Simple Summary
An accurate survival analysis is crucial for disease management in glioblastoma (GBM) patients. Due to the ability of the diffusion MRI techniques of providing a quantitative assessment of GBM tumours, an ever-growing number of studies aimed at investigating the role of diffusion MRI metrics in survival prediction of GBM patients. Since the role of diffusion MRI in prediction and evaluation of survival outcomes has not been fully addressed and results are often controversial or unsatisfactory, we performed this systematic review in order to collect, summarize and evaluate all studies evaluating the role of diffusion MRI metrics in predicting survival in GBM patients. We found that quantitative diffusion MRI metrics provide useful information for predicting survival outcomes in GBM patients, mainly in combination with other clinical and multimodality imaging parameters.
Abstract
Despite advances in surgical and medical treatment of glioblastoma (GBM), the medium survival is about 15 months and varies significantly, with occasional longer survivors and individuals whose tumours show a significant response to therapy with respect to others. Diffusion MRI can provide a quantitative assessment of the intratumoral heterogeneity of GBM infiltration, which is of clinical significance for targeted surgery and therapy, and aimed at improving GBM patient survival. So, the aim of this systematic review is to assess the role of diffusion MRI metrics in predicting survival of patients with GBM. According to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement, a systematic literature search was performed to identify original articles since 2010 that evaluated the association of diffusion MRI metrics with overall survival (OS) and progression-free survival (PFS). The quality of the included studies was evaluated using the QUIPS tool. A total of 52 articles were selected. The most examined metrics were associated with the standard Diffusion Weighted Imaging (DWI) (34 studies) and Diffusion Tensor Imaging (DTI) models (17 studies). Our findings showed that quantitative diffusion MRI metrics provide useful information for predicting survival outcomes in GBM patients, mainly in combination with other clinical and multimodality imaging parameters.
1. Introduction
Glioblastoma (GBM) is classified by World Health Organization as a grade IV astrocytoma and is the most common and fatal primary brain tumour of the central nervous system (CNS) in adults [1]. GBMs can be classified based on their clinical history as primary (pGBM) or secondary (sGMB). pGBMs are the most common (about 90%) and are characterised by the absence of a clinical or pathologic evidence of a less malignant precursor, while sGBMs result from lower grade gliomas that evolved into GBM. pGBM patients tend to be older and have a poorer prognosis than patients with sGBMs [2].
The clinical presentation and symptoms of a patient with a newly diagnosed GBM are highly variable and dependent on the size, location and degree of infiltration of the tumour [3].
The current clinical treatment for newly diagnosed GBM requires a multidisciplinary approach and consists of maximal tumour resection, followed by concurrent chemoradiotherapy with temozolomide (CCRT) and adjuvant temozolomide (TMZ) [4]. However, despite maximal safe surgical resection and multimodality therapy, about 70% of these tumours invariably recurs, with standards of care at recurrence far less well defined than in the newly diagnosed setting [3,5,6]. Despite advances in surgical and medical neuro-oncology, the medium survival is only 15 months after first diagnosis and with standard surgery and chemoradiation treatment [3,7,8,9,10]. However, the overall survival of patients with GBM varies significantly and there are occasional longer survivors and individuals whose tumours show a significant response to therapy with respect to others [11,12]. For this reason, an accurate survival analysis is crucial to support surgical and therapeutic decisions and so for disease management in GBM patients. This has led to the search for various markers which could be predictive of patient outcome or treatment response [11]. Several clinical (e.g., patient age at diagnosis and Karnofsky Performance Status (KPS)) and therapeutic (extent of surgery, radiation therapy and chemotherapy) factors, as well as specific tumour characteristics (e.g., volume and location) and histopathological and genetic markers (Ki67, isocitrate dehydrogenase 1—IDH1—mutation status and O6-methylguanin-DNA-methyltransferase—MGMT—promoter methylation status) have been studied as potential prognostic markers of survival with variable degrees of sensitivity and specificity [12].
However, their assessment currently requires tissue removing through biopsy or surgical procedures. Thus, a non-invasive assessment of GBM prognosis would be particularly important for treatment planning and GBM patient management in the context of personalized medicine. In this scenario, imaging represents an attractive option compared to more invasive approaches based on tissue-derived biomarkers. There is an urgent need to discover imaging biomarkers that can aid in the selection of patients who will likely derive the most benefit from surgical and/or chemotherapy and/or radiotherapy treatment in terms of overall survival (OS) and progression-free survival (PFS), which are the most commonly evaluated clinical endpoints [13]. Conventional magnetic resonance imaging (MRI) employed for GBM investigation include T1-weighted imaging (T1WI), T2WI, fluid attenuation inversion recovery (FLAIR), T2*WI gradient echo and contrast-enhanced T1WI. These sequences can yield information on the gross anatomic structure of GBM, but they provide little functional information. In fact, the poor prognosis of GBM patients is largely attributed to tumour growth and infiltration that are sometimes difficult to detect by conventional MRI, making novel imaging biomarkers important for aiding in both tumour spatial localization and patient survival prediction [14,15,16].
Given the lack of standardization for their use in GBM clinical practice, advanced imaging techniques such as Positron Emission Tomography (PET), in which different radiotracers are injected to target metabolic and molecular profiles, Perfusion Weighted Imaging (PWI), which provides information on tissue perfusion and microvascular permeability, and diffusion MRI techniques were investigated for GBM management [17,18,19,20].
Diffusion MRI-derived parameters can provide a quantitative assessment of the intratumoral heterogeneity of GBM infiltration, adding new microstructural insights on apparent normal peritumoral white matter that are not detectable by conventional MRI. So, there is increasing interest in these techniques as a biomarker of prognosis in GBM patients.
Diffusion Weighted Imaging (DWI) and Diffusion Tensor Imaging (DTI) are the most commonly used diffusion techniques for GBM applications. In particular, they hold great promise for the improvement of GBM diagnosis and identification of histological classification, prediction of treatment response and monitoring GBM recurrence [20,21,22]. DWI is sensitive to water proton motion at the cellular level and can provide information concerning the microscopic structural environment of neoplasm [21,23]. DTI, other than measuring the magnitude of water molecule movement, such as DWI, is also able to quantify the orientation of the diffusion and has been shown to be sensitive in detecting tumour infiltration [21,23,24].
However, more advanced diffusion imaging techniques, such as non-Gaussian diffusion techniques (e.g., Diffusion Kurtosis Imaging (DKI), Intravoxel Incoherent Motion (IVIM), Stretched Exponential (SE) and Restriction Spectrum Imaging (RSI) were also investigated for GBM applications [25,26,27,28].
Given the urgent need for finding the optimal and targeted treatment option for GBM patients [29,30], and given the proven power of diffusion MRI techniques for applications such as preoperative grading, postoperative assessment of glial tumours and differentiation of GBM from other tumours types (such as brain metastasis or PCNSL [31,32,33]), several studied investigated the association of diffusion MRI metrics with survival outcomes in patients with GBM. Some of these were performed on newly diagnosed GBM patients, while others on recurrent GBM patients. A wide range of diffusion metrics arising from different diffusion MRI models (DWI, DTI, non-Gaussian DWI models and RSI) were explored.
However, their role in prediction and evaluation of survival outcomes has not been fully addressed and results are often controversial or unsatisfactory.
In the context of precision medicine, the analysis of OS and PFS in patients receiving a certain surgical treatment and/or therapy for newly diagnosed or recurrent GBM may help clinicians to better understand the evolution of this disease in each single patient, thus improving patient care and clinical decision-making. Therefore, the aim of this systematic review is to collect, summarize and discuss all studies evaluating the role of diffusion MRI metrics in predicting survival in GBM patients and raise awareness for future research in this field.
2. Materials and Methods
2.1. Search Strategy and Selection Criteria
A systematic search for all the published studies examining the association of diffusion metrics arising from any diffusion model with OS and/or PFS was conducted. The most relevant scientific electronic databases (PubMed, Cochrane Library, MEDLINE, ScienceDirect and Google Scholar) were comprehensively explored and used to build the search. Only studies published since 2010 were selected. The search strategy included the key terms listed in Supplementary Materials Table S1. The literature search was restricted to English language publications and studies of human subject.
Two reviewers, after having independently screened identified titles and abstracts, assessed the full text of the articles that evaluated at least one diffusion MRI metric in terms of OS or PFS, and were not review articles.
For articles meeting these criteria with full text available, the following further selection criteria had to be fulfilled: adult patients; patients with histopathologically confirmed GBM (newly diagnosed or recurrent); and at least one diffusion MRI metric examined in terms of OS or PFS. Studies were excluded if the patient population included also patients with any type of brain tumours other than GBM.
2.2. Planning and Conducting the Review
After the selection procedure, the selected articles were analysed by two reviewers, and data useful for conducting the systematic review were collected in a predesigned sheet. The extracted data will include the following: study characteristics (first author name, publication year and study design, in particular, prospective/retrospective and cross-sectional/longitudinal and number of patients); patient characteristics (age, diagnosis and treatment); MR examination timepoints, namely, the MR images from which the investigated diffusion metrics derive; clinical outcome (OS and/or PFS); diffusion MRI model/s evaluated; diffusion acquisition details; diffusion MRI metric/s evaluated; information on placement of regions of interest (ROIs), namely, the segmentation method and MRI sequence on which the ROIs were placed on; performing the statistical analysis; and the main findings.
The articles were classified and analysed according to the diffusion model involved and the diagnosis (newly diagnosed GBM or recurrent). If more than one diffusion model was investigated in the same study, each model was treated as belonging to a separated study.
This systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (see Supplementary Materials Table S2 for the PRISMA checklist).
2.3. Quality Assessment
The quality of the individual studies was assessed using the QUality In Prognosis Studies (QUIPS) tool [34,35]. According to QUIPS, six domains are critical for assessing biases in prognostic studies: selection of study participants, study attrition, prognostic factor measurement, outcome measurement and study confounding and statistical analysis and reporting. For each of these six domains, the responses “yes”, “partial”, “no” or “unsure” for three up to seven items within each domain are combined to assess the risk of bias. An overall rating for each domain is assigned as “high”, “moderate” or “low” risk of bias. The QUIPS assessment for each study was independently completed by two reviewers and discrepancies were resolved by discussion.
