic significance of FLT3 in CML progression. Leukemia Res. 2010, 34, 995–1001. [Google Scholar] [CrossRef]
- Garg, S.; Reyes-Palomares, A.; He, L.; Bergeron, A.; Lavallee, V.P.; Lemieux, S.; Gendron, P.; Rohde, C.; Xia, J.; Jagdhane, P.; et al. Hepatic leukemia factor is a novel leukemic stem cell regulator in DNMT3A, NPM1, and FLT3-ITD triple-mutated AML. Blood 2019, 134, 263–276. [Google Scholar] [CrossRef]
- Lee, B.H.; Tothova, Z.; Levine, R.L.; Anderson, K.; Buza-Vidas, N.; Cullen, D.E.; McDowell, E.P.; Adelsperger, J.; Frohling, S.; Huntly, B.J.; et al. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell 2007, 12, 367–380. [Google Scholar] [CrossRef]
- Li, L.; Piloto, O.; Nguyen, H.B.; Greenberg, K.; Takamiya, K.; Racke, F.; Huso, D.; Small, D. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood 2008, 111, 3849–3858. [Google Scholar] [CrossRef]
- Skayneh, H.; Jishi, B.; Hleihel, R.; Hamieh, M.; Darwiche, N.; Bazarbachi, A.; El Sabban, M.; El Hajj, H. A Critical Review of Animal Models Used in Acute Myeloid Leukemia Pathophysiology. Genes (Basel) 2019, 10, 614. [Google Scholar] [CrossRef]
- Zorko, N.A.; Bernot, K.M.; Whitman, S.P.; Siebenaler, R.F.; Ahmed, E.H.; Marcucci, G.G.; Yanes, D.A.; McConnell, K.K.; Mao, C.; Kalu, C.; et al. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood 2012, 120, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, M.C.; Kim, Y.M.; Krivtsov, A.V.; Wright, R.D.; Feng, Z.; Agarwal, J.; Kung, A.L.; Armstrong, S.A. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: Development of a model for rapid therapeutic assessment. Leukemia 2008, 22, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, S.; Li, L.; Slape, C.; Nguyen, B.; Novak, R.; Duffield, A.; Huso, D.; Desiderio, S.; Borowitz, M.J.; Aplan, P.; et al. Knock-in of a FLT3/ITD mutation cooperates with a NUP98-HOXD13 fusion to generate acute myeloid leukemia in a mouse model. Blood 2012, 119, 2883–2894. [Google Scholar] [CrossRef] [PubMed]
- Mupo, A.; Celani, L.; Dovey, O.; Cooper, J.L.; Grove, C.; Rad, R.; Sportoletti, P.; Falini, B.; Bradley, A.; Vassiliou, G.S. A powerful molecular synergy between mutant Nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia 2013, 27, 1917–1920. [Google Scholar] [CrossRef]
- Mallardo, M.; Caronno, A.; Pruneri, G.; Raviele, P.R.; Viale, A.; Pelicci, P.G.; Colombo, E. NPMc+ and FLT3_ITD mutations cooperate in inducing acute leukaemia in a novel mouse model. Leukemia 2013, 27, 2248–2251. [Google Scholar] [CrossRef]
- Rau, R.; Magoon, D.; Greenblatt, S.; Li, L.; Annesley, C.; Duffield, A.S.; Huso, D.; McIntyre, E.; Clohessy, J.G.; Reschke, M.; et al. NPMc+ cooperates with Flt3/ITD mutations to cause acute leukemia recapitulating human disease. Exp. Hematol. 2014, 42, 101–113.e5. [Google Scholar] [CrossRef]
- Poitras, J.L.; Heiser, D.; Li, L.; Nguyen, B.; Nagai, K.; Duffield, A.S.; Gamper, C.; Small, D. Dnmt3a deletion cooperates with the Flt3/ITD mutation to drive leukemogenesis in a murine model. Oncotarget 2016, 7, 69124–69135. [Google Scholar] [CrossRef]
- Mendelson, A.; Frenette, P.S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 2014, 20, 833–846. [Google Scholar] [CrossRef]
