The Warburg Effect 97 Years after Its Discovery
Simple Summary
Abstract
1. Introduction
2. The Deregulation of the Oxidative Metabolism in Cancer
3. The Genes Involved
4. The Genetic Susceptibility to Cancer
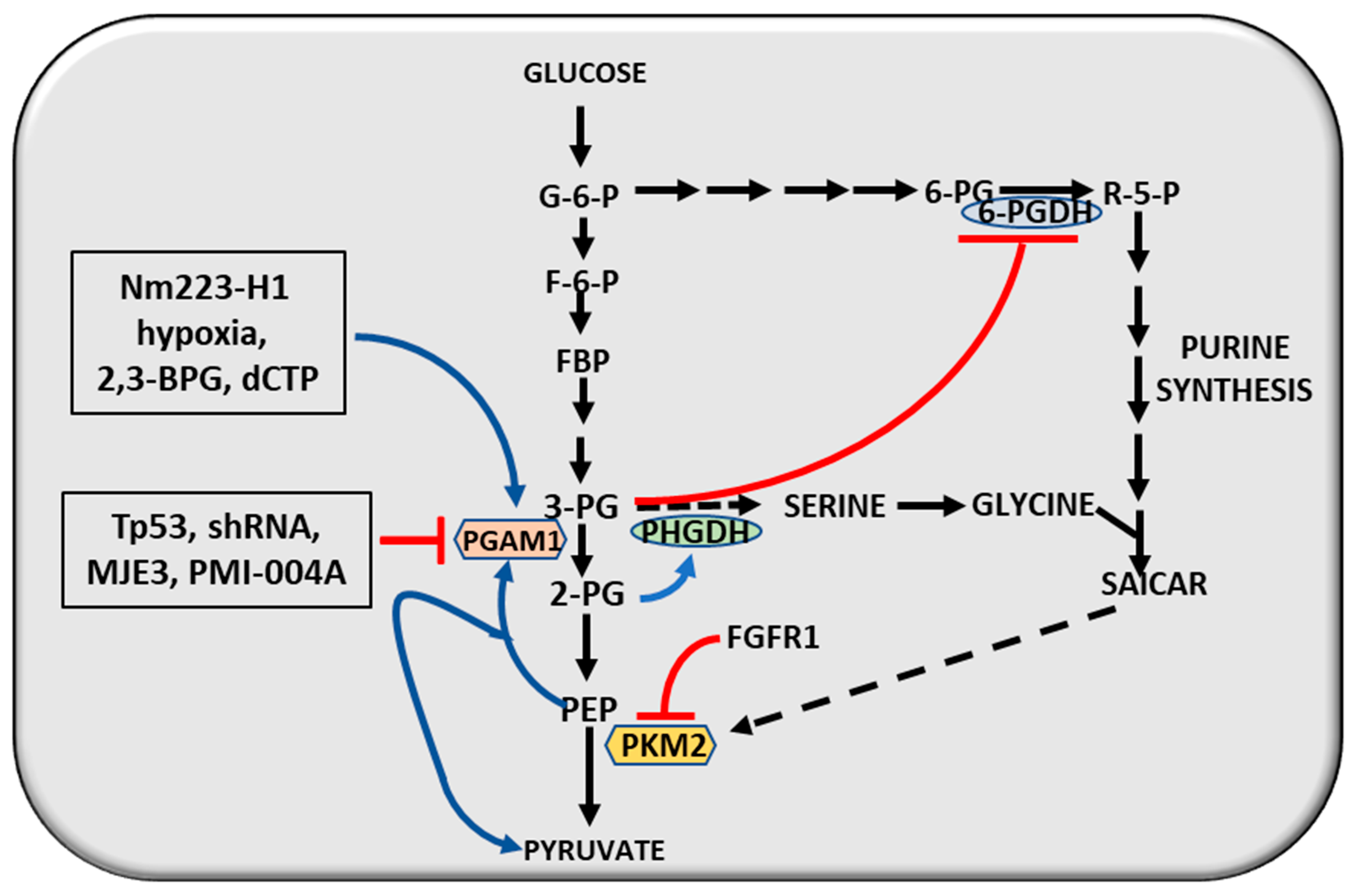
5. The Molecular Correlations
6. Cancer Cell Mitochondria
7. Defective Mitochondrial Biogenesis
8. Interactions between Glycolysis and Mitochondrial Oxidative Activity: The Reverse Warburg Effect
9. The Warburg Effect and Tumor Therapy
9.1. Therapeutic Effect of the Glycolysis Inhibition
9.2. Intracellular Alkalinity of Cancer Cells
9.3. Glutaminolysis and Cancer Therapy
9.4. Nampt Inhibitors
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| α-KG | α-ketoglutarate |
| ACLY | ATP citrate lyase |
| 2,3-BPG | 2,3-biphosphoghycerae |
| 6-PGDH | 6-phosphogluconate dehydrogenase |
| ACR | acceptor control ratio |
| AMPK | AMP-activated protein kinase |
| BHMT | betaine-homocysteine methyl-transferase |
| CDK2 | Cyclin-dependent kinase |
| CDK4 | Cyclin-dependent kinase 4 |
| DHA-P | dihydroxyacetone phosphate |
| dCTP | deoxycytidine triphosphate |
| 2-DG | 2-deoxyglucose |
| 2dP-GLY | 2-diphosphoglycerate |
| 3PG | 3-phosphoglycerate |
| Dusp1 | Dual-specificity phosphatase 1 |
| E2f1 | E2F transcription factor1 |
| FASN | fatty acid synthetase |
| F6P | fructose-6-phosphate |
| FBP | fructose-1,6-biphosphate |
| FBPase | fructose biphosphatase |
| FGFR1 | phosphotyrosine-mediated growth signal |
| FASN | fatty acid synthetase |
| GA-3-P | glyceraldhydre-3-phosphate |
| GDH | glutamate dehydrogenase |
| G6P | glucose-6-phosphate |
| GNMT | glycine N-methyltransferase |
| GPI | gluose-6-phosphate isomerase |
| HCC | hepatocellular carcinoma |
| HDAC3 | histone deacetylase |
| HER2 | ERB B2 |
| HIF-1 | hypoxia-inducible factor-1 |
| HK | hexokinase |
| JMJD5 | Jumonji C domain-containing dioxygenase |
| LKB1 | serine/threonine protein kinase |
| MCT1–4 | monocarboxylate transporters 1-4 |
| MDR | multidrug resistance |
| ME | malic enzyme |
| MEK5 | mitogen-activated protein kinase 5 |
| mtDNA | mitochondrial DNA |
| MTHFD2 | methylenetetrahydrofolate dehydrogenase 2 |
| mTOR | mechanistic target of rapamycin |
| Nm23-H1 | non-metastatic protein 23, homolog 1 |
| OAA | oxalacetic acid |
| PDH | pyruvic dehydrogenase |
| PDK1 | pyruvate dehydrogenase kinase 1 |
| PEP | phosphoenolpyruvate |
| PFK | phosphofructokinase |
| PFK1 | phosphofructokinase muscle type |
| PFK15 | [1-(4-pyridinyl)-3-(2-quinolinyl)-2-propen-1-one] |
| PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| PGAM1 | phosphoglycerate mutase 1 |
| PGC-1α | peroxisome proliferator-activated receptor gamma coactivator-1 alpha |
| PHGDH | phosphoglycerate dehydrogenase |
| PGK | phosphoglycerate kinase |
| PI3K | phosphatdilisitol-3-kinase catalytic gamma |
| PK | phosphoglycerate kinase |
| PKM2 | phosphoglycerate kinase M2 |
| 3PO | (3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one) |
| PTEN | phosphatase and tensin homolog |
| PyrCx | pyruvate carboxylase |
| ROS | reactive oxygen species |
| SAICAR | succinylaminoimidazolecarboxamide |
| SAM | S-adenosylmethionine |
| Sirt1 | sirtuin1 |
| SIRT4 | sirtuin 4 TIGAR, TP53-induced glycolysis and apoptosis regulator |
| VDAC | voltage-dependent anion channel |
| VEGF | vascular endothelial growth factor |
| YAP | yes associated protein |
References
- Warburg, O. Versuche an überlebendem carcinom-gewebe. Klin. Wochenschr. 1923, 2, 776–777. [Google Scholar] [CrossRef]
- Warburg, O. Über den stoffwechsel der carcinomzelle. Naturwissenschaften 1924, 12, 1131–1137. [Google Scholar] [CrossRef]
- Elwood, J.; Lin, Y.; Cristofalo, V.; Weinhouse, S.; Morris, H.P. Glucose utilization in homogenates of the Morris hepatoma 5123 and related tumors. Cancer Res. 1963, 23, 906–913. [Google Scholar]
- Weinhouse, S. Glycolysis, respiration, and anomalous gene expression in experimental hepatomas. G.H.A. Clowes memorial lecture. Cancer Res. 1972, 32, 2007–2016. [Google Scholar] [PubMed]
- Crabtree, H.G. Observations on the carbohydrate metabolism of tumors. Biochem. J. 1929, 23, 536–545. [Google Scholar] [CrossRef]
- Chance, B.; Hess, B. Spectroscopic evidence of metabolic control. Science 1959, 129, 700–708. [Google Scholar] [CrossRef]
- Chance, B.; Garfinkel, D.; Higgins, J.; Hess, B. Metabolic control mechanisms. A solution for the equations representing interaction between glycolysis and respiration in ascites tumor cells. J. Biol. Chem. 1960, 235, 2426–2439. [Google Scholar] [PubMed]
- Schreiber, J.R.; Balcavage, W.X.; Morris, H.P.; Pedersen, P.L. Enzymatic and spectral analysis of cytochrome oxidase in adult and fetal rat liver and Morris hepatoma 3924A. Cancer Res. 1970, 30, 2497–2501. [Google Scholar] [PubMed]
- Jiang, F.; Ryan, M.T.; Schlame, M.; Zhao, M.; Gu, Z.; Klingenberg, M.; Pfanner, N.; Greenberg, M.L. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 2000, 275, 22387–22394. [Google Scholar] [CrossRef] [PubMed]
- Claypool, S.M.; Oktay, Y.; Boontheung, P.; Loo, J.A.; Koehler, C.M. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008, 182, 937–950. [Google Scholar] [CrossRef]
- Kiebish, M.A.; Han, X.; Cheng, H.; Chuang, J.H.; Seyfried, T.N. Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: Lipidomic evidence supporting the Warburg theory of cancer. J. Lipid Res. 2008, 49, 2545–2556. [Google Scholar] [CrossRef]
- Shidara, Y.; Yamagata, K.; Kanamori, T.; Nakano, K.; Kwong, J.Q.; Manfredi, G.; Oda, H.; Ohta, S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005, 65, 1655–1663. [Google Scholar] [CrossRef]
- Park, S.Y.; Chang, I.; Kim, J.Y.; Kang, S.W.; Park, S.-H.; Singh, K.; Lee, M.-S. Resistance of mitochondrial DNA-depleted cells against cell death: Role of mitochondrial superoxide dismutase. J. Biol. Chem. 2004, 279, 7512–7520. [Google Scholar] [CrossRef]
- Pelicano, H.; Xu, R.H.; Du, M.; Feng, L.; Sasaki, S.; Carew, J.S.; Hu, Y.; Ramdas, L.; Hu, L.J.; Keating, M.J.; et al. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J. Cell. Biol. 2006, 175, 913–923. [Google Scholar] [CrossRef]
- Tseng, L.M.; Yin, P.H.; Chi, C.W.; Hsu, C.Y.; Wu, C.W.; Lee, L.M.; Wei, Y.H.; Lee, H.C. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 2006, 45, 629–638. [Google Scholar] [CrossRef]
- Lee, H.C.; Yin, P.P.; Lin, J.C.; Wu, C.C.; Chen, C.Y.; Wu, C.W.; Chi, C.W.; Tam, T.T.; Wei, Y.H. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann. N. Y. Acad. Sci. 2005, 1042, 109–122. [Google Scholar] [CrossRef]
- Wu, C.W.; Yin, P.H.; Hung, W.Y.; Li, A.F.-Y.; Li, S.-H.; Ch, C.H.; Wei, Y.H.; Lee, H.C. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer 2005, 44, 19–28. [Google Scholar] [CrossRef]
- Naito, A.; Cook, C.C.; Mizumachi, T.; Wang, M.; Xie, C.H.; Evans, T.T.; Kelly, T.; Higuchi, M. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Sci. 2008, 99, 584–1588. [Google Scholar] [CrossRef]
- Lee, H.C.; Li, S.H.; Lin, J.C.; Wu, C.C.; Yeh, D.C.; Wei, Y.H. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat. Res. 2004, 547, 71–78. [Google Scholar] [CrossRef]
- Lee, W.; Choi, H.I.; Kim, M.J.; Park, S.Y. Depletion of mitochondrial DNA up-regulates the expression of MDR1 gene via an increase in mRNA stability. Exp. Mol. Med. 2008, 40, 109–117. [Google Scholar] [CrossRef]
- Ferraresi, R.; Troiano, L.; Pinti, M.; Roat, E.; Lugli, E.; Quaglino, D.; Taverna, D.; Bellizzi, D.; Passarino, G.; Cossarizza, A. Resistance of mtDNA-depleted cells to apoptosis. Cytometry A 2008, 73, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S. Contribution of somatic mutations in the mitochondrial genome to the development of cancer and tolerance against anticancer drugs. Oncogene 2006, 25, 4768–4776. [Google Scholar] [CrossRef] [PubMed]