3. Results
3.1. Study Selection
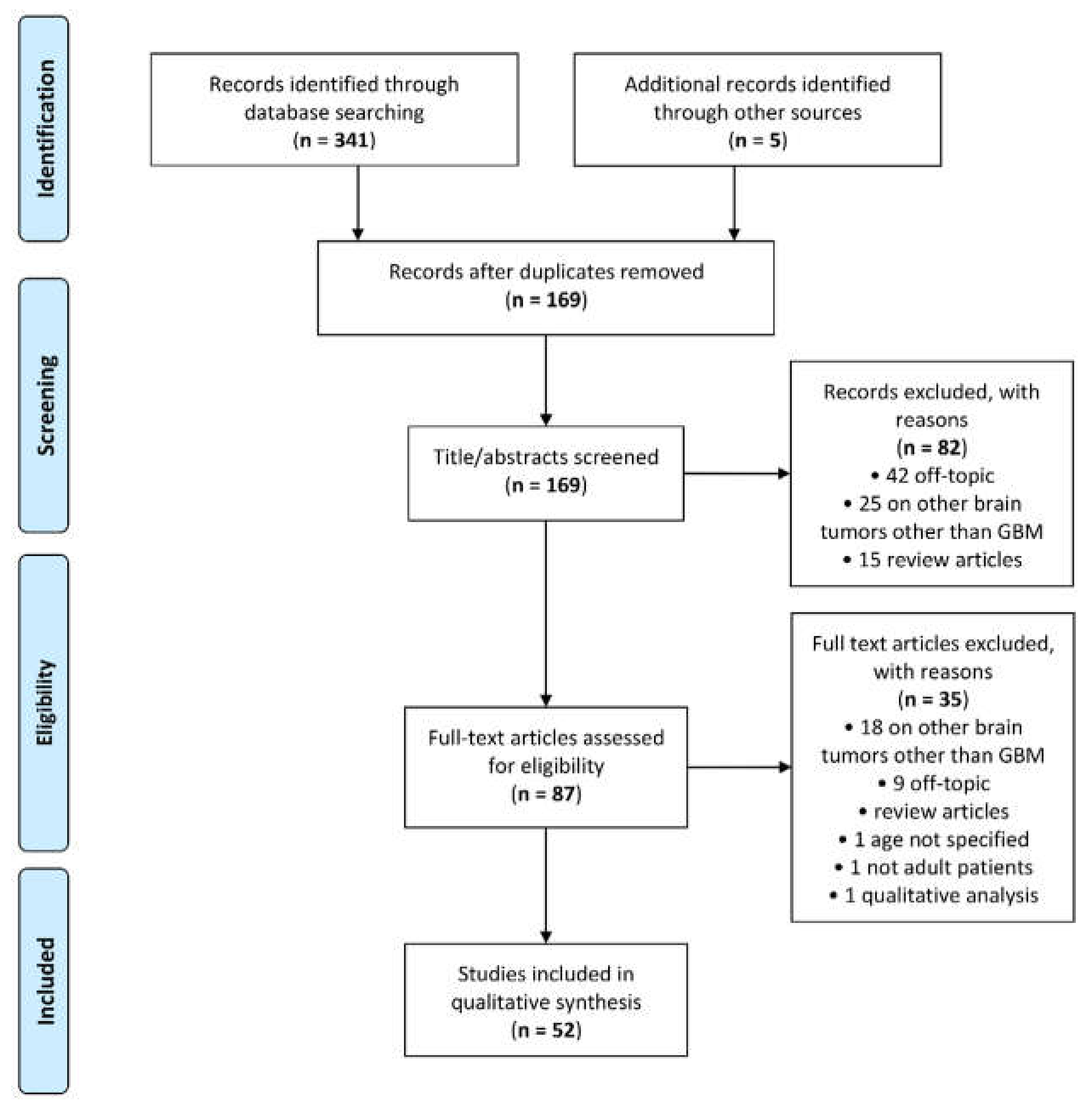
A total of 341 articles was retrieved by the scientific electronic databases search. Five additional articles were found through article references, bringing the total number of records suitable for further evaluation to 346. After removal of the duplicates, there were 169 articles left for investigation. By scanning the title and abstract of these records, 82 records were excluded because they clearly did not match the inclusion criteria (15 review articles, 42 off-topic and 25 including patients with other brain tumours other than GBM). A total of 87 articles were evaluated on their full text. Of these articles, 35 records were excluded based on the inclusion criteria (1 was excluded because it involved non-adult patients [36]; 1 was excluded because it did not specify patient age [37]; 1 was excluded because it performed prognostic analysis based on qualitative visual examinations [38]; 5 were excluded since they were review articles; 18 were excluded since they involved also other types of brain tumour other than GBM; and 9 were not in the field of interest). Finally, 52 records were included for qualitative synthesis. The PRISMA flow diagram of the included studies according to the inclusion and exclusion criteria is reported in Figure 1.
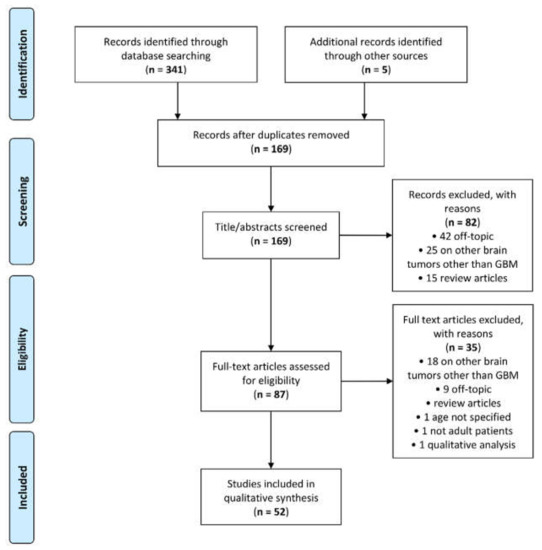
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram.
3.2. Characteristics of the Included Studies
The study characteristics of the 52 articles selected for this review are described in Table 1.

Table 1.
Characteristics of the included studies. Abbreviations: P/R = Prospective/Retrospective; DWI = Diffusion Weighted Imaging; DTI = Diffusion Tensor Imaging; DKI = Diffusion Kurtosis Imaging; IVIM = Intravoxel Incoherent Motion; SE = Stretched Exponential; RSI = Restriction Spectrum Imaging; SO = Survival Outcome; ROI = Region of Interest; OS = Overall Survival; PFS = Progression-Free Survival; NAWM = Normal Appearing White Matter; CEL = Contrast Enhancing Lesion; NEL = Non-Enhancing Lesion; EOR = Extent of Resection; KPS = Karnofsky Performance Score; 2-GMM = 2 Gaussian Mixture Model; 4-GMM = 4 Gaussian Mixture Model; ND/R = Newly Diagnosed/Recurrent; RT = Radio Therapy; TMZ = Temozolomide; BV = Bevacizumab; FS = Field Strength; EPI = Echo Planar Imaging; FOV = Field of View; TR = Relaxation Time; TE = Echi Time; Seq = Sequence; ST = Slice Thickness.
All the selected studies were targeted to adults and the median number (±absolute deviation) of individuals was 53 ± 44.27. Most study designs were retrospective (43/52).
Most of the selected papers (39 studies) involved patients with newly diagnosed GBM (ND-GBM), while 13 studies were specific for recurrent GBM (R-GBM). However, it should be highlighted that in studies involving ND-GBM patients, it is frequently so that several patients present recurrence during clinical history (until death in case of OS analysis). Concerning patient treatment, in studies on ND-GBM patients, most patients received the standard treatment, including chemotherapy and radiotherapy after surgical resection. Among studies on R-GBM patients, only two study involved patients who underwent a second surgery [39,40], while the remaining 11 involved patients receiving chemotherapy and/or radiotherapy treatments. However, many studies did not refer to a uniform treatment for all patients though TMZ and bevacizumab (BV) were the most used chemotherapeutic drugs. See Table 1 for more details on treatments of patients.
Management timepoints at the MR examination vary across studies. Concerning ND-GBM patients, most studies used the preoperative study setting (26/39), while the remaining study investigated metrics arising from postoperative MRI examinations, often considering multiple timepoints. All studies on R-GBM patients investigated the parameters arising from pretreatment and/or posttreatment after recurrence.
