High G2M Pathway Score Pancreatic Cancer is Associated with Worse Survival, Particularly after Margin-Positive (R1 or R2) Resection
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
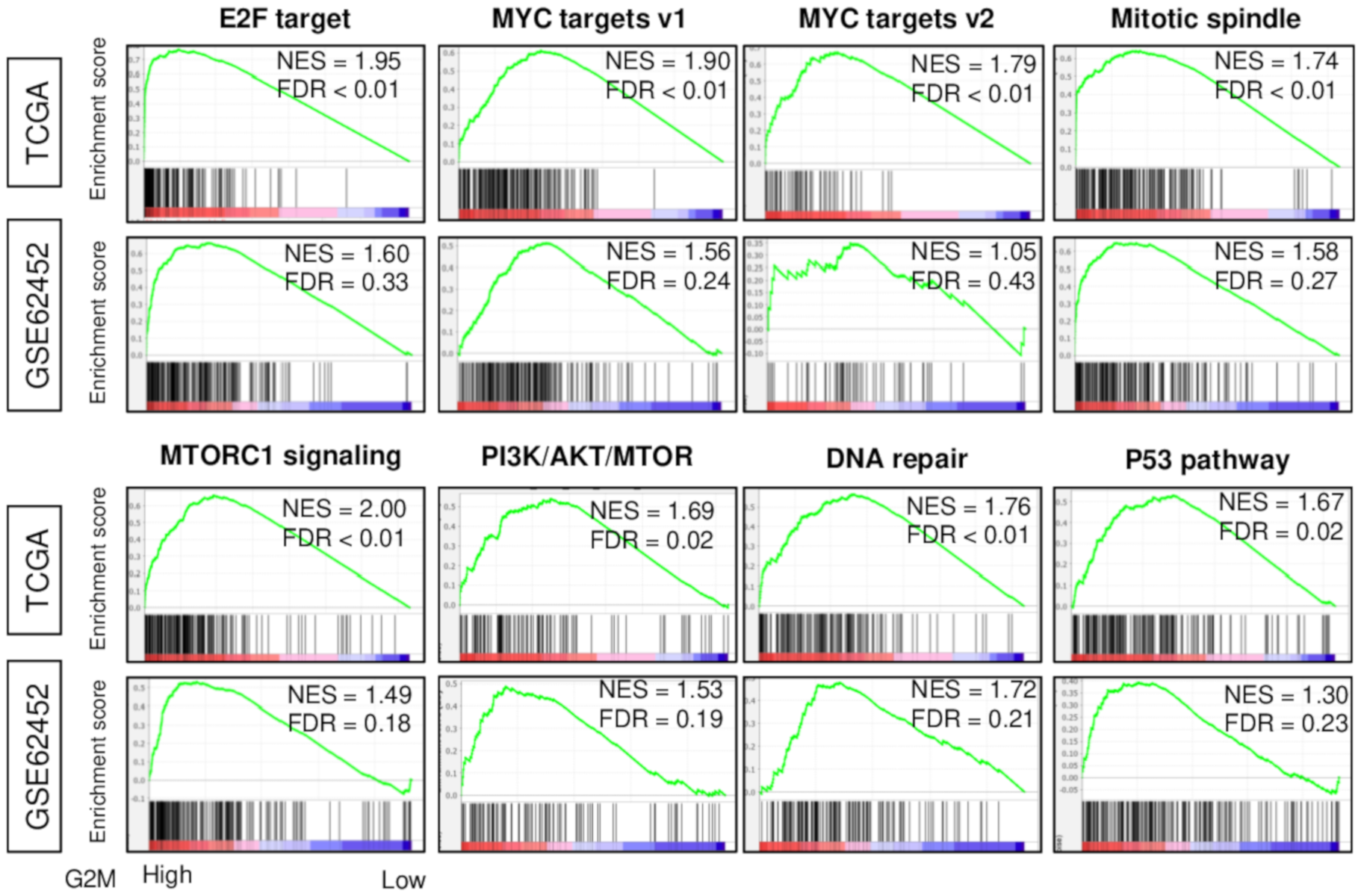
2.1. High G2M Pathway Score Reflects Increased Cell Proliferation in Pancreatic Cancer
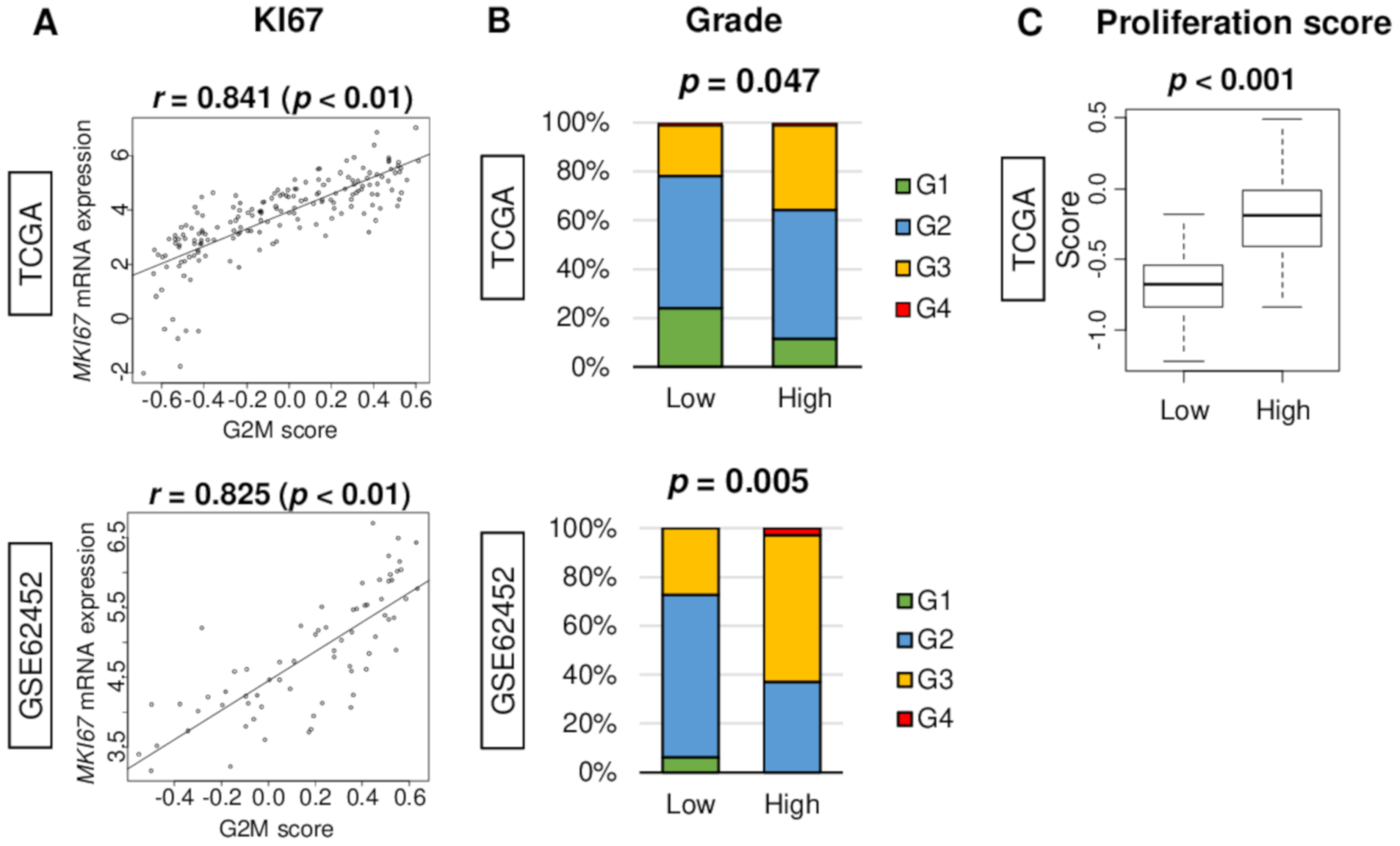
2.2. G2M Pathway Score Associates