- Amuthan, G.; Biswas, G.; Zhang, S.Y.; Klein-Szanto, A.; Vijayasarathy, A.; Avadhani, N.G. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001, 20, 1910–1920. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.K.; Pedersen, P.L. Functional significance of mitochondrial bound hexokinase in tumor cell metabolism. Evidence for preferential phosphorylation of glucose by intramitochondrially generated ATP. J. Biol. Chem. 1988, 263, 17422–17428. [Google Scholar]
- Nakashima, R.A.; Mangan, P.S.; Colombini, M.; Pedersen, P.L. Hexokinase receptor complex in hepatoma mitochondria: Evidence from N,N′-dicyclohexylcarbodiimide-labeling studies for the involvement of the pore-forming protein VDAC. Biochemistry 1986, 25, 1015–1021. [Google Scholar] [CrossRef]
- Klepinin, A.; Ounpuu, L.; Mado, K.; Truu, L.; Chekulayev, V.; Puurand, M.; Shevchuk, I.; Tepp, K.; Planken, A.; Kaambre, T. The complexity of mitochondrial outer membrane permeability and VDAC regulation by associated proteins. J. Bioenerg. Biomembr. 2018, 50, 339–354. [Google Scholar] [CrossRef]
- Wigfield, S.M.; Winter, S.C.; Giatromanolaki, A.; Taylor, J.; Koukourakis, M.L.; Harris, A.L. PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Br. J. Cancer 2008, 98, 1975–1984. [Google Scholar] [CrossRef]
- Tanner, L.B.; Goglia, A.G.; Wei, M.H.; Sehgal, T.; Lance, R.P.; Junyoung, O.P.; White, E.; Toettcher, J.E.; Rabinowitz, J.D. Four Key Steps Control Glycolytic Flux in Mammalian Cells. Cell Syst. 2018, 7, 49–62. [Google Scholar] [CrossRef]
- Sun, J.; Li, J.; Guo, Z.; Sun, L.; Juan, C.; Zhou, Y.; Gu, H.; Yu, Y.; Hu, Q.; Kan, Q.; et al. Overexpression of Pyruvate Dehydrogenase E1α Subunit Inhibits Warburg Effect and Induces Cell Apoptosis Through Mitochondria-Mediated Pathway in Hepatocellular Carcinoma. Oncol. Res. 2019, 27, 407–414. [Google Scholar] [CrossRef]
- Isidoro, A.; Martínez, M.; Fernández, P.L.; Ortega, A.D.; Santamaría, G.; Chamorro, M.; Reed, J.C.; Cuezva, J.M. Alteration of the bioenergetic phenotype of mitochondria is a hallmark of breast, gastric, lung and oesophageal cancer. Biochem. J. 2004, 378, 17–20. [Google Scholar] [CrossRef]
- Mathupala, S.P.; Ko, Y.H.; Pedersen, P.L. Hexokinase-2 bound to mitochondria: Cancer’s stygian link to the “Warburg Effect” and a pivotal target for effective therapy. Semin. Cancer Biol. 2009, 19, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Ruiz, R.; Rigoulet, M.; Devin, A. The Warburg and Crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta 2011, 1807, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Hu, X.; Chakravarty, H.; Yang, Z.; Tam, K.Y. Identification of Novel Pyruvate Dehydrogenase Kinase 1 (PDK1) Inhibitors by Kinase Activity-Based High-Throughput Screening for Anticancer Therapeutics. ACS Comb. Sci. 2018, 20, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Liloglou, T.; Tegos, C.; Spandidos, D. T24 h-ras gene-expression increases the activity of phosphoglycerate kinase, enolase and pyruvate-kinase and decreases the activity of adenosine-deaminase in fibroblast cells. Oncol. Rep. 1994, 1, 1193–1197. [Google Scholar] [CrossRef]
- David, C.J.; Chen, M.; Assanah, M.; Canoll, P.; Manley, J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010, 463, 364–368. [Google Scholar] [CrossRef]
- Shim, H.; Dolde, C.; Lewis, B.C.; Wu, C.S.; Dang, G.; Jungmann, R.A.; Dalla-Favera, D.; Dang, C.V. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA 1997, 94, 6658–6663. [Google Scholar] [CrossRef]
- Hsieh, A.L.; Walton, Z.E.; Altman, B.J.; Stine, Z.E.; Dang, C.V. MYC and metabolism on the path to cancer. Semin. Cell Dev. Biol. 2015, 43, 11–21. [Google Scholar]
- Calvisi, D.F.; Frau, M.; Tomasi, M.L.; Feo, F.; Pascale, R.M. Deregulation of signaling pathways in prognostic subtypes of hepatocellular carcinoma: Novel insight from interspecies comparison. Biochim. Biophys. Acta 2012, 1826, 215–237. [Google Scholar]
- Liu, R.; Li, Y.; Tian, L.; Shi, H.; Wang, J.; Liang, Y.; Sun, B.; Wang, S.; Zhou, M.; Wu, L.; et al. Gankyrin drives metabolic reprogramming to promote tumorigenesis, metastasis and drug resistance through activating β-catenin/c-Myc signaling in human hepatocellular carcinoma. Cancer Lett. 2019, 443, 34–46. [Google Scholar] [CrossRef]
- Yu, P.; Wilhelm, K.; Dubrac, A.; Tung, J.K.; Alves, T.C.; Fang, J.S.; Xie, X.; Zhu, J.; Chen, Z.; De Smet, F.; et al. FGF-dependent metabolic control of vascular development. Nature 2017, 545, 224–228. [Google Scholar] [CrossRef]
- Dimri, M.; Humphries, A.; Laknaur, A.; Elattar, S.; Lee, T.J.; Sharma, A.; Kolhe, R.; Satyanarayana, A. NAD(P)H Quinone Dehydrogenase 1 Ablation Inhibits Activation of the Phosphoinositide 3-Kinase/Akt Serine/Threonine Kinase and Mitogen-Activated Protein Kinase/Extracellular Signal-Regulated Kinase Pathways and Blocks Metabolic Adaptation in Hepatocellular Carcinoma. Hepatology 2020, 71, 549–568. [Google Scholar] [PubMed]
- Xie, Y.; Shi, X.; Sheng, K.; Han, S.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia. Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Simon-Molas, H.; Arnedo-Pac, C.; Fontova, P.; Vidal-Alabró, A.; Castaño, E.; Rodríguez-García, A.; Navarro-Sabaté, A.; Lloberas, N.; Manzano, A.; Bartrons, R. PI3K-Akt signaling controls PFKFB3 expression during human T-lymphocyte activation. Mol. Cell Biochem. 2018, 448, 187–197. [Google Scholar] [CrossRef]
- Villalobos, P.; Soto, F.; Baez, M.; Babul, J. Regulatory network of the allosteric ATP inhibition of E. coli phosphofructokinase-2 studied by hybrid dimers. Biochimie 2016, 128, 209–216. [Google Scholar] [CrossRef]
- Mailloux, R.J.; Appanna, V.D. Aluminum toxicity triggers the nuclear translocation of HIF-1alpha and promotes anaerobiosis in Hepatocytes. Toxicol Vitro 2007, 21, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Marín-Hernández, A.; Gallardo-Pérez, J.C.; Ralph, S.J.; Rodríguez-Enríquez, S.; Moreno-Sánchez, R. HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Rev. Med. Chem. 2009, 9, 1084–1101. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.C.; Quintin, J.; Cramer, R.A.; Shepardson, K.M.; Saeed, S.; Kumar, V.; Giamarellos-Bourboulis, E.J.; Martens, J.H.A.; Rao, N.A.; Aghajanirefah, A.; et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014, 345, 1250684. [Google Scholar] [CrossRef]
- Dang, C.V.; Semenza, G.L. Oncogenic alterations of metabolism. Trends Biochem. Sci. 1999, 24, 68–72. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Kim, J.W.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006, 3, 187–197. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, Z. Glucose Transporter Regulation in Cancer: A Profile and the Loops. Crit. Rev. Eukaryot. Gene Expr. 2016, 26, 223–238. [Google Scholar]
- Singh, D.; Arora, R.; Kaur, P.; Singh, B.; Mannan, R.; Arora, S. Overexpression of hypoxia-inducible factor and metabolic pathways: Possible targets of cancer. Cell Biosci. 2017, 7, 62. [Google Scholar] [CrossRef]
- Li, F.L.; Liu, J.P.; Bao, R.X.; Yan, G.Q.; Feng, X.; Xu, Y.-P.; Sun, Y.-P.; Yan, W.; Ling, Z.-Q.; Xiong, Y.; et al. Acetylation accumulates PFKFB3 in cytoplasm to promote glycolysis and protects cells from cisplatin-induced apoptosis. Nat. Commun. 2018, 9, 508. [Google Scholar] [CrossRef]
- Houddane, A.; Bultot, L.; Novellasdemunt, L.; Johanns, M.; Gueuning, M.A.; Vertommen, D.; Coulie, P.G.; Bartrons, R.; Hue, L.; Rider, M.H. Role of Akt/PKB and PFKFB isoenzymes in the control of glycolysis, cell proliferation and protein synthesis in mitogen-stimulated thymocytes. Cell Signal. 2017, 34, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Liu, R.; Li, J.; Zhang, C.; Wang, Y.; Cai, Q.; Qian, X.; Xia, Y.; Zheng, Y.; Piao, Y.; et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat. Commun. 2017, 8, 949. [Google Scholar] [CrossRef] [PubMed]