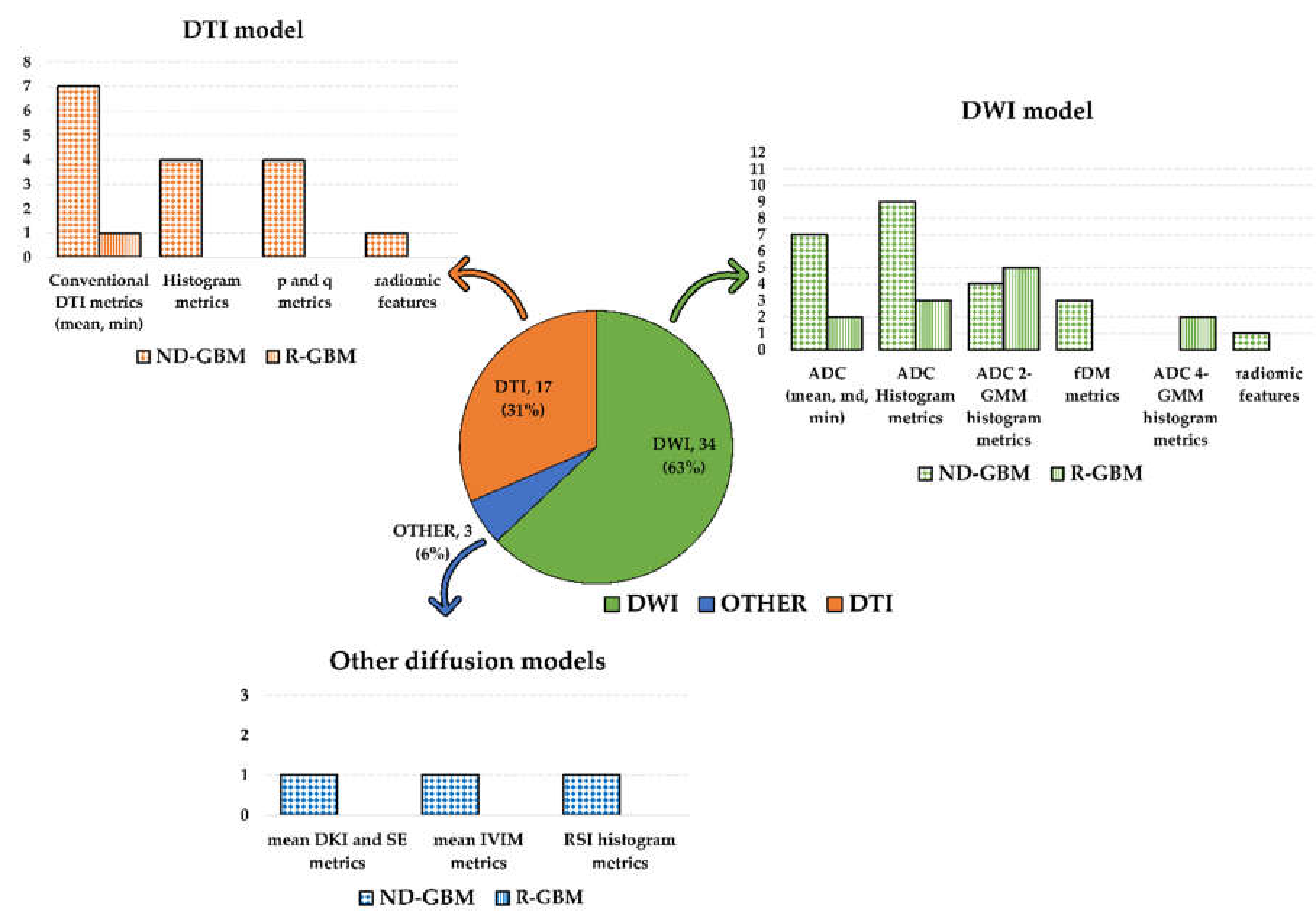
Most of the selected papers examined metrics associated with the standard DWI model (34 studies). The second most examined metrics were associated with the DTI model (17 studies). Among the studies investigating the DWI model, one investigated DWI and RSI [28], while one DWI together with DKI and SE [26]. The remaining study investigated the IVIM model [25]. Refer to Figure 2 for a graphic visualization of the obtained results according to the diffusion models and metrics investigated in the selected studies.
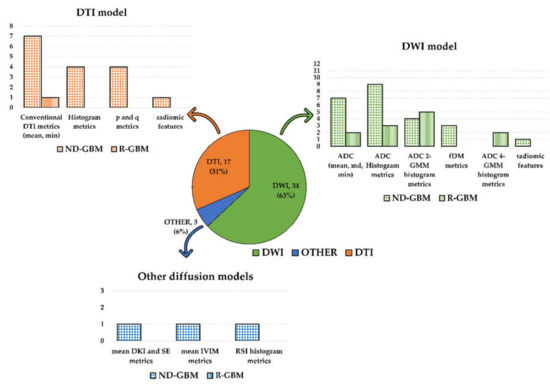
Figure 2.
Graphic summary of the systematic review results according to the diffusion models and metrics investigated in the selected studies. The pie chart shows the number of included studies according to the diffusion MRI model (DWI in green; DTI in orange; other diffusion models in blue). Number and percentage of studies included in each of the three groups were reported. The bar plots show the diffusion metrics investigated in each group. Dotted bars count studies on newly diagnosed GBM patients, while the bars filled with vertical lines count studies on recurrent GBM patients. The study by Wen et al. [55] was counted three times in the DWI model histogram since they investigated both histogram and ADC 2-GMM and fDM metrics.
3.3. Association of Diffusion MR Imaging Metrics with OS and PFS
3.3.1. DWI Metrics
A total of 34 studies investigated the association of DWI metrics with survival endpoints. Among them, 22 involved ND-GBM patients, while the remaining 12 concern R-GBM patients. Concerning studies involving ND-GBM patients, several of them investigated the power of the ADC mean, median and minimum (min) values in tumoral regions, sometimes considering also its normalized value for the normal-appearing white matter (NAWM) or normal contralateral brain [12,26,44,50,53,56,63]. Romano et al. [44] found that the min ADC values in enhancing the tumour component on T1-weighted images were associated with PFS and OS and so could be used as a preoperative parameter to predict patient survival. In particular, patients with higher ADC min values survived longer than patients with lower ADC min values. Similar results were found by Nakamura et al. [50] and Shankar et al. [63]. However, the latter considered only OS as a survival outcome and normalized the ADC min value as the diffusion metric, which showed a positive correlation with OS. Moreover, according to the survival analysis performed by Elson et al. [56] in the post-operative, pre-radiotherapy setting, the normalized ADC min associated with the hyperintense T2/FLAIR regions was a strong predictor of PFS and OS in patients with ND-GBM. Conversely, Coban et al. [12] found no significant difference in the min ADC between patients having a short and long OS. Omuro et al. [53] found that preoperative lower normalized ADC in the maximal portion of the T1 contrast-enhancing tumour was associated with prolonged OS, but not PFS. In Chakoyan et al. [26], patients with large, positive changes in their post-treatment median ADC have a higher OS compared to stable or decreasing ADC changes. Other studies on ND-GBM patients investigated the association with OS and PFS of the ADC histogram metrics (normalized or not) obtained performing standard histogram analysis [28,41,48,55,57,62,64,75,76] or using a double Gaussian mixture modelling for a histogram of the ADC intensities within the considered region [42,55,61,69]. Among the studies investigating the standard ADC histogram parameters, six studies investigated the association of the standard ADC histogram metrics with survival outcomes in a postoperative setting [28,41,55,57,64,76]. Li et al. [41], in a postoperative setting, found that the median and 10th percentile of the normalized ADC values observed pre-and post-treatment were not associated with OS and PFS in a time-independent analysis, but they were significantly correlated with OS according to a time-dependent survival analysis. Wen et al. [55] found an association between the ADC 10th and 50th percentile with both OS and PFS within the T2-enhanced lesions at 2 months post treatment, while in a study by Lee et al. [57], any association with PFS was found between the normalized ADC histogram metrics within the T1 contrast-enhancing lesions. Unfavourable results were also found by Van der Hoorn et al. [64], who evaluated the association of the histogram parameters of the post-treatment changes in normalized ADC within the periresectional area with OS and PFS. In this study, the increase in ADC value post-treatment in comparison to pre-treatment did not predict an increase in PFS or OS neither in the univariate nor multivariate survival analysis. Krishnan et al. [28] also found no association of ADC histogram metrics within the T1 contrast-enhanced and T2 FLAIR hyperintensity volume with OS and PFS. The remaining four studies investigating the power of the standard ADC histogram parameters for predicting survival in ND-GBM patients were performed in a preoperative setting [48,62,75,76]. In a study by Sunwoo et al. [48], PFS was positively associated with the mean ADC value in enhancing the tumour volumes segmented on the postcontrast T1-weighted presurgical scans, but not with the ADC 5th percentile in these regions. The univariate survival analysis performed by Burth et al. [62] showed a positive association of the ADC 10th percentile in T1-enhancing tumour volumes and hyperintense signal changes on T2 FLAIR with OS, but not with PFS. However, in multivariable analysis, the diffusion-derived MRI parameters did not predict survival. In a study by Kim et al. [76], lower ADC histogram parameters were significant predictors of poor OS. In a longitudinal study by Rulseh et al. [75], the ADC histogram analysis was performed using a whole-brain approach. Results showed that serial standardized median ADC and p85 values correlated with PFS and OS, respectively. Four studies investigated the survival prediction power of the histogram metrics obtained applying double-Gaussian mixture modelling (2-GMM) to ADC histograms in regions of interest [42,55,61,69]. A study by Pope et al. [42] performed on preoperative ADC images showed that the ADC mean values for the lower peak (ADCL) of the 2-GMM distribution can stratify PFS in ND-GBM patients with newly diagnosed GBM treated with BV. Moreover, although no statistically significant findings were found, the BV-treated patients with low ADCL also tended to have better OS. In the preoperative study by Kondo et al. [69], promising results also were reported on the association between broader and lower values in the ADCL and PFS and OS. The remaining two studies were performed in a postoperative setting and showed conflicting results [55,61]. Chang et al. ND-GBM patients with low ADCL after treatment have shorter PFS and OS than those with higher ADCL (within contrast-enhancing tumour regions on T1 subtraction images). The ADC mean values for the higher peak (ADCH) of the 2-GMM were not useful for stratifying survival. Conversely, according to Wen et al. [55], no ADC parameters from the 2-GMM fitting in T1 contrast-enhancing lesions were found to be associated with either OS or PFS.
Three studies investigated the power of functional diffusion maps (fDMs)-associated parameters for stratifying survival in ND-GBM patients [43,49,55]. Two of these were by Ellingson et al. [43,49] and showed that the fDMs and probabilistic fDMs metrics obtained considering pre- and post-treatment ADC maps were promising PFS and OS predictors. In a study by Wen et al. [55], the fDM metrics were associated with OS and PFS within the T1 contrast-enhancing lesions, but not in the T2-enhanced lesions. Only Park et al. [83] used a radiomic approach for building a multiparametric MR model able to predict OS in ND-GBM patients and incorporating ADC histogram skewness in its equation.