- Crane, G.M.; Jeffery, E.; Morrison, S.J. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 2017, 17, 573–590. [Google Scholar] [CrossRef]
- Agarwal, P.; Bhatia, R. Influence of Bone Marrow Microenvironment on Leukemic Stem Cells: Breaking Up an Intimate Relationship. Adv. Cancer Res. 2015, 127, 227–252. [Google Scholar]
- Krause, D.S.; Scadden, D.T. A hostel for the hostile: The bone marrow niche in hematologic neoplasms. Haematologica 2015, 100, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
- Mead, A.J.; Neo, W.H.; Barkas, N.; Matsuoka, S.; Giustacchini, A.; Facchini, R.; Thongjuea, S.; Jamieson, L.; Booth, C.A.G.; Fordham, N.; et al. Niche-mediated depletion of the normal hematopoietic stem cell reservoir by Flt3-ITD-induced myeloproliferation. J. Exp. Med. 2017, 214, 2005–2021. [Google Scholar] [CrossRef]
- Noack, M.; Miossec, P. Selected cytokine pathways in rheumatoid arthritis. Semin. Immunopathol. 2017, 39, 365–383. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Broxmeyer, H.E.; Pelus, L.M. Flt3 ligand and the Flt3 receptor regulate hematopoietic cell migration by modulating the SDF-1alpha(CXCL12)/CXCR4 axis. Blood 2005, 105, 3117–3126. [Google Scholar] [CrossRef] [PubMed]
- Voermans, C.; van Heese, W.P.; de Jong, I.; Gerritsen, W.R.; van Der Schoot, C.E. Migratory behavior of leukemic cells from acute myeloid leukemia patients. Leukemia 2002, 16, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Grundler, R.; Brault, L.; Gasser, C.; Bullock, A.N.; Dechow, T.; Woetzel, S.; Pogacic, V.; Villa, A.; Ehret, S.; Berridge, G.; et al. Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J. Exp. Med. 2009, 206, 1957–1970. [Google Scholar] [CrossRef]
- Onishi, C.; Mori-Kimachi, S.; Hirade, T.; Abe, M.; Taketani, T.; Suzumiya, J.; Sugimoto, T.; Yamaguchi, S.; Kapur, R.; Fukuda, S. Internal tandem duplication mutations in FLT3 gene augment chemotaxis to Cxcl12 protein by blocking the down-regulation of the Rho-associated kinase via the Cxcl12/Cxcr4 signaling axis. J. Biol. Chem. 2014, 289, 31053–31065. [Google Scholar] [CrossRef]
- Alonso, S.; Su, M.; Jones, J.W.; Ganguly, S.; Kane, M.A.; Jones, R.J.; Ghiaur, G. Human bone marrow niche chemoprotection mediated by cytochrome P450 enzymes. Oncotarget 2015, 6, 14905–14912. [Google Scholar] [CrossRef]
- Chang, Y.T.; Hernandez, D.; Alonso, S.; Gao, M.; Su, M.; Ghiaur, G.; Levis, M.J.; Jones, R.J. Role of CYP3A4 in bone marrow microenvironment-mediated protection of FLT3/ITD AML from tyrosine kinase inhibitors. Blood Adv. 2019, 3, 908–916. [Google Scholar] [CrossRef]
- Chen, F.; Ishikawa, Y.; Akashi, A.; Naoe, T.; Kiyoi, H. Co-expression of wild-type FLT3 attenuates the inhibitory effect of FLT3 inhibitor on FLT3 mutated leukemia cells. Oncotarget 2016, 7, 47018–47032. [Google Scholar] [CrossRef]