- Gumińska, M.; Wazewska-Czyzewska, M. Enzymatic pattern of glucose metabolic pathways in pyruvate kinase-deficient erythrocytes. Clin. Chim. Acta 1975, 64, 165–172. [Google Scholar] [CrossRef]
- Fukuda, R.; Zhang, H.; Kim, J.W. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 2007, 129, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Bosch-Marce, M.; Shimoda, L.A.; Yee, S.T.; Baek, J.H.; Wesley, J.B.; Gonzalez, F.J.; Semenza, G.L. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008, 283, 10892–10903. [Google Scholar] [CrossRef]
- Wang, D.; Wang, Q.; Yan, G.; Qia, Y.; Zhu, B.; Liu, B.; Tang, C. Hypoxia induces lactate secretion and glycolytic efflux by downregulating mitochondrial pyruvate carrier levels in human umbilical vein endothelial cells. Mol. Med. Rep. 2018, 18, 1710–1717. [Google Scholar] [CrossRef]
- Nawaz, M.H.; Ferreira, J.C.; Nedyalkova, L.; Zhu, H.; Carrasco-López, C.; Kirmizialtin, S.; Rabeh, V.M. The catalytic inactivation of the N-half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation. Biosci. Rep. 2018, 38, BSR20171666. [Google Scholar] [CrossRef] [PubMed]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef] [PubMed]
- San-Millán, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2017, 38, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Halestrap, A.P. The monocarboxylate transporter family–structure and functional characterization. IUBMB Life 2012, 64, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Végran, F.; Boidot, R.; Michiels, C.; Sonveaux, P.; Feron, O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011, 71, 2550–2560. [Google Scholar] [CrossRef]
- Porporato, P.E.; Payen, V.L.; De Saedeleer, C.J.; Préat, V.; Thissen, J.-P.; Feron, O.; Sonveaux, P. Lactate stimulates angiogenesis and accelerates the healing of superficial and ischemic wounds in mice. Angiogenesis 2012, 15, 581–592. [Google Scholar] [CrossRef]
- Dhup, S.; Dadhich, R.K.; Porporato, P.E.; Préat, V.; Thissen, J.-P.; Feron, O.; Sonveaux, P. Multiple biological activities of lactic acid in cancer: Influences on tumor growth, angiogenesis and metastasis. Curr. Pharm. Des. 2012, 18, 1319–1330. [Google Scholar] [CrossRef]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef]
- Zhai, X.; Yang, Y.; Wan, J.; Zhu, R.; Wu, Y. Inhibition of LDH-A by oxamate induces G2/M arrest, apoptosis and increases radiosensitivity in nasopharyngeal carcinoma cells. Oncol. Rep. 2013, 30, 2983–2991. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Z.; Chen, Z.; Chen, R.; Zhao, D.; Zhou, Y.; Qiao, L. Effects of the suppression of lactate dehydrogenase A on the growth and invasion of human gastric cancer cells. Oncol Rep. 2015, 33, 157–162. [Google Scholar] [CrossRef]
- Le, A.; Cooper, C.L.; Gouw, A.M.; Dinavahi, R.; Maitra, A.; Deck, L.M.; Royer, R.E.; Vander Jagt, D.L.; Semenza, G.L.; Dang, C.V. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 2037–2042. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Yuan, X.; Wei, M.; Wu, J.; Qin, Z.-H. The diverse role of TIGAR in cellular homeostasis and cancer. Free Radic. Res. 2018, 52, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, A.; Samsó, P.; Fontova, P.; Simon-Molas, H.; Manzano, A.; Castaño, E.; Rosa, J.L.; Martinez-Outshoorn, U.; Ventura, F.; Navarro-Sabaté, A.; et al. R.TGF-β1 targets Smad, p38 MAPK, and PI3K/Akt signaling pathways to induce PFKFB3 gene expression and glycolysis in glioblastoma cells. FEBS J. 2017, 284, 3437–3454. [Google Scholar] [CrossRef] [PubMed]
- Soukupova, J.; Malfettone, A.; Hyroššová, P.; Hernández-Alvarez, M.-I.; Peñuelas-Haro, I.; Bertran, E.; Junza, A.; Capellades, J.; Giannelli, G.; Yanes, O.; et al. Role of the Transforming Growth Factor-β in regulating hepatocellular carcinoma oxidative metabolism. Sci. Rep. 2017, 7, 12486. [Google Scholar] [CrossRef]
- Scatena, R.; Bottoni, P.; Pontoglio, A.; Giardina, B. Revisiting the Warburg effect in cancer cells with proteomics. The emergence of new approaches to diagnosis, prognosis and therapy. Proteomics. Clin. Appl. 2010, 4, 143–158. [Google Scholar] [CrossRef]
- Zhou, W.; Liotta, L.A.; Petricoin, E.F. The Warburg Effect and Mass Spectrometry-based Proteomic Analysis. Cancer Genomics. Proteomics. 2017, 14, 211–218. [Google Scholar]
- Matsumoto, M.; Matsuzaki, F.; Oshikawa, K.; Goshima, N.; Mori, M.; Kawamura, Y.; Ogawa, K.; Fukuda, E.; Nakatsumi, H.; Natsume, T.; et al. A large-scale targeted proteomics assay resource based on an in vitro human proteome. Nat. Methods. 2017, 14, 251–258. [Google Scholar] [CrossRef]
- Moore, L.E.; Pfeiffer, R.M.; Zhang, Z.; Lu, K.H.; Fung, E.T.; Bast, R.C., Jr. Proteomic biomarkers in combination with CA 125 for detection of epithelial ovarian cancer using prediagnostic serum samples from the Prostate, Lung, Colorectal, and Ovarian (PLCO). Cancer Screen. Trial. 2012, 118, 91–100. [Google Scholar]
- Zhong, H.; De Marzo, A.M.; Laughner, E.; Lu, K.H.; Fung, E.T.; Bast, R.C., Jr. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999, 59, 5830–5835. [Google Scholar]
- Unwin, R.D.; Craven, R.A.; Harnden, P.; Hanrahan, S.; Totty, N.; Knowles, M.; Eardley, I.; Selby, P.J.; Banks, R.E. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 2003, 3, 1620–1632. [Google Scholar] [CrossRef]
- Courtnay, R.; Ngo, D.C.; Malik, N.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Almozyan, S.; Colak, D.; Mansour, F.; Alaiya, A.; Al-Harazi, O.; Qattan, A.; Al-Mohanna, F.; Al-Alwan, M.; Ghebeh, N. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int. J. Cancer. 2017, 141, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Denko, N.C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 2008, 8, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Pascale, R.M.; Simile, M.M.; Peitta, G.; Seddaiu, M.A.; Feo, F.; Calvisi, D.F. Experimental Models to Define the Genetic Predisposition to Liver Cancer. Cancers 2019, 11, 1450. [Google Scholar] [CrossRef] [PubMed]
- Casabona, D.; Frau, M.; Seddaiu, M.A.; Feo, F. Cell cycle deregulation in liver lesions of rats with and without genetic predisposition to hepatocarcinogenesis. Hepatology 2002, 35, 1341–1350. [Google Scholar]
- De Miglio, M.R.; Simile, M.M.; Muroni, M.R.; Pusceddu, S.; Calvisi, D.; Carru, A.; Seddaiu, M.A.; Daino, L.; Deiana, L.; Pascale, R.M.; et al. Correlation of c-myc overexpression and amplification with progression of preneoplastic liver lesions to malignancy in the poorly susceptible Wistar rat strain. Mol. Carcinog. 1999, 25, 21–29. [Google Scholar] [CrossRef]
- Calvisi, D.F.; Pinna, F.; Pellegrino, R.; Sanna, V.; Sini, M.; Daino, L.; Simile, M.M.; De Miglio, M.R.; Frau, M.; Tomasi, M.L.; et al. Ras-driven proliferation and apoptosis signaling during rat liver carcinogenesis is under genetic control. Int. J. Cancer 2008, 123, 2057–2064. [Google Scholar] [CrossRef]
- Ranzani, G.N.; Pellegata, N.S.; Previderè, C.; Saragoni, A.; Vio, A.; Maltoni, M.; Amadori, D. Heterogeneous protooncogene amplification correlates with tumor progression and presence of metastases in gastric cancer patients. Cancer Res. 1990, 50, 7811–7814. [Google Scholar]
- Frau, M.; Simile, M.M.; Tomasi, M.L.; Demartis, M.I.; Daino, L.M.E.; Seddaiu, M.A.; Brozzetti, S.; Feo, C.F.; Massarelli, G.; Solinas, G.; et al. An expression signature of phenotypic resistance to hepatocellular carcinoma identified by cross-species gene expression analysis. Cell Oncol. 2012, 35, 163–173. [Google Scholar] [CrossRef]
- Feo, F.; Canuto, R.A.; Garcea, R. Acceptor control ratio of mitochondria. Factors affecting it in Morris hepatoma 5123 and Yoshida hepatoma AH-130. Eur. J. Cancer 1973, 9, 203–214. [Google Scholar] [CrossRef]