Concerning studies involving R-GBM patients, all but two studies [39,40] evaluated the predictive value of the ADC metrics in stratifying PFS and OS for R-GBM patients following a certain radio- and/or chemo-therapeutic treatment after the occurrence of first or second recurrence. The most investigated chemotherapeutic regimen was BV. Only two studies [39,40] investigated the ADC metrics survival prediction power for R-GBM patients undergoing a second surgery. Most of them used ADC metrics arising from the histogram analysis based on fitting a double-component Gaussian mixture model to the obtained ADC values [40,45,52,70,77]. In particular, Pope et al. [45] found that low ADCL within tumour regions placed on pre-treatment contrast-enhancing T1-weighted images at baseline was associated with worse OS and PFS in BV-treated R-GBM patients. Similar results were obtained by Ellingson et al. [52]. Differently, Buemi et al. [77] found ADCL in T1 contrast-enhancing tumour regions useful only for stratifying PFS, but not OS in R-GBM patients prior to BV treatment. In the same study, ADCL in T2/FLAIR abnormalities was unable to stratify both PFS and OS. In a subsequent study by Ellingson et al. [70], ADCL was predictive of OS in patients with recurrent GBM treated with anti-VEGF monotherapy at first or second recurrence. Patel et al. [40] also investigated the utility of ADCL, concluding that patients with low ADCL have a survival benefit when surgically excised, whereas large tumours with high ADCL may be best treated with BV. Three studies investigated the survival predictive power of the ADC monomodal histogram parameters [39,58,78], of which two involved BV-treated patients [58,78]. In the first it was found that, unlike the normalized ADC 5th percentile that was also evaluated, the volume of low-ADC lesions in enhancing pre- and post-BV scans predicted shorter OS [58], while in the second [78], using a machine learning approach, the ADC histogram metrics in combination with perfusion metrics arising from pre- and post-treatment images were found to be useful for BV response assessment in terms of PFS and OS. Among the histogram features inspected by Zolal et al. [39] in R-GBM patients undergoing their second surgery, the ADC histogram skewness was an independent prognostic factor for OS and PFS after the second surgery.
Two studies [54,65], rather than using a mono or bi-component curve-fitting histogram analysis as done in the previously described studies, used a four-component one. Rahman et al. [54] evaluated ADC within the volume of contrast-enhancing and non-enhancing T2/FLAIR lesions in BV R-GBM patients, revealing a significant association between the histogram parameters and OS and PFS after performing uni- and multivariable Cox-regression analyses. Chang et al. [65], in a similar study setting, used machine learning approach based on a multimodal approach, including ADC texture and four-component histogram metrics, and developed a valid predictive model for OS. The remaining two studies on R-GBM patients investigated the power of normalized ADC mean and min values in enhancing and non-enhancing tumoral regions [71,84]. Galla et al. [71] found that in R-GBM patients treated with superselective intra-arterial cerebral infusion of BV, the change in normalized ADC mean and min after treatment is predictive of OS. Song et al. [84] found that normalized ADC can be used as an imaging biomarker to determine PFS in R-GBM patients treated with immune checkpoint inhibitors.
3.3.2. DTI Metrics
A total of 17 studies investigated the association of DTI metrics with survival endpoints, of which only 1 involved R-GBM patients, while the remaining involved ND-GBM patients. Amongst the latter, seven studies investigated the power of the mean and min values of diffusion tensor metrics in the tumoral regions, sometimes considering also their normalized value for normal-appearing brain tissue, in a preoperative setting [11,47,59,67,73,79,81]. The most investigated features were mean diffusivity (MD) and fractional anisotropy (FA). Jamjoom et al. [59] found that lower min MD within enhancing tumour regions predicted shorted OS. Similar behaviour was observed in a study by Heiland et al. [73] for mean MD. Conversely, the mean MD and values were found not predictive for OS in studies by Huber et al. and Mohan et al. [67,81] and, normalized for normal contralateral brain tissue, predictive for OS only in the univariate analysis performed by Zikou et al. [47], but not in the multivariate one. Controversial results were also observed concerning FA. In a study by Heiland et al. [73], a lower FA in the contrast-enhancing region was significantly associated with better OS. Moreover, Flores-Alvares et al. [79] found significant association between the peritumoral oedema measurement of FA with OS. Mohan et al. [81] found that FA in corpus callosum infiltrations was able to be a prognostic marker for prediction of OS in ND-GBM patients. On the other hand, in studies by Saksena et al. and Huber et al. [11,67], respectively considering tumour on FLAIR signal abnormality and contrast enhancing zones, FA showed promising results in terms of PFS and OS prediction only in univariate, but not multivariate survival analysis. Finally, although there is a detected trend towards better OS for patients with lower FA values, no significant results were found by Zikou et al. [47].
Four studies performed histogram analysis of DTI metrics [59,60,66,74]. In a preoperative setting, higher MD gradient histogram values of the tumour boundary predicted shorter OS in a study by Jamjoom et al. [59]. Promising results in terms of prediction of both OS and PFS were also showed by Choi et al. [66]. Specifically, lower MD histogram parameters were significant predictors of poor OS and PFS, according to univariate analysis. Multivariable models with MD parameters had significantly higher performances that those without MD parameters for OS and PFS prediction. However, conflicting results were obtained in a later study performed by the same group [74] since any feature from the ADC histogram parameter were useful to build a predictive model based on MRI radiomic features and clinical and genetic parameters, which lead to better performance in terms of OS and PFS when compared with models containing clinical and genetic profiles alone. In a postoperative setting, Wen et al. [60] compared the survival predictive power of the DTI histogram metrics in two differently treated cohorts, showing MD and radial eigenvalues association with OS and PFS at different timepoints. Moreover, in the same study, a volumetric analysis on MD low values was performed and volumetric diffusion parameters were also associated with OS and PFS at different timepoints. A volumetric analysis on MD low values was also performed in a study by Boonzaier et al. [72], who found that regions of low MD (implying high cellularity) and high relative cerebral blood volume (rCBV) (implying vascularity) were correlated with PFS and OS in non-enhancing GBM regions.
The remaining 5 of 16 studies on ND-GBM patients assessed the predictive power in terms of the OS and PFS of the tensor isotropic and anisotropic components (respectively p and q) [24,51,68,80,82]. In a study by Mohsen et al. and two by Yan et al. [51,68,82], volumetric analyses were performed on postoperative p and q maps. Mohsen et al. [51], considering visive abnormality volumes on p and q maps, assessed that a minimal invasive pattern predicts a higher PFS. Similarly, the size of the abnormal q regions was correlated with OS and PFS in studies by Yan et al. [68,82]. Li et al., in two separate studies [24,80], evaluated the association with survival outcomes of the histogram and joint histogram features from the normalized DTI-p and q maps values within the enhancing and non-enhancing tumour zones. In the first study, the joint histogram features, in particular the proportion of the non-enhancing tumour subregion with the decreased isotropic diffusion and increased anisotropic diffusion, were associated with OS and PFS in multivariate models [24]. Similar results on q were found for histogram analysis performed in the second study by Li et al. [80]. In the same study, the p and q histogram metrics showed a significantly incremental value in predicting OS and PFS with respect to the clinical variables alone.
Only Paldino et al. [46], in a longitudinal study, investigated the survival predictive power of the DTI metrics in R-GBM patients. In particular, changes in the MD mean and FA between pre- and post-treatment were investigated in the T1 contrast-enhancing zones and abnormalities on FLAIR images, resulting in significant results in terms of OS and PFS only for changes in the MD mean within abnormalities on FLAIR images.
3.3.3. Other Models Metrics
Only three of the selected studies investigated the association with survival outcomes of the diffusion parameters arising from models different from DWI an DTI [25,26,28]. Puig et al. [25] investigated IVIM metrics within contrast-enhancing and non-enhancing regions, finding that the pseudo-diffusion coefficient and perfusion fraction in the contrast-enhancing regions were significant predictors of 6 months survival. Krishnan et al. [28] performed a survival analysis of the volumetric and intensities metrics arising from the RSI diffusion technique, comparing them with those arising from DWI. While none of ADC metrics were associated with OS and PFS, the RSI volume fraction and 90th percentile within the FLAIR hyperintensity tumour regions were associated with PFS and OS. Lastly, Chakhoyan et al. [26] showed that, in comparison with ADC (which show significant value in predicting OS), any diffusion metrics arising from the DKI and SE models provided additional prognostic value.