- Parmar, A.; Marz, S.; Rushton, S.; Holzwarth, C.; Lind, K.; Kayser, S.; Dohner, K.; Peschel, C.; Oostendorp, R.A.; Gotze, K.S. Stromal niche cells protect early leukemic FLT3-ITD+ progenitor cells against first-generation FLT3 tyrosine kinase inhibitors. Cancer Res. 2011, 71, 4696–4706. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.J.; Sugita, M.; Koblish, H.; Perl, A.E.; Carroll, M. Hematopoietic cytokines mediate resistance to targeted therapy in FLT3-ITD acute myeloid leukemia. Blood Adv. 2019, 3, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Dumas, P.Y.; Naudin, C.; Martin-Lanneree, S.; Izac, B.; Casetti, L.; Mansier, O.; Rousseau, B.; Artus, A.; Dufossee, M.; Giese, A.; et al. Hematopoietic niche drives FLT3-ITD acute myeloid leukemia resistance to quizartinib via STAT5-and hypoxia-dependent upregulation of AXL. Haematologica 2019, 104, 2017–2027. [Google Scholar] [CrossRef] [PubMed]
- Traer, E.; Martinez, J.; Javidi-Sharifi, N.; Agarwal, A.; Dunlap, J.; English, I.; Kovacsovics, T.; Tyner, J.W.; Wong, M.; Druker, B.J. FGF2 from Marrow Microenvironment Promotes Resistance to FLT3 Inhibitors in Acute Myeloid Leukemia. Cancer Res. 2016, 76, 6471–6482. [Google Scholar] [CrossRef] [PubMed]
- Antar, A.I.; Otrock, Z.K.; Jabbour, E.; Mohty, M.; Bazarbachi, A. FLT3 inhibitors in acute myeloid leukemia: Ten frequently asked questions. Leukemia 2020, 34, 682–696. [Google Scholar] [CrossRef] [PubMed]
- Mosquera Orgueira, A.; Bao Perez, L.; Mosquera Torre, A.; Peleteiro Raindo, A.; Cid Lopez, M.; Diaz Arias, J.A.; Ferreiro Ferro, R.; Antelo Rodriguez, B.; Gonzalez Perez, M.S.; Albors Ferreiro, M.; et al. FLT3 inhibitors in the treatment of Acute Myeloid Leukemia: Current status and future perspectives. Minerva Med. 2020. [Google Scholar] [CrossRef]
- Levis, M. Midostaurin approved for FLT3-mutated AML. Blood 2017, 129, 3403–3406. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Dohner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Davis, M.I.; Hunt, J.P.; Herrgard, S.; Ciceri, P.; Wodicka, L.M.; Pallares, G.; Hocker, M.; Treiber, D.K.; Zarrinkar, P.P. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1046–1051. [Google Scholar] [CrossRef]
- Naqvi, K.; Ravandi, F. FLT3 inhibitor quizartinib (AC220). Leuk. Lymphoma 2019, 60, 1866–1876. [Google Scholar] [CrossRef]
- Klaeger, S.; Heinzlmeir, S.; Wilhelm, M.; Polzer, H.; Vick, B.; Koenig, P.A.; Reinecke, M.; Ruprecht, B.; Petzoldt, S.; Meng, C.; et al. The target landscape of clinical kinase drugs. Science 2017, 358. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Dong, Z.; Liu, J.; Peery, R.C.; Zhang, S.; Liu, J.Y.; Zhang, J.T. Effective Targeting of the Survivin Dimerization Interface with Small-Molecule Inhibitors. Cancer Res. 2016, 76, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Pinto, A.; Colon-Bolea, P.; Casar, B.; Jones, M.; Agudo-Ibanez, L.; Vidal, R.; Tenbaum, S.P.; Nuciforo, P.; Valdizan, E.M.; et al. Small Molecule Inhibition of ERK Dimerization Prevents Tumorigenesis by RAS-ERK Pathway Oncogenes. Cancer Cell 2015, 28, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Gelain, A.; Porta, F.; Asai, A.; Martinelli, A.; Tuccinardi, T. Identification of a new STAT3 dimerization inhibitor through a pharmacophore-based virtual screening approach. J. Enzym. Inhib. Med. Chem. 2016, 31, 1011–1017. [Google Scholar] [CrossRef]