- Frau, M.; Feo, F.; Pascale, R.M. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J. Hepatol. 2013, 59, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Gerbracht, U.; Eigenbrodt, E.; Simile, M.M.; Pascale, R.M.; Gaspa, L.; Daino, L.; A Seddaiu, M.; De Miglio, M.R.; Nufris, A.; Feo, F. Effect of S-adenosyl-L-methionine on the development of preneoplastic foci and the activity of some carbohydrate metabolizing enzymes in the liver, during experimental hepatocarcinogenesis. Anticancer. Res. 1993, 13, 1965–1972. [Google Scholar] [PubMed]
- Weber, G.; Lea, M.A. The molecular correlation concept of neoplasia. Adv. Enzym. Regul. 1966, 4, 115–145. [Google Scholar] [CrossRef]
- Lu, J.; Tan, M.; Cai, Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2014, 356, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg effect: Essential part of metabolic reprogramming and central contributor to cancer progression. Int. J. Radiat. Boil. 2019, 95, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, T.; Schuster, S.; Bonhoeffer, S.; Döring, V.; Mootz, H.D.; Nangle, L.A.; Hendrickson, T.L.; De Crécy-Lagard, V.; Schimmel, P.; Marlière, P. Cooperation and Competition in the Evolution of ATP-Producing Pathways. Science 2001, 292, 504–507. [Google Scholar] [CrossRef]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Gaude, E.; Hilvo, M.; Frezza, C. The Metabolic Alterations of Cancer Cells. Enzym. Eng. Evol. Gen. Methods 2014, 542, 1–23. [Google Scholar]
- Yi, W.; Clark, P.M.; Mason, D.E.; Keenan, M.C.; Hill, C.; Goddard, W.A.; Peters, E.C.; Driggers, E.M.; Hsieh-Wilson, L.C. Phosphofructokinase 1 Glycosylation Regulates Cell Growth and Metabolism. Science 2012, 337, 975–980. [Google Scholar] [CrossRef]
- Kilbur, D.G.; Lilly, M.D.; Webb, F.C. The energetics of mammalian cell growth. J. Cell Sci. 2011, 4, 645–654. [Google Scholar]
- Lincet, H.; Icard, P. How do glycolytic enzymes favour cancer cell proliferation by nonmetabolic functions? Oncogene 2015, 34, 3751–3759. [Google Scholar] [CrossRef] [PubMed]
- Krüger, A.; Grüning, N.-M.; Wamelink, M.M.; Kerick, M.; Kirpy, A.; Parkhomchuk, D.; Bluemlein, K.; Schweiger, M.-R.; Soldatov, A.; Lehrach, H.; et al. The Pentose Phosphate Pathway Is a Metabolic Redox Sensor and Regulates Transcription During the Antioxidant Response. Antioxid. Redox Signal. 2011, 15, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Chaneton, B.; Hillmann, P.; Zheng, L.M.; Martin, A.C.L.; Maddocks, O.D.K.; Chokkathukalam, A.; Coyle, J.E.; Jankevics, A.; Holding, F.P.; Vousden, H.K.; et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 2012, 491, 458–462, Erratum in 2013, 496, 386. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Chen, X.; Ma, J.; Peng, H.; Wang, F.; Zha, X.; Wang, Y.; Jing, Y.; Yang, H.; Chen, R.; et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc. Natl. Acad. Sci. USA 2011, 108, 4129–4134. [Google Scholar] [CrossRef]
- Luo, W.; Hu, H.; Chang, R.; Zhong, J.; Knabel, M.; O’Meally, R.; Cole, R.N.; Pandey, A.; Semenza, G.L. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell 2011, 145, 732–744. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, L.Z.; Yin, Y.; He, J.; Li, Q.; Qian, X.; You, Y.; Lu, Z.; Peiper, S.C.; Shu, Y.; et al. Regulatory circuit of PKM2/NF-κB/miR-148a/152-modulated tumor angiogenesis and cancer progression. Oncogene 2015, 34, 5482–5493. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Ojo, D.; Yan, J.; Tang, D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015, 356, 184–191. [Google Scholar] [CrossRef]
- Luo, W.; Semenza, G.L. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol. Metab. 2012, 23, 560–566. [Google Scholar] [CrossRef]
- Yang, W.; Zheng, Y.; Xia, Y.; Ji, H.; Chen, X.; Guo, F.; Lyssiotis, C.A.; Aldape, K.; Cantley, L.C.; Lu, Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012, 14, 1295–1304. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Gupta, V.; Gopinath, P.; Mazurek, S.; Bamezai, R.N.K. Pyruvate kinase M2 and cancer: An updated assessment. FEBS Lett. 2014, 588, 2685–2692. [Google Scholar] [CrossRef]
- Hoshino, A.; Hirst, J.A.; Fujii, H. Regulation of Cell Proliferation by Interleukin-3-induced Nuclear Translocation of Pyruvate Kinase. J. Boil. Chem. 2007, 282, 17706–17711. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-J.; Hsieh, Y.-J.; Cheng, W.-C.; Lin, C.-P.; Lin, Y.-S.; Yang, S.-F.; Chen, C.-C.; Izumiya, Y.; Yu, J.-S.; Kung, H.-J.; et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1-mediated glucose metabolism. Proc. Natl. Acad. Sci. USA 2013, 111, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, H.K.; Han, Y.-M.; Kim, J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int. J. Biochem. Cell Boil. 2008, 40, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Chen, M.; Wei, W.; Zhang, X.; Zhang, M.; Yao, Y.; Lv, Y.; Ling, T.; Wang, L.; Zou, X. Crosstalk of mTOR/PKM2 and STAT3/c-Myc signaling pathways regulate the energy metabolism and acidic microenvironment of gastric cancer. J. Cell Biochem. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Geng, L.; Zeng, L.; Liu, F.; Huang, Q. AKT2 contributes to increase ovarian cancer cell migration and invasion through the AKT2-PKM2-STAT3/NF-κB axis. Cell Signal. 2018, 45, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, H.; Yang, J.J.; Liu, X.; Liu, Z.-R.; Jenny, J.Y. Pyruvate Kinase M2 Regulates Gene Transcription by Acting as a Protein Kinase. Mol. Cell 2012, 45, 598–609. [Google Scholar] [CrossRef]
- Luo, W.; Semenza, G.L. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget 2011, 2, 551–556. [Google Scholar] [CrossRef]
- Yang, W.; Xia, Y.; Hawke, D.; Li, X.; Liang, J.; Xing, D.; Aldape, K.; Hunter, T.; Yung, W.A.; Lu, Z. PKM2 Phosphorylates Histone H3 and Promotes Gene Transcription and Tumorigenesis. Cell 2012, 150, 685–696. [Google Scholar] [CrossRef]
- Israelsen, W.J.; Dayton, T.L.; Davidson, S.M.; Fiske, B.P.; Hosios, A.M.; Bellinger, G.; Li, J.; Yu, Y.; Sasaki, M.; Horner, J.W.; et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 2013, 155, 397–409. [Google Scholar] [CrossRef]
- Matsuda, S.; Adachi, J.; Ihara, M.; Tanuma, N.; Shima, H.; Kakizuka, A.; Ikura, M.; Ikura, T.; Matsuda, T. Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic Acids Res. 2015, 44, 636–647. [Google Scholar] [CrossRef]
- Chen, J.; Chung, F.; Yang, G.; Pu, M.; Gao, H.; Jiang, W.; Yin, H.; Capka, V.; Kasibhatla, S.; Laffitte, B.; et al. Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget 2013, 4, 2502–2511. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Guo, S.; Li, Q.; Yang, L.; Xia, Z.; Zhang, L.; Huang, Z.; Zhang, N. Phosphoglycerate dehydrogenase induces glioma cells proliferation and invasion by stabilizing forkhead box M1. J. Neurooncol. 2013, 111, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Chakravarthy, S.; Tokuda, J.M.; Pollack, L.; Bowman, G.D.; Lee, Y.-S. Succinyl-5-aminoimidazole-4-carboxamide-1-ribose 5′-Phosphate (SAICAR) Activates Pyruvate Kinase Isoform M2 (PKM2) in Its Dimeric Form. Biochemistry 2016, 55, 4731–4736. [Google Scholar] [CrossRef]
- Kondoh, H.; E Lleonart, M.; Gil, J.; Wang, J.; Degan, P.; Peters, G.; Martinez, D.; Carnero, A.; Beach, D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005, 65, 177–185. [Google Scholar] [PubMed]
- Hallows, W.C.; Yu, W.; Denu, J.M. Regulation of Glycolytic Enzyme Phosphoglycerate Mutase-1 by Sirt1 Protein-mediated Deacetylation. J. Boil. Chem. 2011, 287, 3850–3858. [Google Scholar] [CrossRef]
- Hitosugi, T.; Zhou, L.; Elf, S.; Fan, J.; Kang, H.; Seo, J.H.; Shan, C.; Dai, Q.; Zhang, L.; Xie, J.; et al. Phosphoglycerate Mutase 1 Coordinates Glycolysis and Biosynthesis to Promote Tumor Growth. Cancer Cell 2012, 22, 585–600. [Google Scholar] [CrossRef]