3.4. Quality Assessment
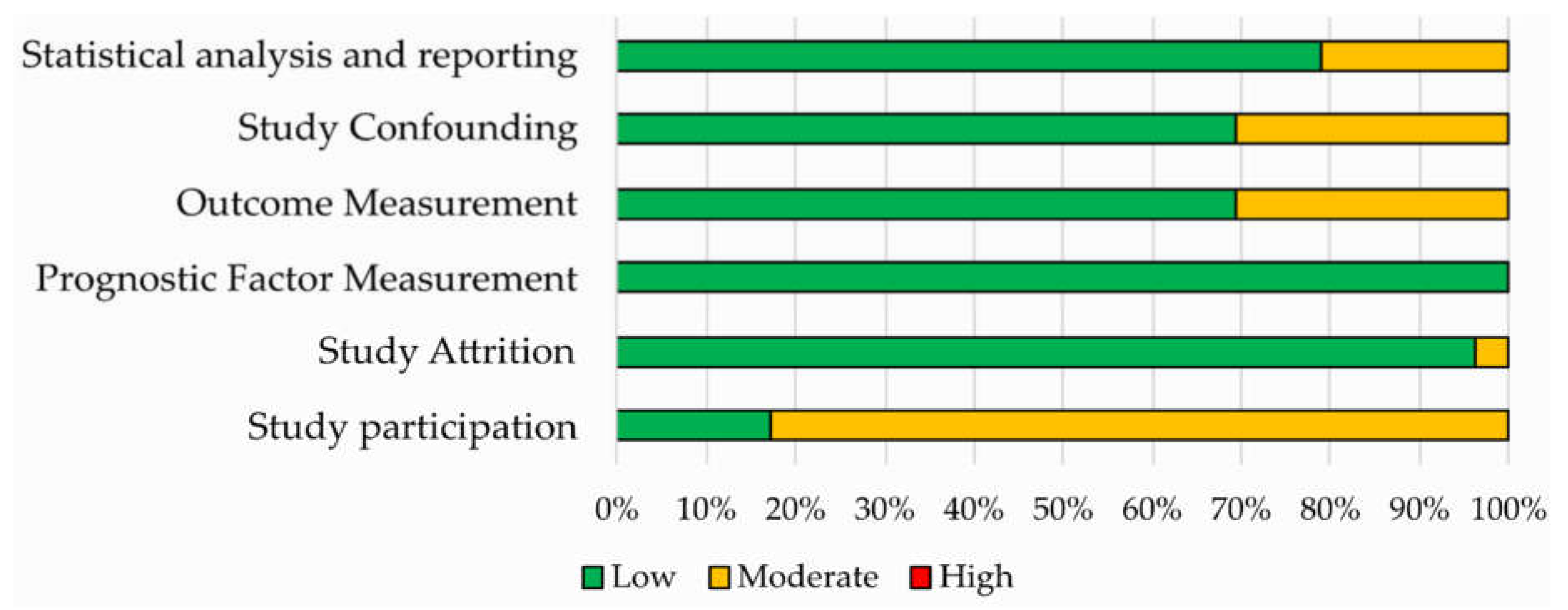
Results of the QUIPS assessment are shown in Figure 3 and reported in Supplementary Materials Table S3. The risk of bias was ranked low or moderate across all the studies for all the six QUIPS domains. Most studies displayed a low risk of bias in the domains of study attrition, outcome measurement, study confounding and statistical analysis and reporting. All studies were judged to be at low risk of bias for study attrition, while a higher percentage of studies with moderate risk of bias was found concerning the study participation domain.
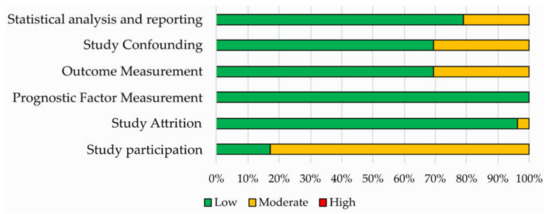
Figure 3.
Risk of bias assessment according to the six domains of the Quality in Prognostic Studies (QUIPS) tool for the 52 studies included in the systematic review.
4. Discussions
In this systematic review, we aimed at investigating the role of the diffusion MR biomarkers in predicting survival outcomes in GBM patients. In the last decade, although diffusion imaging is not yet a primary modality in GBM management, an ever-growing number of studies aimed at investigating the role of diffusion MR metrics in OS and PFS prediction. This is mainly due to the ability of the diffusion MRI techniques (with respect to traditional morphological MRI) of providing a quantitative assessment of intratumoral heterogeneity of GBM infiltration, which is of clinical significance for targeted surgery and therapy, and are crucial for improving GBM patient survival. However, the role of diffusion MRI metrics in prediction and evaluation of survival outcomes has not been fully addressed and results are often controversial or unsatisfactory. In this scenario, our systematic review can provide important new insights and help to reach a common view on the use of diffusion MRI metrics for GBM prognosis. After appropriate inclusion and exclusion criteria, we examined 52 studies from 2010 onwards, evaluating the association of diffusion MRI metrics with OS and PFS in ND-GBM and R-GBM patients. We found that the most investigated diffusion metrics were those associated with DWI and DTI, and only few studies considered different diffusion models (non-Gaussian DWI models and RSI). The main findings and conclusions of the selected studies varied from each other, often showing inconsistencies and not a clear idea about the actual usefulness and the effective prognostic power of the diffusion MR biomarkers.
The most investigated metrics were the minimum and mean ADC value and its histogram metrics arising from uni- or bi-variate histogram analysis for DWI, and MD and FA for DTI, either as mean values or as histogram parameters. These metrics, obtained from different tumour zones (e.g., T1 contrast enhancement and non-enhancement and FLAIR signal abnormality), showed prognostic significance in a different study setting for both ND- and R-GBM patients [54,59,63,66,69,76]. However, there is no shortage of studies showing adverse results [12,47,57,64]. A lower number of studies investigated metrics arising from different approaches, such as four-component histogram analysis, fDM maps analysis, radiomics analysis and DTI isotropic and anisotropic components [24,43,49,51,68,74,80,82,83].
The most interesting finding emerged from our analysis is that, in most studies, the diffusion metrics could not be identified as independent prognostic parameters besides established clinical factors (like age, KPS, MGMT promoter methylation status and extent of resection) and other quantitative parameters arising from other MRI techniques (such as rCBV from perfusion MRI or DCE–MRI-derived metrics), also performing analysis of radiomic features [62,72,74,76,83,84]. In this context, it should be mentioned that the recently introduced radiomics approach may contribute significantly towards survival prediction and stratification of GBM patients [74,83], even if further studies specific for GBM patients are required.
Characteristics of the included studies, such as patient treatment, study aim and setting, diffusion sequence parameters, areas on which ROIs were placed, diffusion metrics for the same diffusion model, analysis methods and the OS and PFS definition, were highly variable across studies, preventing us from performing a meta-analysis. Among the just-mentioned sources of variability, segmentation approaches used for ROI delineation remained tricky considering that the T1-contrast enhancement sequence highlights more perfused tumoral regions, reducing the intratumoral heterogeneity sampling. On the other side, ROI outlining based on FLAIR sequences are affected by peritumoural oedema that can reduce the significance of the findings due to the inclusion of unaffected components in the analyses [19,85]. The variability in patient treatment is particularly noticeable in studies on R-GBM patients, and this is largely due to the lack of a standard of care and the limited efficacy of the therapeutic options, which also justify the larger presence of clinical trials among studies on R-GBM than those on ND-GBM [86].
Moreover, since most of included studies were retrospective, they are supposed to have more bias and should be validated through prospective studies [87].
To our knowledge, this is the first systematic review aiming at summarizing the role of diffusion MR imaging metrics in predicting survival in GBM patients. To date, systematic studies exploring diffusion metrics applied to GBM patients aimed at evaluating the power of DWI and DTI for differentiating GBM from brain metastasis or PCNSL, or from lower grade gliomas [32,33,88]. However, analysis of survival outcomes was out of the purpose of these works. Oltra-Sastre et al. [89] systematically reviewed the multiparametric MR imaging biomarkers associated with clinical outcomes, including the survival outcomes. However, this study was not focused on diffusion metrics and considered studies on gliomas of any WHO grade.
Nowadays, the greatest drawback of diffusion MRI techniques is the lack of standardization [23,90,91]. Continued research efforts and standardization of acquisition parameters and analytic methods, possibly with automation, are required to arrive at the most effective approach that can be applied across institutions. Moreover, due to the ever-growing application of artificial intelligence techniques in the context of medical imaging, it could be interesting to further explore the association between diffusion metrics and GBM survival outcomes using machine learning and deep learning techniques, as done by Chang et al. and Petrova et al. for DWI metrics [65,78] or by Zacharaki et al. and Nie et al. [92,93] for DTI metrics, even if the latter two works were not specific for GBM patients.
5. Conclusions
In conclusion, although further work is required to optimize the methodology associated with the acquisition and analysis of diffusion MR parameters, it seems that diffusion metrics may improve the prediction of survival outcomes in GBM patients, in particular in combination with clinical parameters and conventional or radiomic imaging features arising from multimodal techniques.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/10/2858/s1, Table S1: Key terms used in literature search, Table S2: PRISMA Checklist, Table S3: Quality assessment using QUIPS tool.