- Takahashi, K.; Mernaugh, R.L.; Friedman, D.B.; Weller, R.; Tsuboi, N.; Yamashita, H.; Quaranta, V.; Takahashi, T. Thrombospondin-1 acts as a ligand for CD148 tyrosine phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 1985–1990. [Google Scholar] [CrossRef]
- Takahashi, K.; Sumarriva, K.; Kim, R.; Jiang, R.; Brantley-Sieders, D.M.; Chen, J.; Mernaugh, R.L.; Takahashi, T. Determination of the CD148-Interacting Region in Thrombospondin-1. PLoS ONE 2016, 11, e0154916. [Google Scholar] [CrossRef]
- Bloch, E.; Sikorski, E.L.; Pontoriero, D.; Day, E.K.; Berger, B.W.; Lazzara, M.J.; Thevenin, D. Disrupting the transmembrane domain-mediated oligomerization of protein tyrosine phosphatase receptor J inhibits EGFR-driven cancer cell phenotypes. J. Biol. Chem. 2019, 294, 18796–18806. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Muller, J.P.; Bauer, R.; Bohmer, S.A.; Lassig, J.; Cerny-Reiterer, S.; Sperr, W.R.; Valent, P.; Maurer, B.; Moriggl, R.; et al. NOX4-driven ROS formation mediates PTP inactivation and cell transformation in FLT3ITD-positive AML cells. Leukemia 2016, 30, 473–483. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Moloney, J.N.; Bohmer, F.D.; Cotter, T.G. NOX-driven ROS formation in cell transformation of FLT3-ITD-positive AML. Exp. Hematol. 2016, 44, 1113–1122. [Google Scholar] [CrossRef]
- Williams, A.B.; Li, L.; Nguyen, B.; Brown, P.; Levis, M.; Small, D. Fluvastatin inhibits FLT3 glycosylation in human and murine cells and prolongs survival of mice with FLT3/ITD leukemia. Blood 2012, 120, 3069–3079. [Google Scholar] [CrossRef]
- Tsitsipatis, D.; Jayavelu, A.K.; Muller, J.P.; Bauer, R.; Schmidt-Arras, D.; Mahboobi, S.; Schnoder, T.M.; Heidel, F.; Bohmer, F.D. Synergistic killing of FLT3ITD-positive AML cells by combined inhibition of tyrosine-kinase activity and N-glycosylation. Oncotarget 2017, 8, 26613–26624. [Google Scholar] [CrossRef]
- Wingelhofer, B.; Maurer, B.; Heyes, E.C.; Cumaraswamy, A.A.; Berger-Becvar, A.; de Araujo, E.D.; Orlova, A.; Freund, P.; Ruge, F.; Park, J.; et al. Pharmacologic inhibition of STAT5 in acute myeloid leukemia. Leukemia 2018, 32, 1135–1146. [Google Scholar] [CrossRef]
- Minami, Y.; Kiyoi, H.; Yamamoto, Y.; Yamamoto, K.; Ueda, R.; Saito, H.; Naoe, T. Selective apoptosis of tandemly duplicated FLT3-transformed leukemia cells by Hsp90 inhibitors. Leukemia 2002, 16, 1535–1540. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Nishiuchi, R.; Li, Q.; Kumar, A.R.; Hudson, W.A.; Kersey, J.H. FLT3 expressing leukemias are selectively sensitive to inhibitors of the molecular chaperone heat shock protein 90 through destabilization of signal transduction-associated kinases. Clin. Cancer Res. 2003, 9, 4483–4493. [Google Scholar] [PubMed]
- George, P.; Bali, P.; Cohen, P.; Tao, J.; Guo, F.; Sigua, C.; Vishvanath, A.; Fiskus, W.; Scuto, A.; Annavarapu, S.; et al. Cotreatment with 17-allylamino-demethoxygeldanamycin and FLT-3 kinase inhibitor PKC412 is highly effective against human acute myelogenous leukemia cells with mutant FLT-3. Cancer Res. 2004, 64, 3645–3652. [Google Scholar] [CrossRef] [PubMed]