- Evans, M.J.; Saghatelian, A.; Sorensen, E.J.; Cravatt, B.F. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat. Biotechnol. 2005, 23, 1303–1307. [Google Scholar] [CrossRef]
- Chang, C.-J.; Yin, P.-H.; Yang, D.-M.; Wang, C.-H.; Hung, W.-Y.; Chi, C.-W.; Wei, Y.-H.; Lee, H.-C. Mitochondrial dysfunction-induced amphiregulin upregulation mediates chemo-resistance and cell migration in HepG2 cells. Cell. Mol. Life Sci. 2009, 66, 1755–1765. [Google Scholar] [CrossRef]
- Medes, G.; A Spirtes, M.; Weinhouse, S. The estimation of fatty acid synthesis in rat liver slices. J. Boil. Chem. 1953, 205, 401–408. [Google Scholar]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011, 481, 380–384. [Google Scholar] [CrossRef]
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniëls, V.W.; Machiels, J.; et al. De novo Lipogenesis Protects Cancer Cells from Free Radicals and Chemotherapeutics by Promoting Membrane Lipid Saturation. Cancer Res. 2010, 70, 8117–8126. [Google Scholar] [CrossRef] [PubMed]
- Warmoes, M.; Jaspers, J.E.; Xu, G.; Sampadi, B.K.; Pham, T.V.; Knol, J.C.; Piersma, S.R.; Boven, E.; Jonkers, J.; Rottenberg, S.; et al. Proteomics of Genetically Engineered Mouse Mammary Tumors Identifies Fatty Acid Metabolism Members as Potential Predictive Markers for Cisplatin Resistance. Mol. Cell. Proteom. 2013, 12, 1319–1334. [Google Scholar] [CrossRef] [PubMed]
- Bhutia, Y.D.; Babu, E.; Ramachandran, S.; Ganapathy, V. Amino Acid Transporters in Cancer and Their Relevance to “Glutamine Addiction”: Novel Targets for the Design of a New Class of Anticancer Drugs. Cancer Res. 2015, 75, 1782–1788. [Google Scholar] [CrossRef] [PubMed]
- Felig, P.; Wahren, J.; Räf, L. Evidence of Inter-organ Amino-Acid Transport by Blood Cells in Humans. Proc. Natl. Acad. Sci. USA 1973, 70, 1775–1779. [Google Scholar] [CrossRef]
- Moreadith, R.W.; Lehninger, A.L. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme. J. Boil. Chem. 1984, 259, 6215–6221. [Google Scholar]
- Ward, P.S.; Patel, J.; Wise, D.R.; Abdel-Wahab, O.; Bennett, B.D.; Coller, H.A.; Cross, J.R.; Fantin, V.R.; Hedvat, C.V.; Perl, A.E.; et al. The Common Feature of Leukemia-Associated IDH1 and IDH2 Mutations Is a Neomorphic Enzyme Activity Converting α-Ketoglutarate to 2-Hydroxyglutarate. Cancer Cell 2010, 17, 225–234. [Google Scholar] [CrossRef]
- Fan, J.; Kamphorst, J.J.; Mathew, R.; Chung, M.K.; White, E.; Shlomi, T.; Rabinowitz, J.D. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Boil. 2013, 9, 712. [Google Scholar] [CrossRef]
- Jiang, X.; Sun, Q.; Li, H.; Li, K.; Ren, X. The role of phosphoglycerate mutase 1 in tumor aerobic glycolysis and its potential therapeutic implications. Int. J. Cancer 2013, 135, 1991–1996. [Google Scholar] [CrossRef]
- Kaschnitz, R.; Hatefi, Y.; Morris, H. Oxidative phosphorylation properties of mitochondria isolated from transplanted hepatoma. Biochim. Biophys. Acta Bioenerg. 1976, 449, 224–235. [Google Scholar] [CrossRef]
- Chen, X.; Qian, Y.; Wu, S. The Warburg effect: Evolving interpretations of an established concept. Free. Radic. Boil. Med. 2014, 79, 253–263. [Google Scholar] [CrossRef]
- Schwartz, L.; Supuran, C.; Reshkin, S.J. The Warburg Effect and the Hallmarks of Cancer. Anti-Cancer Agents Med. Chem. 2017, 17, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Daye, D.; Wellen, K.E. Metabolic reprogramming in cancer: Unraveling the role of glutamine in tumorigenesis. Semin. Cell Dev. Boil. 2012, 23, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of pyruvate metabolism and human disease. Cell Mol. Life Sci. 2014, 71, 2577–2604. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. Myc, metabolism, and cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef]
- Jeong, S.M.; Lee, A.; Lee, J.J.; Haigis, M.C. SIRT4 Protein Suppresses Tumor Formation in Genetic Models of Myc-induced B Cell Lymphoma. J. Boil. Chem. 2013, 289, 4135–4144. [Google Scholar] [CrossRef]
- Dolezal, J.M.; Wang, H.; Kulkarni, S.; Jackson, L.; Lu, J.; Ranganathan, S.; Goetzman, E.S.; Bharathi, S.S.; Beezhold, K.; Byersdorfer, C.A.; et al. Sequential adaptive changes in a c-Myc-driven model of hepatocellular carcinoma. J. Boil. Chem. 2017, 292, 10068–10086. [Google Scholar] [CrossRef]
- Morita, M.; Gravel, S.-P.; Hulea, L.; Larsson, O.; Pollak, M.; St-Pierre, J.; Topisirovic, I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 2015, 14, 473–480. [Google Scholar] [CrossRef]
- Tan, Z.; Luo, X.; Xiao, L.; Tang, M.; Bode, A.M.; Dong, Z.; Cao, Y. The Role of PGC1 in Cancer Metabolism and its Therapeutic Implications. Mol. Cancer Ther. 2016, 15, 774–782. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, Y.; Luo, L.; Shi, F.; Zou, J.; Lin, H.; Ying, Y.; Luo, Y.; Zhan, Z.; Liu, P.; et al. AMP-activated protein kinase regulates cancer cell growth and metabolism via nuclear and mitochondria events. J. Cell. Mol. Med. 2019, 23, 3951–3961. [Google Scholar] [CrossRef]
- Berkers, C.R.; Maddocks, O.D.K.; Cheung, E.C.; Mor, I.; Vousden, K.H. Metabolic Regulation by p53 Family Members. Cell Metab. 2013, 18, 617–633. [Google Scholar] [CrossRef]
- Jiang, D.; LaGory, E.L.; Broz, D.K.; Bieging, K.T.; Brady, C.A.; Link, N.; Abrams, J.M.; Giaccia, A.J.; Attardi, L.D. Analysis of p53 transactivation domain mutants reveals Acad11 as a metabolic target important for p53 pro-survival function. Cell Rep. 2015, 10, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Vaseva, A.V.; Moll, U.M. The mitochondrial p53 pathway. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 414–420. [Google Scholar] [CrossRef]
- Dashzeveg, N.; Yoshida, K. Crosstalk between tumor suppressors p53 and PKCδ: Execution of the intrinsic apoptotic pathways. Cancer Lett. 2016, 377, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Bause, A.S.; Haigis, M.C. SIRT3 regulation of mitochondrial oxidative stress. Exp. Gerontol. 2013, 48, 634–639. [Google Scholar] [CrossRef]
- Bell, E.L.; Emerling, B.M.; Ricoult, S.J.; Guarente, L. Sirt3 Suppresses Hypoxia Inducible Factor 1alpha and Tumor Growth by Inhibiting Mitochondrial Ros Production. Oncogene 2011, 30, 2986–2996. [Google Scholar] [CrossRef] [PubMed]
- Finley, L.W.; Carracedo, A.; Lee, J.J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.; Cardoso, S.M.; Clish, C.B.; et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1α Destabilization. Cancer Cell 2011, 19, 416–428. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, M.; Zeng, X.; Yang, J.; Deng, H.; Yi, L.; Mi, M.-T. Resveratrol regulates mitochondrial reactive oxygen species homeostasis through Sirt3 signaling pathway in human vascular endothelial cells. Cell Death Dis. 2014, 5, e1576. [Google Scholar] [CrossRef]
- Montessuit, C.; Thorburn, A. Transcriptional Activation of the Glucose Transporter GLUT1 in Ventricular Cardiac Myocytes by Hypertrophic Agonists. J. Boil. Chem. 1999, 274, 9006–9012. [Google Scholar] [CrossRef]
- Kole, H.K.; Resnick, R.J.; Van Doren, M.; Racker, E. Regulation of 6-phosphofructo-1-k, nase activity Nin ras-transformed rat-1 fibroblasts. Arch. Biochem. Biophys. 1991, 286, 586–590. [Google Scholar] [CrossRef]
- Yalcin, A.; Telang, S.; Clem, B.; Chesney, J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/ fructose-2, 6-bisphosphatases in cancer. Exp. Mol. Pathol. 2009, 86, 174–179. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia and cancer. Cancer Metastasis Rev. 2007, 26, 223–224. [Google Scholar] [CrossRef] [PubMed]
- Csibi, A.; Fendt, S.-M.; Li, C.; Poulogiannis, G.; Choo, A.Y.; Chapski, D.J.; Jeong, S.M.; Dempsey, J.M.; Parkhitko, A.A.; Morrison, T.; et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 2013, 153, 840–854. [Google Scholar] [CrossRef] [PubMed]