Author Contributions
Conceptualization, V.B. and C.C.; methodology, V.B.; validation, S.N. and V.B.; formal analysis, S.N., L.T. and V.B.; investigation, S.N. and L.T.; writing—original draft preparation, V.B.; writing—review and editing, C.C., S.N. and G.C.; visualization, V.B.; supervision, M.S., C.C. and G.C. All authors discussed the results and commented on the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by “Ricerca Corrente” Grant from Italian Ministry of Health (IRCCS SDN).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. The Definition of Primary and Secondary Glioblastoma. Clin. Cancer Res. 2013, 19, 764–772. [Google Scholar] [CrossRef]
- Davis, M. Glioblastoma: Overview of Disease and Treatment. CJON 2016, 20, S2–S8. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Mallick, S.; Benson, R.; Hakim, A.; Rath, G.K. Management of glioblastoma after recurrence: A changing paradigm. J. Egypt. Natl. Cancer Inst. 2016, 28, 199–210. [Google Scholar] [CrossRef]
- Seystahl, K.; Hentschel, B.; Loew, S.; Gramatzki, D.; Felsberg, J.; Herrlinger, U.; Westphal, M.; Schackert, G.; Thon, N.; Tatagiba, M.; et al. Bevacizumab versus alkylating chemotherapy in recurrent glioblastoma. J. Cancer Res. Clin. Oncol. 2020, 146, 659–670. [Google Scholar] [CrossRef]
- Huang, R.Y.; Neagu, M.R.; Reardon, D.A.; Wen, P.Y. Pitfalls in the Neuroimaging of Glioblastoma in the Era of Antiangiogenic and Immuno/Targeted Therapy—Detecting Illusive Disease, Defining Response. Front. Neurol. 2015, 6. [Google Scholar] [CrossRef]
- Alexander, B.M.; Cloughesy, T.F. Adult Glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef]
- Zhao, Y.-H.; Wang, Z.-F.; Pan, Z.-Y.; Péus, D.; Delgado-Fernandez, J.; Pallud, J.; Li, Z.-Q. A Meta-Analysis of Survival Outcomes Following Reoperation in Recurrent Glioblastoma: Time to Consider the Timing of Reoperation. Front. Neurol. 2019, 10, 286. [Google Scholar] [CrossRef]
- Rossignol, J.; Srinageshwar, B.; Dunbar, G.L. Current Therapeutic Strategies for Glioblastoma. Brain Sci. 2019, 10, 15. [Google Scholar] [CrossRef]
- Saksena, S.; Jain, R.; Narang, J.; Scarpace, L.; Schultz, L.R.; Lehman, N.L.; Hearshen, D.; Patel, S.C.; Mikkelsen, T. Predicting survival in glioblastomas using diffusion tensor imaging metrics. J. Magn. Reson. Imaging 2010, 32, 788–795. [Google Scholar] [CrossRef]
- Coban, G.; Mohan, S.; Kural, F.; Wang, S.; O’Rourke, D.M.; Poptani, H. Prognostic Value of Dynamic Susceptibility Contrast-Enhanced and Diffusion-Weighted MR Imaging in Patients with Glioblastomas. Am. J. Neuroradiol. 2015, 36, 1247–1252. [Google Scholar] [CrossRef]
- Madsen, H.; Hellwinkel, J.E.; Graner, M.W. Clinical Trials in Glioblastoma—Designs and Challenges. In Molecular Considerations and Evolving Surgical Management Issues in the Treatment of Patients with a Brain Tumor; Lichtor, T., Ed.; InTech: New York, NY, USA, 2015; ISBN 978-953-51-2031-5. [Google Scholar]
- Gonçalves, F.G.; Chawla, S.; Mohan, S. Emerging MRI Techniques to Redefine Treatment Response in Patients with Glioblastoma: MRI Treatment Response in Glioblastoma. J. Magn. Reson. Imaging 2020. [Google Scholar] [CrossRef]
- Aum, D.J.; Kim, D.H.; Beaumont, T.L.; Leuthardt, E.C.; Dunn, G.P.; Kim, A.H. Molecular and cellular heterogeneity: The hallmark of glioblastoma. FOC 2014, 37, E11. [Google Scholar] [CrossRef]
- Friedmann-Morvinski, D. Glioblastoma Heterogeneity and Cancer Cell Plasticity. Crit. Rev. Oncog. 2014, 19, 327–336. [Google Scholar] [CrossRef]
- Mansoor, N.M.; Thust, S.; Militano, V.; Fraioli, F. PET imaging in glioma: Techniques and current evidence. Nucl. Med. Commun. 2018, 39, 1064–1080. [Google Scholar] [CrossRef]
- Van Dijken, B.R.J.; van Laar, P.J.; Smits, M.; Dankbaar, J.W.; Enting, R.H.; van der Hoorn, A. Perfusion MRI in treatment evaluation of glioblastomas: Clinical relevance of current and future techniques. J. Magn. Reson. Imaging 2019, 49, 11–22. [Google Scholar] [CrossRef]
- Shukla, G.; Alexander, G.S.; Bakas, S.; Nikam, R.; Talekar, K.; Palmer, J.D.; Shi, W. Advanced magnetic resonance imaging in glioblastoma: A review. Chin. Clin. Oncol. 2017, 6, 40. [Google Scholar] [CrossRef]
- Drake-Pérez, M.; Boto, J.; Fitsiori, A.; Lovblad, K.; Vargas, M.I. Clinical applications of diffusion weighted imaging in neuroradiology. Insights Imaging 2018, 9, 535–547. [Google Scholar] [CrossRef]
- Yanagihara, T.K.; Wang, T.J.C. Diffusion-weighted imaging of the brain for glioblastoma: Implications for radiation oncology. Appl. Radiat. Oncol. 2014, 9, 5–13. [Google Scholar]
- Schmainda, K.M. Diffusion-weighted MRI as a biomarker for treatment response in glioma. CNS Oncol. 2012, 1, 169–180. [Google Scholar] [CrossRef]
- Salama, G.R.; Heier, L.A.; Patel, P.; Ramakrishna, R.; Magge, R.; Tsiouris, A.J. Diffusion Weighted/Tensor Imaging, Functional MRI and Perfusion Weighted Imaging in Glioblastoma—Foundations and Future. Front. Neurol. 2018, 8, 660. [Google Scholar] [CrossRef]
- Li, C.; Wang, S.; Yan, J.-L.; Piper, R.J.; Liu, H.; Torheim, T.; Kim, H.; Zou, J.; Boonzaier, N.R.; Sinha, R.; et al. Intratumoral Heterogeneity of Glioblastoma Infiltration Revealed by Joint Histogram Analysis of Diffusion Tensor Imaging. Neurosurgery 2019, 85, 524–534. [Google Scholar] [CrossRef]
- Puig, J.; Sánchez-González, J.; Blasco, G.; Daunis-i-Estadella, P.; Federau, C.; Alberich-Bayarri, Á.; Biarnes, C.; Nael, K.; Essig, M.; Jain, R.; et al. Intravoxel Incoherent Motion Metrics as Potential Biomarkers for Survival in Glioblastoma. PLoS ONE 2016, 11, e0158887. [Google Scholar] [CrossRef]
- Chakhoyan, A.; Woodworth, D.C.; Harris, R.J.; Lai, A.; Nghiemphu, P.L.; Liau, L.M.; Pope, W.B.; Cloughesy, T.F.; Ellingson, B.M. Mono-exponential, diffusion kurtosis and stretched exponential diffusion MR imaging response to chemoradiation in newly diagnosed glioblastoma. J. Neurooncol. 2018, 139, 651–659. [Google Scholar] [CrossRef]
- Khan, U.A.; Rennert, R.C.; White, N.S.; Bartsch, H.; Farid, N.; Dale, A.M.; Chen, C.C. Diagnostic utility of restriction spectrum imaging (RSI) in glioblastoma patients after concurrent radiation-temozolomide treatment: A pilot study. J. Clin. Neurosci. 2018, 58, 136–141. [Google Scholar] [CrossRef]
- Krishnan, A.P.; Karunamuni, R.; Leyden, K.M.; Seibert, T.M.; Delfanti, R.L.; Kuperman, J.M.; Bartsch, H.; Elbe, P.; Srikant, A.; Dale, A.M.; et al. Restriction Spectrum Imaging Improves Risk Stratification in Patients with Glioblastoma. Am. J. Neuroradiol. 2017, 38, 882–889. [Google Scholar] [CrossRef]
- Holland, E.; Ene, C. Personalized Medicine for Gliomas. Surg. Neurol. Int. 2015, 6, 89. [Google Scholar] [CrossRef]
- Nuzzo, S.; Brancato, V.; Affinito, A.; Salvatore, M.; Cavaliere, C.; Condorelli, G. The Role of RNA and DNA Aptamers in Glioblastoma Diagnosis and Therapy: A Systematic Review of the Literature. Cancers 2020, 12, 2173. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, B. Differentiation among Glioblastomas, Primary Cerebral Lymphomas, and Solitary Brain Metastases Using Diffusion-Weighted Imaging and Diffusion Tensor Imaging: A PRISMA-Compliant Meta-analysis. ACS Chem. Neurosci. 2020, 11, 477–483. [Google Scholar] [CrossRef]
- Suh, C.H.; Kim, H.S.; Jung, S.C.; Park, J.E.; Choi, C.G.; Kim, S.J. MRI as a diagnostic biomarker for differentiating primary central nervous system lymphoma from glioblastoma: A systematic review and meta-analysis: Differentiating PCNSL from Glioblastoma. J. Magn. Reson. Imaging 2019, 50, 560–572. [Google Scholar] [CrossRef]
- Lu, X.; Xu, W.; Wei, Y.; Li, T.; Gao, L.; Fu, X.; Yao, Y.; Wang, L. Diagnostic performance of DWI for differentiating primary central nervous system lymphoma from glioblastoma: A systematic review and meta-analysis. Neurol. Sci. 2019, 40, 947–956. [Google Scholar] [CrossRef]
- Hayden, J.A.; Côté, P.; Bombardier, C. Evaluation of the Quality of Prognosis Studies in Systematic Reviews. Ann. Intern. Med. 2006, 144, 427. [Google Scholar] [CrossRef]
- Hayden, J.A.; van der Windt, D.A.; Cartwright, J.L.; Côté, P.; Bombardier, C. Assessing bias in studies of prognostic factors. Ann. Intern. Med. 2013, 158, 280–286. [Google Scholar] [CrossRef]