- Katayama, K.; Noguchi, K.; Sugimoto, Y. Heat shock protein 90 inhibitors overcome the resistance to Fms-like tyrosine kinase 3 inhibitors in acute myeloid leukemia. Oncotarget 2018, 9, 34240–34258. [Google Scholar] [CrossRef]
- He, X.; Zhu, Y.; Lin, Y.C.; Li, M.; Du, J.; Dong, H.; Sun, J.; Zhu, L.; Wang, H.; Ding, Z.; et al. PRMT1-mediated FLT3 arginine methylation promotes maintenance of FLT3-ITD(+) acute myeloid leukemia. Blood 2019, 134, 548–560. [Google Scholar] [CrossRef]
- Guedj, A.; Volman, Y.; Geiger-Maor, A.; Bolik, J.; Schumacher, N.; Kunzel, S.; Baines, J.F.; Nevo, Y.; Elgavish, S.; Galun, E.; et al. Gut microbiota shape ‘inflamm-ageing’ cytokines and account for age-dependent decline in DNA damage repair. Gut 2020, 69, 1064–1075. [Google Scholar] [CrossRef]
- Maifrede, S.; Nieborowska-Skorska, M.; Sullivan-Reed, K.; Dasgupta, Y.; Podszywalow-Bartnicka, P.; Le, B.V.; Solecka, M.; Lian, Z.; Belyaeva, E.A.; Nersesyan, A.; et al. Tyrosine kinase inhibitor-induced defects in DNA repair sensitize FLT3(ITD)-positive leukemia cells to PARP1 inhibitors. Blood 2018, 132, 67–77. [Google Scholar] [CrossRef]
- Li, L.; Zhang, L.; Fan, J.; Greenberg, K.; Desiderio, S.; Rassool, F.V.; Small, D. Defective nonhomologous end joining blocks B-cell development in FLT3/ITD mice. Blood 2011, 117, 3131–3139. [Google Scholar] [CrossRef]
- Doshi, K.A.; Trotta, R.; Natarajan, K.; Rassool, F.V.; Tron, A.E.; Huszar, D.; Perrotti, D.; Baer, M.R. Pim kinase inhibition sensitizes FLT3-ITD acute myeloid leukemia cells to topoisomerase 2 inhibitors through increased DNA damage and oxidative stress. Oncotarget 2016, 7, 48280–48295. [Google Scholar] [CrossRef] [PubMed]
- McCann, K.E. Advances in the use of PARP inhibitors for BRCA1/2-associated breast cancer: Talazoparib. Future Oncol. 2019, 15, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Larrue, C.; Saland, E.; Boutzen, H.; Vergez, F.; David, M.; Joffre, C.; Hospital, M.A.; Tamburini, J.; Delabesse, E.; Manenti, S.; et al. Proteasome inhibitors induce FLT3-ITD degradation through autophagy in AML cells. Blood 2016, 127, 882–892. [Google Scholar] [CrossRef]
- Heydt, Q.; Larrue, C.; Saland, E.; Bertoli, S.; Sarry, J.E.; Besson, A.; Manenti, S.; Joffre, C.; Mansat-De Mas, V. Oncogenic FLT3-ITD supports autophagy via ATF4 in acute myeloid leukemia. Oncogene 2018, 37, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Gallipoli, P.; Giotopoulos, G.; Tzelepis, K.; Costa, A.S.H.; Vohra, S.; Medina-Perez, P.; Basheer, F.; Marando, L.; Di Lisio, L.; Dias, J.M.L.; et al. Glutaminolysis is a metabolic dependency in FLT3(ITD) acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood 2018, 131, 1639–1653. [Google Scholar] [CrossRef]
- Shu, S.; Wu, H.J.; Ge, J.Y.; Zeid, R.; Harris, I.S.; Jovanovic, B.; Murphy, K.; Wang, B.; Qiu, X.; Endress, J.E.; et al. Synthetic Lethal and Resistance Interactions with BET Bromodomain Inhibitors in Triple-Negative Breast Cancer. Mol. Cell. 2020, 78, 1096–1113.e8. [Google Scholar] [CrossRef]
- McDaniel, N.K.; Iida, M.; Nickel, K.P.; Longhurst, C.A.; Fischbach, S.R.; Rodems, T.S.; Kranjac, C.A.; Bo, A.Y.; Luo, Q.; Gallagher, M.M.; et al. AXL Mediates Cetuximab and Radiation Resistance Through Tyrosine 821 and the c-ABL Kinase Pathway in Head and Neck Cancer. Clin. Cancer Res. 2020, 26, 4349–4359. [Google Scholar] [CrossRef]