- Ben-Sahra, I.; Hoxhaj, G.; Ricoult, S.J.; Asara, J.M.; Manning, B.D. Mtorc1 Induces Purine Synthesis through Control of the Mitochondrial Tetrahydrofolate Cycle. Science 2016, 351, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Zeng, J.; Geng, P.; Fang, C.; Wang, Y.; Sun, M.; Wang, C.; Wang, J.; Yin, P.; Hu, C.; et al. Global Metabolic Profiling Identifies a Pivotal Role of Proline and Hydroxyproline Metabolism in Supporting Hypoxic Response in Hepatocellular Carcinoma. Clin. Cancer Res. 2017, 24, 474–485. [Google Scholar] [CrossRef] [PubMed]
- LaGory, E.L.; Wu, C.; Taniguchi, C.M.; Ding, C.-K.C.; Chi, J.-T.; von Eyben, R.; Scott, D.A.; Richardson, A.D.; Giaccia, A.J. Suppression of Pgc-1alpha Is Critical for Reprogramming Oxidative Metabolism in Renal Cell Carcinoma. Cell Rep. 2015, 12, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Halling, J.F.; Jessen, H.; Nøhr-Meldgaard, J.; Buch, B.T.; Christensen, N.M.; Gudiksen, A.; Ringholm, S.; Neufer, P.D.; Prats, C.; Pilegaard, H. PGC-1α regulates mitochondrial properties beyond biogenesis with aging and exercise training. Am. J. Physiol. Metab. 2019, 317, E513–E525. [Google Scholar] [CrossRef]
- LeBleu, V.S.; O’Connell, J.T.; Herrera, K.N.G.; Wikman, H.; Pantel, K.; Haigis, M.C.; De Carvalho, F.M.; Damascena, A.; Chinen, L.T.D.; Rocha, R.M.; et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014, 16, 992–1003. [Google Scholar] [CrossRef]
- Lee, M.; Yoon, J.-H. Metabolic interplay between glycolysis and mitochondrial oxidation: The reverse Warburg effect and its therapeutic implication. World J. Boil. Chem. 2015, 6, 148–161. [Google Scholar] [CrossRef]
- Lisanti, M.P.; Martinez-Outschoorn, U.E.; Chiavarina, B.; Pavlides, S.; Whitaker-Menezes, D.; Tsirigos, A.; Witkiewicz, A.K.; Lin, Z.; Balliet, R.M.; Howell, A.; et al. Understanding the “lethal” drivers of tumor stroma co-evolution: Emerging role(s) for hypoxia, oxidative stress and autophagy/mitophagy in the tumor micro-environment. Cancer Boil. Ther. 2010, 10, 537–542. [Google Scholar] [CrossRef]
- Sonveaux, P.; Végran, F.; Schroeder, T.; Wergin, M.; Verrax, J.; Rabbani, Z.N.; De Saedeleer, C.J.; Kennedy, K.M.; Diepart, C.; Jordan, B.F.; et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Investig. 2008, 118, 3930–3942. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Giatromanolaki, A.; Harris, A.L.; Sivridis, E. Comparison of Metabolic Pathways between Cancer Cells and Stromal Cells in Colorectal Carcinomas: A Metabolic Survival Role for Tumor-Associated Stroma. Cancer Res. 2006, 66, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Koukourakis, M.I.; Giatromanolaki, A.; Sivridis, E.; Gatter, K.C.; Harris, A.L. Pyruvate Dehydrogenase and Pyruvate Dehydrogenase Kinase Expression in Non Small Cell Lung Cancer and Tumor-Associated Stroma. Neoplasia 2005, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yaku, K.; Okabe, K.; Hikosaka, K.; Nakagawa, T. NAD Metabolism in Cancer Therapeutics. Front. Oncol. 2018, 8, 622. [Google Scholar] [CrossRef] [PubMed]
- Pascale, R.M.; Marras, V.; Simile, M.M.; Daino, L.; Pinna, G.; Bennati, S.; Carta, M.; A Seddaiu, M.; Massarelli, G.; Feo, F. Chemoprevention of rat liver carcinogenesis by S-adenosyl-L-methionine: A long-term study. Cancer Res. 1992, 52, 4979–4986. [Google Scholar]
- Terranova, T.; Feo, F.; Gravela, E.; Gariel, L. Die wirkung der 2-desoxyglucose auf energhetischen stoffwechsel nund auf die proteinsynthese von tumorzellen und normalzellen. Z. Krebsforsch 1964, 66, 41–45. [Google Scholar] [CrossRef]
- DeWaal, D.; Nogueira, V.; Terry, A.R.; Patra, K.C.; Jeon, S.-M.; Guzman, G.; Au, J.; Long, C.P.; Antoniewicz, M.R.; Hay, N. Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat. Commun. 2018, 9, 446. [Google Scholar] [CrossRef]
- Bertero, T.; Cottrill, K.A.; Annis, S.; Bhat, B.; Gochuico, B.R.; Osorio, J.C.; Rosas, I.; Haley, K.J.; Corey, K.E.; Chung, R.T. A YAP/TAZ-miR-130/301 molecular circuit exerts systems-level control of fibrosis in a network of human diseases and physiologic conditions. Sci. Rep. 2015, 5, 18277. [Google Scholar] [CrossRef]
- Singh, S.; Pandey, S.; Bhatt, A.N.; Chaudhary, R.; Bhuria, V.; Kalra, N.; Soni, R.; Roy, B.G.; Saluja, D.; Dwarakanath, B. Chronic Dietary Administration of the Glycolytic Inhibitor 2-Deoxy-D-Glucose (2-DG) Inhibits the Growth of Implanted Ehrlich’s Ascites Tumor in Mice. PLoS ONE 2015, 10, e0132089. [Google Scholar] [CrossRef]
- Al-Shammari, A.M.; Abdullah, A.H.; Allami, Z.M.; Yaseen, N.Y. 2-Deoxyglucose and Newcastle Disease Virus Synergize to Kill Breast Cancer Cells by Inhibition of Glycolysis Pathway Through Glyceraldehyde3-Phosphate Downregulation. Front. Mol. Biosci. 2019, 6, 90. [Google Scholar] [CrossRef]
- Maschek, G.; Savaraj, N.; Priebe, W.; Braunschweiger, P.; Hamilton, K.; Tidmarsh, G.F.; De Young, L.R.; Lampidis, T.J. 2-deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo. Cancer Res. 2004, 64, 31–34. [Google Scholar] [CrossRef]
- Enzo, E.; Santinon, G.; Pocaterra, A.; Aragona, M.; Bresolin, S.; Forcato, M.; Grifoni, D.; Pession, A.; Zanconato, F.; Guzzo, G.; et al. Aerobic glycolysis tunes YAP / TAZ transcriptional activity. EMBO J. 2015, 34, 1349–1370. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.H.; Guan, K.-L. Interplay between YAP/TAZ and Metabolism. Cell Metab. 2018, 28, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Simile, M.M.; Latte, G.; Demartis, M.I.; Brozzetti, S.; Calvisi, D.F.; Porcu, A.; Feo, C.F.; Seddaiu, M.A.; Daino, L.; Berasain, C.; et al. Post-translational deregulation of YAP1 is genetically controlled in rat liver cancer and determines the fate and stem-like behavior of the human disease. Oncotarget 2016, 7, 49194–49216. [Google Scholar] [CrossRef] [PubMed]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Clem, B.F.; O’Neal, J.; Tapolsky, G.; Clem, A.L.; Imbert-Fernandez, Y.; Kerr, D.A.; Klarer, A.C.; Redman, R.; Miller, N.M.; Trent, J.O.; et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol. Cancer Ther. 2013, 12, 1461–1470. [Google Scholar] [CrossRef]
- O’Neal, J.; Clem, A.; Reynolds, L.; Dougherty, S.; Imbert-Fernandez, Y.; Telang, S.; Chesney, J.; Clem, B.F. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) suppresses glucose metabolism and the growth of HER2+ breast cancer. Breast Cancer Res. Treat. 2016, 160, 29–40. [Google Scholar] [CrossRef]
- Clem, B.; Telang, S.; Clem, A.; Yalcin, A.; Meier, J.; Simmons, A.; Rasku, M.A.; Arumugam, S.; Dean, W.L.; Eaton, J.; et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol. Cancer Ther. 2008, 7, 110–120. [Google Scholar] [CrossRef]
- Krasnov, G.S.; Dmitriev, A.A.; Snezhkina, A.V.; Kudryavtseva, A.V. Deregulation of glycolysis in cancer: Glyceraldehyde-3-phosphate dehydrogenase as a therapeutic target. Expert Opin. Ther. Targets 2013, 17, 681–693. [Google Scholar] [CrossRef]
- Wang, Y.; Hao, F.; Nan, Y.; Qu, L.; Na, W.; Jia, C.; Chen, X. PKM2 Inhibitor Shikonin Overcomes the Cisplatin Resistance in Bladder Cancer by Inducing Necroptosis. Int. J. Boil. Sci. 2018, 14, 1883–1891. [Google Scholar] [CrossRef]
- Lei, R.; Zhang, S.; Wang, Y.; Dai, S.; Sun, J.; Zhu, C. Metformin Inhibits Epithelial-to-Mesenchymal Transition of Keloid Fibroblasts via the HIF-1α/PKM2 Signaling Pathway. Int. J. Med Sci. 2019, 16, 960–966. [Google Scholar] [CrossRef]
- Granchi, C.; Bertini, S.; Macchia, M.; Minutolo, F. Inhibitors of lactate dehydrogenase isoforms and their therapeutic potentials. Curr. Med. Chem. 2010, 17, 672–697. [Google Scholar] [CrossRef] [PubMed]
- Oshima, N.; Ishida, R.; Kishimoto, S.; Beebe, K.; Brender, J.R.; Yamamoto, K.; Urban, D.; Rai, G.; Johnson, M.S.; Benavides, G.; et al. Dynamic Imaging of LDH Inhibition in Tumors Reveals Rapid In Vivo Metabolic Rewiring and Vulnerability to Combination Therapy. Cell Rep. 2020, 30, 1798.e4–1810.e4. [Google Scholar] [CrossRef] [PubMed]