- Yamasaki, F.; Sugiyama, K.; Ohtaki, M.; Takeshima, Y.; Abe, N.; Akiyama, Y.; Takaba, J.; Amatya, V.J.; Saito, T.; Kajiwara, Y.; et al. Glioblastoma treated with postoperative radio-chemotherapy: Prognostic value of apparent diffusion coefficient at MR imaging. Eur. J. Radiol. 2010, 73, 532–537. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Cloughesy, T.F.; Lai, A.; Mischel, P.S.; Nghiemphu, P.L.; Lalezari, S.; Schmainda, K.M.; Pope, W.B. Graded functional diffusion map-defined characteristics of apparent diffusion coefficients predict overall survival in recurrent glioblastoma treated with bevacizumab. Neuro Oncol. 2011, 13, 1151–1161. [Google Scholar] [CrossRef]
- Deike, K.; Wiestler, B.; Graf, M.; Reimer, C.; Floca, R.O.; Bäumer, P.; Kickingereder, P.; Heiland, S.; Schlemmer, H.-P.; Wick, W.; et al. Prognostic value of combined visualization of MR diffusion and perfusion maps in glioblastoma. J. Neurooncol. 2016, 126, 463–472. [Google Scholar] [CrossRef]
- Zolal, A.; Juratli, T.A.; Linn, J.; Podlesek, D.; Sitoci Ficici, K.H.; Kitzler, H.H.; Schackert, G.; Sobottka, S.B.; Rieger, B.; Krex, D. Enhancing tumor apparent diffusion coefficient histogram skewness stratifies the postoperative survival in recurrent glioblastoma multiforme patients undergoing salvage surgery. J. Neurooncol. 2016, 127, 551–557. [Google Scholar] [CrossRef]
- Patel, K.S.; Everson, R.G.; Yao, J.; Raymond, C.; Goldman, J.; Schlossman, J.; Tsung, J.; Tan, C.; Pope, W.B.; Ji, M.S.; et al. Diffusion Magnetic Resonance Imaging Phenotypes Predict Overall Survival Benefit from Bevacizumab or Surgery in Recurrent Glioblastoma with Large Tumor Burden. Neurosurgery 2020, nyaa135. [Google Scholar] [CrossRef]
- Li, Y.; Lupo, J.M.; Polley, M.-Y.; Crane, J.C.; Bian, W.; Cha, S.; Chang, S.; Nelson, S.J. Serial analysis of imaging parameters in patients with newly diagnosed glioblastoma multiforme. Neuro Oncol. 2011, 13, 546–557. [Google Scholar] [CrossRef]
- Pope, W.B.; Lai, A.; Mehta, R.; Kim, H.J.; Qiao, J.; Young, J.R.; Xue, X.; Goldin, J.; Brown, M.S.; Nghiemphu, P.L.; et al. Apparent Diffusion Coefficient Histogram Analysis Stratifies Progression-Free Survival in Newly Diagnosed Bevacizumab-Treated Glioblastoma. Am. J. Neuroradiol. 2011, 32, 882–889. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Cloughesy, T.F.; Zaw, T.; Lai, A.; Nghiemphu, P.L.; Harris, R.; Lalezari, S.; Wagle, N.; Naeini, K.M.; Carrillo, J.; et al. Functional diffusion maps (fDMs) evaluated before and after radiochemotherapy predict progression-free and overall survival in newly diagnosed glioblastoma. Neuro Oncol. 2012, 14, 333–343. [Google Scholar] [CrossRef]
- Romano, A.; Calabria, L.F.; Tavanti, F.; Minniti, G.; Rossi-Espagnet, M.C.; Coppola, V.; Pugliese, S.; Guida, D.; Francione, G.; Colonnese, C.; et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: Correlation with MGMT promoter methylation status. Eur. Radiol. 2013, 23, 513–520. [Google Scholar] [CrossRef]
- Pope, W.B.; Qiao, X.J.; Kim, H.J.; Lai, A.; Nghiemphu, P.; Xue, X.; Ellingson, B.M.; Schiff, D.; Aregawi, D.; Cha, S.; et al. Apparent diffusion coefficient histogram analysis stratifies progression-free and overall survival in patients with recurrent GBM treated with bevacizumab: A multi-center study. J. Neurooncol. 2012, 108, 491–498. [Google Scholar] [CrossRef]
- Paldino, M.J.; Desjardins, A.; Friedman, H.S.; Vredenburgh, J.J.; Barboriak, D.P. A change in the apparent diffusion coefficient after treatment with bevacizumab is associated with decreased survival in patients with recurrent glioblastoma multiforme. BJR 2012, 85, 382–389. [Google Scholar] [CrossRef]
- Zikou, A.K.; Alexiou, G.A.; Kosta, P.; Goussia, A.; Astrakas, L.; Tsekeris, P.; Voulgaris, S.; Malamou-Mitsi, V.; Kyritsis, A.P.; Argyropoulou, M.I. Diffusion tensor and dynamic susceptibility contrast MRI in glioblastoma. Clin. Neurol. Neurosurg. 2012, 114, 607–612. [Google Scholar] [CrossRef]
- Sunwoo, L.; Choi, S.H.; Park, C.-K.; Kim, J.W.; Yi, K.S.; Lee, W.J.; Yoon, T.J.; Song, S.W.; Kim, J.E.; Kim, J.Y.; et al. Correlation of apparent diffusion coefficient values measured by diffusion MRI and MGMT promoter methylation semiquantitatively analyzed with MS-MLPA in patients with glioblastoma multiforme. J. Magn. Reson. Imaging 2013, 37, 351–358. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Cloughesy, T.F.; Lai, A.; Nghiemphu, P.L.; Liau, L.M.; Pope, W.B. Quantitative probabilistic functional diffusion mapping in newly diagnosed glioblastoma treated with radiochemotherapy. Neuro Oncol. 2013, 15, 382–390. [Google Scholar] [CrossRef]
- Nakamura, H.; Murakami, R.; Hirai, T.; Kitajima, M.; Yamashita, Y. Can MRI-derived factors predict the survival in glioblastoma patients treated with postoperative chemoradiation therapy? Acta Radiol. 2013, 54, 214–220. [Google Scholar] [CrossRef]
- Mohsen, L.A.; Shi, V.; Jena, R.; Gillard, J.H.; Price, S.J. Diffusion tensor invasive phenotypes can predict progression-free survival in glioblastomas. Br. J. Neurosurg. 2013, 27, 436–441. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Sahebjam, S.; Kim, H.J.; Pope, W.B.; Harris, R.J.; Woodworth, D.C.; Lai, A.; Nghiemphu, P.L.; Mason, W.P.; Cloughesy, T.F. Pretreatment ADC Histogram Analysis Is a Predictive Imaging Biomarker for Bevacizumab Treatment but Not Chemotherapy in Recurrent Glioblastoma. Am. J. Neuroradiol. 2014, 35, 673–679. [Google Scholar] [CrossRef]
- Omuro, A.; Beal, K.; Gutin, P.; Karimi, S.; Correa, D.D.; Kaley, T.J.; DeAngelis, L.M.; Chan, T.A.; Gavrilovic, I.T.; Nolan, C.; et al. Phase II Study of Bevacizumab, Temozolomide, and Hypofractionated Stereotactic Radiotherapy for Newly Diagnosed Glioblastoma. Clin. Cancer Res. 2014, 20, 5023–5031. [Google Scholar] [CrossRef]
- Rahman, R.; Hamdan, A.; Zweifler, R.; Jiang, H.; Norden, A.D.; Reardon, D.A.; Mukundan, S.; Wen, P.Y.; Huang, R.Y. Histogram analysis of apparent diffusion coefficient within enhancing and nonenhancing tumor volumes in recurrent glioblastoma patients treated with bevacizumab. J. Neurooncol. 2014, 119, 149–158. [Google Scholar] [CrossRef]
- Wen, Q.; Jalilian, L.; Lupo, J.M.; Molinaro, A.M.; Chang, S.M.; Clarke, J.; Prados, M.; Nelson, S.J. Comparison of ADC metrics and their association with outcome for patients with newly diagnosed glioblastoma being treated with radiation therapy, temozolomide, erlotinib and bevacizumab. J. Neurooncol. 2015, 121, 331–339. [Google Scholar] [CrossRef]
- Elson, A.; Bovi, J.; Siker, M.; Schultz, C.; Paulson, E. Evaluation of absolute and normalized apparent diffusion coefficient (ADC) values within the post-operative T2/FLAIR volume as adverse prognostic indicators in glioblastoma. J. Neurooncol. 2015, 122, 549–558. [Google Scholar] [CrossRef]
- Lee, E.K.; Choi, S.H.; Yun, T.J.; Kang, K.M.; Kim, T.M.; Lee, S.-H.; Park, C.-K.; Park, S.-H.; Kim, I.H. Prediction of Response to Concurrent Chemoradiotherapy with Temozolomide in Glioblastoma: Application of Immediate Post-Operative Dynamic Susceptibility Contrast and Diffusion-Weighted MR Imaging. Korean J. Radiol. 2015, 16, 1341. [Google Scholar] [CrossRef]
- Zhang, M.; Gulotta, B.; Thomas, A.; Kaley, T.; Karimi, S.; Gavrilovic, I.; Woo, K.M.; Zhang, Z.; Arevalo-Perez, J.; Holodny, A.I.; et al. Large-volume low apparent diffusion coefficient lesions predict poor survival in bevacizumab-treated glioblastoma patients. Neuro Oncol. 2016, 18, 735–743. [Google Scholar] [CrossRef]
- Jamjoom, A.A.B.; Rodriguez, D.; Rajeb, A.T.; Manita, M.A.; Shah, K.A.; Auer, D.P. Magnetic resonance diffusion metrics indexing high focal cellularity and sharp transition at the tumour boundary predict poor outcome in glioblastoma multiforme. Clin. Radiol. 2015, 70, 1400–1407. [Google Scholar] [CrossRef]
- Wen, Q.; Jalilian, L.; Lupo, J.M.; Li, Y.; Roy, R.; Molinaro, A.M.; Chang, S.M.; Prados, M.; Butowski, N.; Clarke, J.; et al. Association of Diffusion and Anatomic Imaging Parameters with Survival for Patients with Newly Diagnosed Glioblastoma Participating in Two Different Clinical Trials. Transl. Oncol. 2015, 8, 446–455. [Google Scholar] [CrossRef]
- Chang, W.; Pope, W.B.; Harris, R.J.; Hardy, A.J.; Leu, K.; Mody, R.R.; Nghiemphu, P.L.; Lai, A.; Cloughesy, T.F.; Ellingson, B.M. Diffusion MR Characteristics Following Concurrent Radiochemotherapy Predicts Progression-Free and Overall Survival in Newly Diagnosed Glioblastoma. Tomography 2015, 1, 37–43. [Google Scholar] [CrossRef]
- Burth, S.; Kickingereder, P.; Eidel, O.; Tichy, D.; Bonekamp, D.; Weberling, L.; Wick, A.; Löw, S.; Hertenstein, A.; Nowosielski, M.; et al. Clinical parameters outweigh diffusion- and perfusion-derived MRI parameters in predicting survival in newly diagnosed glioblastoma. Neuro Oncol. 2016, 18, 1673–1679. [Google Scholar] [CrossRef]