- Brand, T.M.; Iida, M.; Stein, A.P.; Corrigan, K.L.; Braverman, C.M.; Luthar, N.; Toulany, M.; Gill, P.S.; Salgia, R.; Kimple, R.J.; et al. AXL mediates resistance to cetuximab therapy. Cancer Res. 2014, 74, 5152–5164. [Google Scholar] [CrossRef]
- Zhang, Z.; Lee, J.C.; Lin, L.; Olivas, V.; Au, V.; LaFramboise, T.; Abdel-Rahman, M.; Wang, X.; Levine, A.D.; Rho, J.K.; et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 2012, 44, 852–860. [Google Scholar] [CrossRef]
- Park, I.K.; Trotta, R.; Yu, J.; Caligiuri, M.A. Axl/Gas6 pathway positively regulates FLT3 activation in human natural killer cell development. Eur. J. Immunol. 2013, 43, 2750–2755. [Google Scholar] [CrossRef]
- Park, I.K.; Mundy-Bosse, B.; Whitman, S.P.; Zhang, X.; Warner, S.L.; Bearss, D.J.; Blum, W.; Marcucci, G.; Caligiuri, M.A. Receptor tyrosine kinase Axl is required for resistance of leukemic cells to FLT3-targeted therapy in acute myeloid leukemia. Leukemia 2015, 29, 2382–2389. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Oudin, M.J.; Sullivan, R.J.; Wang, S.J.; Meyer, A.S.; Im, H.; Frederick, D.T.; Tadros, J.; Griffith, L.G.; Lee, H.; et al. Reduced Proteolytic Shedding of Receptor Tyrosine Kinases Is a Post-Translational Mechanism of Kinase Inhibitor Resistance. Cancer Discov. 2016, 6, 382–399. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Schubert, M.L.; Wang, L.; He, B.; Neuber, B.; Dreger, P.; Muller-Tidow, C.; Schmitt, M. Chimeric Antigen Receptor (CAR) T Cell Therapy in Acute Myeloid Leukemia (AML). J. Clin. Med. 2019, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Jetani, H.; Garcia-Cadenas, I.; Nerreter, T.; Thomas, S.; Rydzek, J.; Meijide, J.B.; Bonig, H.; Herr, W.; Sierra, J.; Einsele, H.; et al. CAR T-cells targeting FLT3 have potent activity against FLT3(-)ITD(+) AML and act synergistically with the FLT3-inhibitor crenolanib. Leukemia 2018, 32, 1168–1179. [Google Scholar] [CrossRef]
- Garz, A.K.; Wolf, S.; Grath, S.; Gaidzik, V.; Habringer, S.; Vick, B.; Rudelius, M.; Ziegenhain, C.; Herold, S.; Weickert, M.T.; et al. Azacitidine combined with the selective FLT3 kinase inhibitor crenolanib disrupts stromal protection and inhibits expansion of residual leukemia-initiating cells in FLT3-ITD AML with concurrent epigenetic mutations. Oncotarget 2017, 8, 108738–108759. [Google Scholar] [CrossRef][Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, J.P.; Schmidt-Arras, D. Novel Approaches to Target Mutant FLT3 Leukaemia. Cancers 2020, 12, 2806. https://doi.org/10.3390/cancers12102806
Müller JP, Schmidt-Arras D. Novel Approaches to Target Mutant FLT3 Leukaemia. Cancers. 2020; 12(10):2806. https://doi.org/10.3390/cancers12102806
Chicago/Turabian StyleMüller, Jörg P., and Dirk Schmidt-Arras. 2020. "Novel Approaches to Target Mutant FLT3 Leukaemia" Cancers 12, no. 10: 2806. https://doi.org/10.3390/cancers12102806
APA StyleMüller, J. P., & Schmidt-Arras, D. (2020). Novel Approaches to Target Mutant FLT3 Leukaemia. Cancers, 12(10), 2806. https://doi.org/10.3390/cancers12102806