- Xintaropoulou, C.; Ward, C.; Wise, A.; Marston, H.; Turnbull, A.; Langdon, S.P. A comparative analysis of inhibitors of the glycolysis pathway in breast and ovarian cancer cell line models. Oncotarget 2015, 6, 25677–25695. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, A.; E Purkey, H.; Hitz, A.; Robarge, K.; Peterson, D.; Labadie, S.; Kwong, M.; Hong, R.; Gao, M.; Del Nagro, C.; et al. Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition. Nat. Methods 2016, 12, 779–786. [Google Scholar] [CrossRef]
- Yang, Y.; Su, D.; Zhao, L.; Zhang, D.; Xu, J.; Wan, J.; Fan, S.; Chen, M. Different effects of LDH-A inhibition by oxamate in non-small cell lung cancer cells. Oncotarget 2014, 5, 11886–11896. [Google Scholar] [CrossRef]
- Li, L.; Kang, L.; Zhao, W.; Feng, Y.; Liu, W.; Wang, T.; Mai, H.; Huang, J.; Chen, S.; Liang, Y.; et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017, 400, 89–98. [Google Scholar] [CrossRef]
- Le Floch, R.; Chiche, J.; Marchiq, I.; Naiken, T.; Ilc, K.; Murray, C.M.; Critchlow, S.E.; Roux, D.; Simon, M.-P.; Pouysségur, J. CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 16663–16668. [Google Scholar] [CrossRef]
- Doherty, J.R.; Yang, C.; Scott, K.E.; Cameron, M.D.; Fallahi, M.; Li, W.; Hall, M.A.; Amelio, A.L.; Mishra, J.K.; Li, F.; et al. Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer Res. 2014, 74, 908–920. [Google Scholar] [CrossRef]
- Nath, K.; Guo, L.; Nancolas, B.; Nelson, D.S.; Shestov, A.A.; Lee, S.-C.; Roman, J.; Zhou, R.; Leeper, D.B.; Halestrap, A.P.; et al. Mechanism of antineoplastic activity of lonidamine. Biochim. Biophys. Acta Bioenerg. 2016, 1866, 151–162. [Google Scholar] [CrossRef]
- Colen, C.B.; Seraji-Bozorgzad, N.; Marples, B.; Galloway, M.P.; Sloan, A.E.; Mathupala, S.P. Metabolic remodeling of malignant gliomas for enhanced sensitization during radiotherapy: An in vitro study. Neurosurgery 2006, 59, 1313–1323. [Google Scholar] [CrossRef]
- Wike-Hooley, J.L.; Haveman, J.; Reinhold, H.S. The relevance of tumour pH to the treatment of malignant disease. Radiother. Oncol. 1984, 2, 343–366. [Google Scholar] [CrossRef]
- Yabu, M.; Shime, H.; Hara, H.; Saito, H.; Matsumoto, M.; Seya, T.; Akazawa, T.; Inoue, N. IL-23-dependent and -independent enhancement pathways of IL-17A production by lactic acid. Int. Immunol. 2011, 23, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Shime, H.; Yabu, M.; Akazawa, T.; Kodama, K.; Matsumoto, M.; Seya, T.; Inoue, N. Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J. Immunol. 2008, 180, 7175–7183. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Wilson, M.C. The monocarboxylate transporter family—Role and regulation. IUBMB Life 2012, 64, 109–119. [Google Scholar] [CrossRef]
- Pelicano, H.; Martin, D.S.; Xu, R.H.; Huang, P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006, 25, 4633–4646. [Google Scholar] [CrossRef] [PubMed]
- Cassim, S.; Vučetić, M.; Ždralević, M.; Pouyssegur, J. Warburg and Beyond: The Power of Mitochondrial Metabolism to Collaborate or Replace Fermentative Glycolysis in Cancer. Cancers 2020, 12, 1119. [Google Scholar] [CrossRef]
- Ždralević, M.; Brand, A.; Di Ianni, L.; Dettmer, K.; Reinders, J.; Singer, K.; Peter, K.; Schnell, A.; Bruss, C.; Decking, S.-M.; et al. Double genetic disruption of lactate dehydrogenases A and B is required to ablate the “Warburg effect” restricting tumor growth to oxidative metabolism. J. Boil. Chem. 2018, 293, 15947–15961. [Google Scholar] [CrossRef]
- Alfarouk, K.O.; Verduzco, D.; Rauch, C.; Muddathir, A.K.; Adil, H.H.B.; Elhassan, G.O.; Ibrahim, M.E.; Orozco, J.D.P.; Cardone, R.A.; Reshkin, S.J.; et al. Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question. Oncoscience 2014, 1, 777–802. [Google Scholar] [CrossRef]
- Reshkin, S.J.; Cardone, R.A.; Harguindey, S. Na+-H+ exchanger, pH regulation and cancer. Recent Pat. Anti-Cancer Drug Discov. 2013, 8, 85–99. [Google Scholar] [CrossRef]
- Brisson, L.; Reshkin, S.J.; Goré, J.; Roger, S. pH regulators in invadosomal functioning: Proton delivery for matrix tasting. Eur. J. Cell Boil. 2012, 91, 847–860. [Google Scholar] [CrossRef]
- Brisson, L.; Gillet, L.; Calaghan, S.C.; Besson, P.; Le Guennec, J.-Y.; Roger, S.; Gore, J. NaV1.5 enhances breast cancer cell invasiveness by increasing NHE1-dependent H+ efflux in caveolae. Oncogene 2010, 30, 2070–2076. [Google Scholar] [CrossRef] [PubMed]
- Harguindey, S.; Arranz, J.L.; Wahl, M.L.; Orive, G.; Reshkin, S.J. Proton transport inhibitors as potentially selective anticancer drugs. Anticancer. Res. 2009, 29, 2127–2136. [Google Scholar] [PubMed]
- Doppler, W.; Jaggi, R.; Groner, B. Induction of v-mos and activated Ha-ras oncogene expression in quiescent NIH 3T3 cells causes intracellular alkalinisation and cell-cycle progression. Gene 1987, 54, 147–153. [Google Scholar] [CrossRef]
- Wang, H.; Long, X.; Wang, D.; Lou, M.; Zou, D.; Chen, R.; Nian, W.; Zhou, Q. Increased expression of Na+/H+ exchanger isoform 1 predicts tumor aggressiveness and unfavorable prognosis in epithelial ovarian cancer. Oncol Lett. 2018, 16, 6713–6720. [Google Scholar] [CrossRef]
- Atwal, K.S.; O’Neil, S.V.; Ahmad, S.; Doweyko, L.; Kirby, M.; Dorso, C.R.; Chandrasena, G.; Chen, B.-C.; Zhao, R.; Zahler, R. Synthesis and biological activity of 5-aryl-4-(4-(5-methyl-1H-imidazol-4-yl)piperidin-1-yl)pyrimidine analogs as potent, highly selective, and orally bioavailable NHE-1 inhibitors. Bioorganic Med. Chem. Lett. 2006, 16, 4796–4799. [Google Scholar] [CrossRef]
- Robinson, M.M.; McBryant, S.J.; Tsukamoto, T.; Rojas, C.; Ferraris, D.; Hamilton, S.K.; Hansen, J.C.; Curthoys, N.P. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). Biochem. J. 2007, 406, 407–414. [Google Scholar] [CrossRef]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H.; et al. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B Cells. Cell Metab. 2012, 15, 110–121. [Google Scholar] [CrossRef]
- Xiang, Y.; Stine, Z.E.; Xia, J.; Lu, Y.; O’Connor, R.S.; Altman, B.J.; Hsieh, A.L.; Gouw, A.M.; Thomas, A.G.; Gao, P.; et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Investig. 2015, 125, 2293–2306. [Google Scholar] [CrossRef]
- Colombo, S.L.; Palacios-Callender, M.; Frakich, N.; Carcamo, S.; Kovacs, I.; Tudzarova, S.; Moncada, S. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc. Natl. Acad. Sci, USA 2011, 108, 21069–21074. [Google Scholar] [CrossRef]
- Boysen, G.; Jamshidi-Parsian, A.; Davis, M.A.; Siegel, E.R.; Simecka, C.M.; Kore, R.A.; Dings, R.P.M.; Griffin, R.J. Glutaminase inhibitor CB-839 increases radiation sensitivity of lung tumor cells and human lung tumor xenografts in mice. Int. J. Radiat. Boil. 2019, 95, 436–442. [Google Scholar] [CrossRef]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Meng, Y.; Li, L.; Xu, P.; Wang, J.; Li, Z.; Bian, J. Overview of the Development of Glutaminase Inhibitors: Achievements and Future Directions. J. Med. Chem. 2018, 62, 1096–1115. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Moss, T.; Mangala, L.S.; Marini, J.; Zhao, H.; Wahlig, S.; Armaiz-Pena, G.; Jiang, D.; Achreja, A.; Win, J.; et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol. Syst. Biol. 2014, 10, 728. [Google Scholar] [CrossRef]
- Wang, Q.; Hardie, R.-A.; Hoy, A.J.; van Geldermalsen, M.; Gao, D.; Fazli, L.; Sadowski, M.C.; Balaban, S.; Schreuder, M.; Nagarajah, R.; et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 2015, 236, 278–289. [Google Scholar] [CrossRef]