- Shankar, J.J.S.; Bata, A.; Ritchie, K.; Hebb, A.; Walling, S. Normalized Apparent Diffusion Coefficient in the Prognostication of Patients with Glioblastoma Multiforme. Can. J. Neurol. Sci. 2016, 43, 127–133. [Google Scholar] [CrossRef]
- Van der Hoorn, A.; Yan, J.-L.; Larkin, T.J.; Boonzaier, N.R.; Matys, T.; Price, S.J. Posttreatment Apparent Diffusion Coefficient Changes in the Periresectional Area in Patients with Glioblastoma. World Neurosurg. 2016, 92, 159–165. [Google Scholar] [CrossRef]
- Chang, K.; Zhang, B.; Guo, X.; Zong, M.; Rahman, R.; Sanchez, D.; Winder, N.; Reardon, D.A.; Zhao, B.; Wen, P.Y.; et al. Multimodal imaging patterns predict survival in recurrent glioblastoma patients treated with bevacizumab. Neuro Oncol. 2016, 18, 1680–1687. [Google Scholar] [CrossRef]
- Choi, Y.S.; Ahn, S.S.; Kim, D.W.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Rim, T.H.; Lee, S.-K. Incremental Prognostic Value of ADC Histogram Analysis over MGMT Promoter Methylation Status in Patients with Glioblastoma. Radiology 2016, 281, 175–184. [Google Scholar] [CrossRef]
- Huber, T.; Bette, S.; Wiestler, B.; Gempt, J.; Gerhardt, J.; Delbridge, C.; Barz, M.; Meyer, B.; Zimmer, C.; Kirschke, J.S. Fractional Anisotropy Correlates with Overall Survival in Glioblastoma. World Neurosurg. 2016, 95, 525–534.e1. [Google Scholar] [CrossRef]
- Yan, J.-L.; van der Hoorn, A.; Larkin, T.J.; Boonzaier, N.R.; Matys, T.; Price, S.J. Extent of resection of peritumoral diffusion tensor imaging–detected abnormality as a predictor of survival in adult glioblastoma patients. JNS 2017, 126, 234–241. [Google Scholar] [CrossRef]
- Kondo, M.; Uchiyama, Y. Apparent diffusion coefficient histogram analysis for prediction of prognosis in glioblastoma. J. Neuroradiol. 2018, 45, 236–241. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Gerstner, E.R.; Smits, M.; Huang, R.Y.; Colen, R.; Abrey, L.E.; Aftab, D.T.; Schwab, G.M.; Hessel, C.; Harris, R.J.; et al. Diffusion MRI Phenotypes Predict Overall Survival Benefit from Anti-VEGF Monotherapy in Recurrent Glioblastoma: Converging Evidence from Phase II Trials. Clin. Cancer Res. 2017, 23, 5745–5756. [Google Scholar] [CrossRef]
- Galla, N.; Chiang, G.; Chakraborty, S.; Singh, R.; John Tsiouris, A.; Boockvar, J.; Kovanlikaya, I. Apparent diffusion coefficient changes predict survival after intra-arterial bevacizumab treatment in recurrent glioblastoma. Neuroradiology 2017, 59, 499–505. [Google Scholar] [CrossRef]
- Boonzaier, N.R.; Larkin, T.J.; Matys, T.; van der Hoorn, A.; Yan, J.-L.; Price, S.J. Multiparametric MR Imaging of Diffusion and Perfusion in Contrast-enhancing and Nonenhancing Components in Patients with Glioblastoma. Radiology 2017, 284, 180–190. [Google Scholar] [CrossRef]
- Heiland, D.H.; Simon-Gabriel, C.P.; Demerath, T.; Haaker, G.; Pfeifer, D.; Kellner, E.; Kiselev, V.G.; Staszewski, O.; Urbach, H.; Weyerbrock, A.; et al. Integrative Diffusion-Weighted Imaging and Radiogenomic Network Analysis of Glioblastoma multiforme. Sci. Rep. 2017, 7, 43523. [Google Scholar] [CrossRef]
- Bae, S.; Choi, Y.S.; Ahn, S.S.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Lee, S.-K. Radiomic MRI Phenotyping of Glioblastoma: Improving Survival Prediction. Radiology 2018, 289, 797–806. [Google Scholar] [CrossRef]
- Rulseh, A.M.; Vymazal, J. Whole brain apparent diffusion coefficient measurements correlate with survival in glioblastoma patients. J. Neurooncol. 2020, 146, 157–162. [Google Scholar] [CrossRef]
- Kim, B.S.; Kim, S.T.; Kim, J.H.; Seol, H.J.; Nam, D.-H.; Shin, H.J.; Lee, J.-I.; Kong, D.-S. Apparent Diffusion Coefficient as a Predictive Biomarker for Survival in Patients with Treatment-Naive Glioblastoma Using Quantitative Multiparametric Magnetic Resonance Profiling. World Neurosurg. 2019, 122, e812–e820. [Google Scholar] [CrossRef]
- Buemi, F.; Guzzardi, G.; Del Sette, B.; Sponghini, A.P.; Matheoud, R.; Soligo, E.; Trisoglio, A.; Carriero, A.; Stecco, A. Apparent diffusion coefficient and tumor volume measurements help stratify progression-free survival of bevacizumab-treated patients with recurrent glioblastoma multiforme. Neuroradiol. J. 2019, 32, 241–249. [Google Scholar] [CrossRef]
- Petrova, L.; Korfiatis, P.; Petr, O.; LaChance, D.H.; Parney, I.; Buckner, J.C.; Erickson, B.J. Cerebral blood volume and apparent diffusion coefficient–Valuable predictors of non-response to bevacizumab treatment in patients with recurrent glioblastoma. J. Neurol. Sci. 2019, 405, 116433. [Google Scholar] [CrossRef]
- Flores-Alvarez, E.; Durand-Muñoz, C.; Cortes-Hernandez, F.; Muñoz-Hernandez, O.; Moreno-Jimenez, S.; Roldan-Valadez, E. Clinical Significance of Fractional Anisotropy Measured in Peritumoral Edema as a Biomarker of Overall Survival in Glioblastoma: Evidence Using Correspondence Analysis. Neurol. India 2019, 67, 1074. [Google Scholar] [CrossRef]
- Li, C.; Wang, S.; Serra, A.; Torheim, T.; Yan, J.-L.; Boonzaier, N.R.; Huang, Y.; Matys, T.; McLean, M.A.; Markowetz, F.; et al. Multi-parametric and multi-regional histogram analysis of MRI: Modality integration reveals imaging phenotypes of glioblastoma. Eur. Radiol. 2019, 29, 4718–4729. [Google Scholar] [CrossRef]
- Mohan, S.; Wang, S.; Coban, G.; Kural, F.; Chawla, S.; O’Rourke, D.M.; Poptani, H. Detection of occult neoplastic infiltration in the corpus callosum and prediction of overall survival in patients with glioblastoma using diffusion tensor imaging. Eur. J. Radiol. 2019, 112, 106–111. [Google Scholar] [CrossRef]
- Yan, J.-L.; Li, C.; Boonzaier, N.R.; Fountain, D.M.; Larkin, T.J.; Matys, T.; van der Hoorn, A.; Price, S.J. Multimodal MRI characteristics of the glioblastoma infiltration beyond contrast enhancement. Adv. Neurol. Disord. 2019, 12. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, H.S.; Jo, Y.; Yoo, R.-E.; Choi, S.H.; Nam, S.J.; Kim, J.H. Radiomics prognostication model in glioblastoma using diffusion- and perfusion-weighted MRI. Sci. Rep. 2020, 10, 4250. [Google Scholar] [CrossRef]
- Song, J.; Kadaba, P.; Kravitz, A.; Hormigo, A.; Friedman, J.; Belani, P.; Hadjipanayis, C.; Ellingson, B.M.; Nael, K. Multiparametric MRI for early identification of therapeutic response in recurrent glioblastoma treated with immune checkpoint inhibitors. Neuro Oncol. 2020, noaa066. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Wen, P.Y.; van den Bent, M.J.; Cloughesy, T.F. Pros and cons of current brain tumor imaging. Neuro Oncol. 2014, 16, vii2–vii11. [Google Scholar] [CrossRef]
- Nam, J.Y.; de Groot, J.F. Treatment of Glioblastoma. JOP 2017, 13, 629–638. [Google Scholar] [CrossRef]
- Norvell, D. Study types and bias—Don’t judge a study by the abstract’s conclusion alone. Evid. Based Spine-Care J. 2010, 1, 7–10. [Google Scholar] [CrossRef]
- Wang, Q.; Lei, D.; Yuan, Y.; Xiong, N. Accuracy of ADC derived from DWI for differentiating high-grade from low-grade gliomas: Systematic review and meta-analysis. Medicine 2020, 99, e19254. [Google Scholar] [CrossRef]
- Oltra-Sastre, M.; Fuster-Garcia, E.; Juan-Albarracin, J.; Sáez, C.; Perez-Girbes, A.; Sanz-Requena, R.; Revert-Ventura, A.; Mocholi, A.; Urchueguia, J.; Hervas, A.; et al. Multi-parametric MR Imaging Biomarkers Associated to Clinical Outcomes in Gliomas: A Systematic Review. CMIR 2019, 15, 933–947. [Google Scholar] [CrossRef]
- Pujol, S.; Wells, W.; Pierpaoli, C.; Brun, C.; Gee, J.; Cheng, G.; Vemuri, B.; Commowick, O.; Prima, S.; Stamm, A.; et al. The DTI Challenge: Toward Standardized Evaluation of Diffusion Tensor Imaging Tractography for Neurosurgery: The DTI Challenge on Tractography for Neurosurgery. J. Neuroimaging 2015, 25, 875–882. [Google Scholar] [CrossRef]
- Langen, K.-J.; Galldiks, N.; Hattingen, E.; Shah, N.J. Advances in neuro-oncology imaging. Nat. Rev. Neurol. 2017, 13, 279–289. [Google Scholar] [CrossRef]
- Zacharaki, E.I.; Morita, N.; Bhatt, P.; O’Rourke, D.M.; Melhem, E.R.; Davatzikos, C. Survival Analysis of Patients with High-Grade Gliomas Based on Data Mining of Imaging Variables. Am. J. Neuroradiol. 2012, 33, 1065–1071. [Google Scholar] [CrossRef]
- Nie, D.; Zhang, H.; Adeli, E.; Liu, L.; Shen, D. 3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2016; Volume 9901, pp. 212–220. ISBN 978-3-319-46722-1. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).