- Wang, Y.; Bai, C.; Ruan, Y.; Liu, M.; Chu, C.; Qiu, L.; Yang, C.; Li, B. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat. Commun. 2019, 10, 201. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; Kroemer, G. Ammonia: A Diffusible Factor Released by Proliferating Cells That Induces Autophagy. Sci. Signal. 2010, 3, pe19. [Google Scholar] [CrossRef]
- Perez-Escuredo, J.; Dadhich, R.K.; Dhup, S.; Cacace, A.; Van Hée, V.F.; De Saedeleer, C.J.; Sboarina, M.; Rodriguez, F.; Fontenille, M.-J.; Brisson, L.; et al. Lactate promotes glutamine uptake and metabolism in oxidative cancer cells. Cell Cycle 2016, 15, 72–83. [Google Scholar] [CrossRef]
- Cheong, H.; Lindsten, T.; Wu, J.; Lu, C.; Thompson, C.B. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl. Acad. Sci. USA 2011, 108, 11121–11126. [Google Scholar] [CrossRef]
- Eng, C.H.; Yu, K.; Lucas, J.; White, E.; Abraham, R.T. Ammonia Derived from Glutaminolysis Is a Diffusible Regulator of Autophagy. Sci. Signal. 2010, 3, ra31. [Google Scholar] [CrossRef]
- Ko, Y.-H.; Lin, Z.; Flomenberg, N.; Pestell, R.G.; Howell, A.; Sotgia, F.; Lisanti, M.P.; Martinez-Outschoorn, U.E. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: Implications for preventing chemotherapy resistance. Cancer Boil. Ther. 2011, 12, 1085–1097. [Google Scholar] [CrossRef]
- Sellers, K.; Fox, M.P.; Bousamra, M.; Slone, S.P.; Higashi, R.M.; Miller, N.M.; Wang, Y.; Yan, J.; Yuneva, M.O.; Deshpande, R.; et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Investig. 2015, 125, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Ganji, S.; Hulsey, K.; Madan, A.; Kovacs, Z.; Dimitrov, I.; Zhang, S.; Pichumani, K.; Mendelsohn, D.; Mickey, B.; et al. A comparative study of short- and long-TE ¹H MRS at 3 T for in vivo detection of 2-hydroxyglutarate in brain tumors. NMR Biomed. 2013, 26, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Marin-Valencia, I.; Yang, C.; Mashimo, T.; Cho, S.; Baek, H.; Yang, X.L.; Rajagopalan, K.N.; Maddie, M.; Vemireddy, V.; Zhao, Z.I.; et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012, 15, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Hensley, C.T.; Faubert, B.; Yuan, Q.; Lev-Cohain, N.; Jin, E.; Kim, J.; Jiang, L.; Ko, B.; Skelton, R.; Loudat, L.; et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 2016, 164, 681–694. [Google Scholar] [CrossRef]
- Jacque, N.; Ronchett, A.M.; Larrue, C.; Meunier, G.; Birsen, R.; Willems, L.; Saland, E.; Decroocq, J.; Trovati, M.T.; Lambert, M.; et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015, 126, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Putluri, N.; Long, W.; Zhang, B.; Wang, J.; Kaushik, A.; Arnold, J.M.; Bhowmik, S.K.; Stashi, E.; Brennan, C.A.; et al. Coactivator SRC-2-dependent metabolic reprogramming mediates prostate cancer survival and metastasis. J. Clin. Investig. 2015, 125, 1174–1188. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Jeon, H.M.; Ju, M.K.; Jeong, E.K.; Kim, C.H.; Park, H.G.; Han, S.I.; Kang, H.S. Dlx-2 and glutaminase upregulate epithelial-mesenchymal transition and glycolytic switch. Oncotarget 2016, 7, 7925–7939. [Google Scholar] [CrossRef]
- Yaku, K.; Okabe, K.; Nakagawa, T. NAD metabolism: Implications in aging and longevity. Ageing Res. Rev. 2018, 47, 1–17. [Google Scholar] [CrossRef]
- Hasmann, M.; Schemainda, I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003, 63, 7436–7442. [Google Scholar]
- Tan, B.; Young, D.A.; Lu, Z.-H.; Wang, T.; Meier, T.I.; Shepard, R.L.; Roth, K.; Zhai, Y.; Huss, K.; Kuo, M.-S.; et al. Pharmacological Inhibition of Nicotinamide Phosphoribosyltransferase (NAMPT), an Enzyme Essential for NAD+ Biosynthesis, in Human Cancer Cells. J. Boil. Chem. 2012, 288, 3500–3511. [Google Scholar] [CrossRef]
- Tan, B.; Dong, S.; Shepard, R.L.; Kays, L.; Roth, K.D.; Geeganage, S.; Kuo, M.-S.; Zhao, G. Inhibition of Nicotinamide Phosphoribosyltransferase (NAMPT), an Enzyme Essential for NAD+ Biosynthesis, Leads to Altered Carbohydrate Metabolism in Cancer Cells. J. Boil. Chem. 2015, 290, 15812–15824. [Google Scholar] [CrossRef] [PubMed]
- Del Nagro, C.; Xiao, Y.; Rangell, L.; Reichelt, M.; O’Brien, T. Depletion of the Central Metabolite NAD Leads to Oncosis-mediated Cell Death. J. Boil. Chem. 2014, 289, 35182–35192. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, I.; Bouchard, E.D.J.; Beiggi, S.; Poeppl, A.G.; Johnston, J.B.; Gibson, S.; Banerji, V. On-Target Effect of FK866, a Nicotinamide Phosphoribosyl Transferase Inhibitor, by Apoptosis-Mediated Death in Chronic Lymphocytic Leukemia Cells. Clin. Cancer Res. 2014, 20, 4861–4872. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.J.; Ito, D.; Rees, M.G.; Seashore-Ludlow, B.; Puyang, X.; Ramos, A.H.; Cheah, J.H.; Clemons, P.A.; Warmuth, M.; Zhu, P.; et al. NAMPT Is the Cellular Target of STF-31-Like Small-Molecule Probes. ACS Chem. Boil. 2014, 9, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Revollo, J.R.; Korner, A.; Mills, K.F.; Satoh, A.; Wang, T.; Garten, A.; Dasgupta, B.; Sasaki, Y.; Wolberger, C.; Townsend, R.R.; et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007, 6, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Frederick, D.W.; Loro, E.; Liu, L.; Davila, A., Jr.; Chellappa, K.; Silverman, I.M.; Frederick, D.W.; Loro, E.; Liu, L.; Davila, A., Jr.; et al. Loss of NAD homeostasis leads to progressive and reversibledegeneration of skeletal muscle. Cell Metab. 2016, 24, 269–282. [Google Scholar] [CrossRef]
- Lin, J.B.; Kubota, S.; Ban, N.; Yoshida, M.; Santeford, A.; Séne, A.; Nakamura, R.; Zapata, N.; Kubota, M.; Tsubota, K.; et al. NAMPT-Mediated NAD(+) Biosynthesis Is Essential for Vision In Mice. Cell Rep. 2016, 17, 69–85. [Google Scholar] [CrossRef]
- Hara, N.; Yamada, K.; Shibata, T.; Osago, H.; Hashimoto, T.; Tsuchiya, M. Elevation of Cellular NAD Levels by Nicotinic Acid and Involvement of Nicotinic Acid Phosphoribosyltransferase in Human Cells. J. Boil. Chem. 2007, 282, 24574–24582. [Google Scholar] [CrossRef]
- Tateishi, K.; Wakimoto, H.; Iafrate, A.J.; Tanaka, S.; Loebel, F.; Lelic, N.; Wiederschain, D.; Bedel, O.; Deng, G.; Zhang, B.; et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015, 28, 773–784. [Google Scholar] [CrossRef]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, Ş.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Weinhouse, S. Hepatomas. Science 1967, 158, 542–543. [Google Scholar] [CrossRef] [PubMed]
- Wick, A.N.; Drury, D.R.; I Nakada, H.; Wolfe, J.B. Localization of the primary metabolic block produced by 2-deoxyglucose. J. Boil. Chem. 1957, 224, 963–969. [Google Scholar]
- Zheng, X.; Han, H.; Liu, G.; Ma, Y.; Pan, R.; Sang, L.; Li, R.; Yang, L.; Marks, J.R.; Wang, W.; et al. Lnc RNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J. 2017, 36, 3325–3335. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, H.; Li, Y.; Xia, D.; Yang, L.; Ma, Y.; Li, H. The role of YAP/TAZ activity in cancer metabolic reprogramming. Mol. Cancer 2018, 17, 134. [Google Scholar] [CrossRef]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pascale, R.M.; Calvisi, D.F.; Simile, M.M.; Feo, C.F.; Feo, F. The Warburg Effect 97 Years after Its Discovery. Cancers 2020, 12, 2819. https://doi.org/10.3390/cancers12102819
Pascale RM, Calvisi DF, Simile MM, Feo CF, Feo F. The Warburg Effect 97 Years after Its Discovery. Cancers. 2020; 12(10):2819. https://doi.org/10.3390/cancers12102819
Chicago/Turabian StylePascale, Rosa Maria, Diego Francesco Calvisi, Maria Maddalena Simile, Claudio Francesco Feo, and Francesco Feo. 2020. "The Warburg Effect 97 Years after Its Discovery" Cancers 12, no. 10: 2819. https://doi.org/10.3390/cancers12102819
APA StylePascale, R. M., Calvisi, D. F., Simile, M. M., Feo, C. F., & Feo, F. (2020). The Warburg Effect 97 Years after Its Discovery. Cancers, 12(10), 2819. https://doi.org/10.3390/cancers12102819