with Clinical Parameters of Cell Proliferation in Pancreatic Cancer
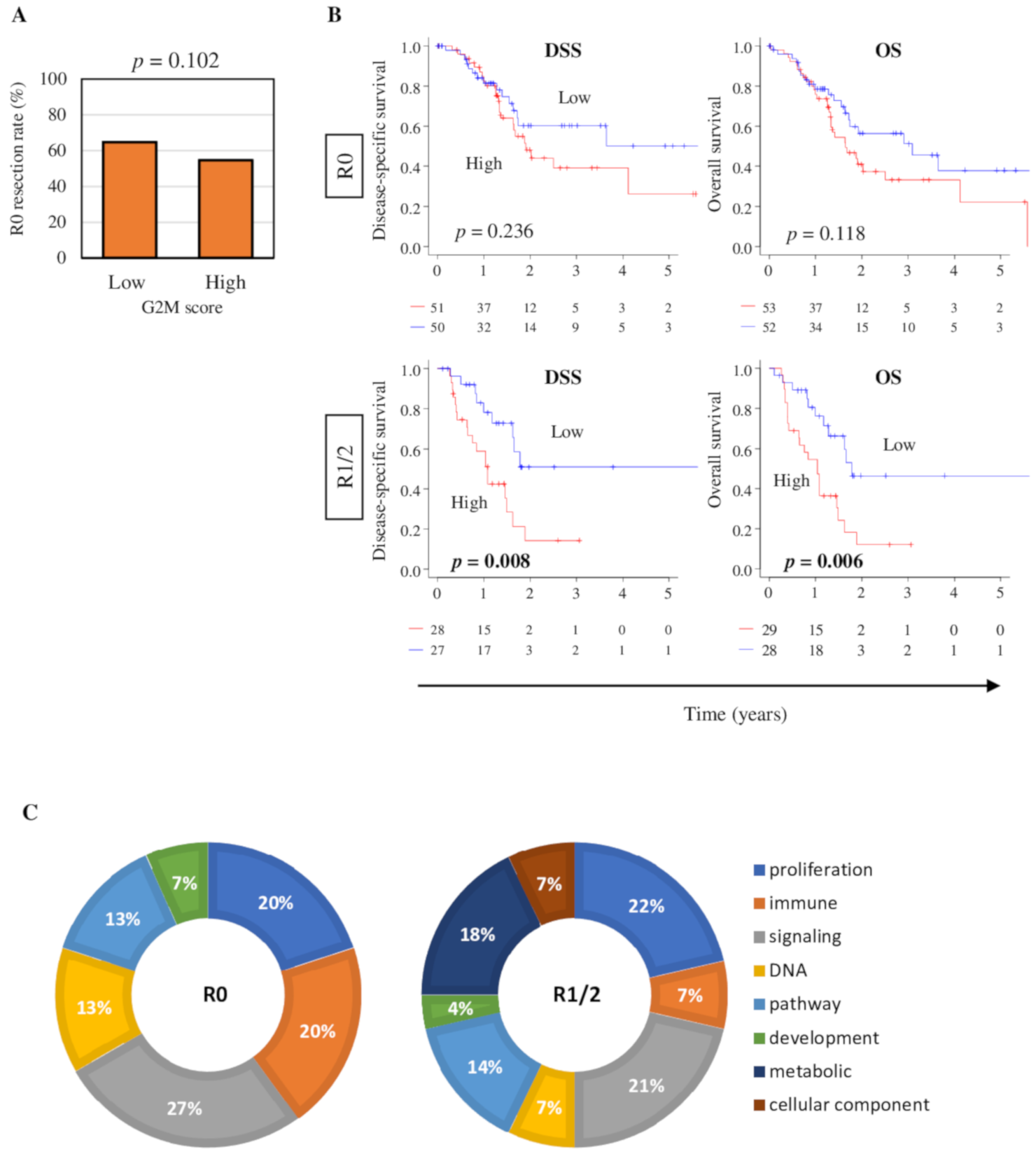
2.3. Clinical Characteristics of the High G2M Pathway Score Pancreatic Cancer Patients
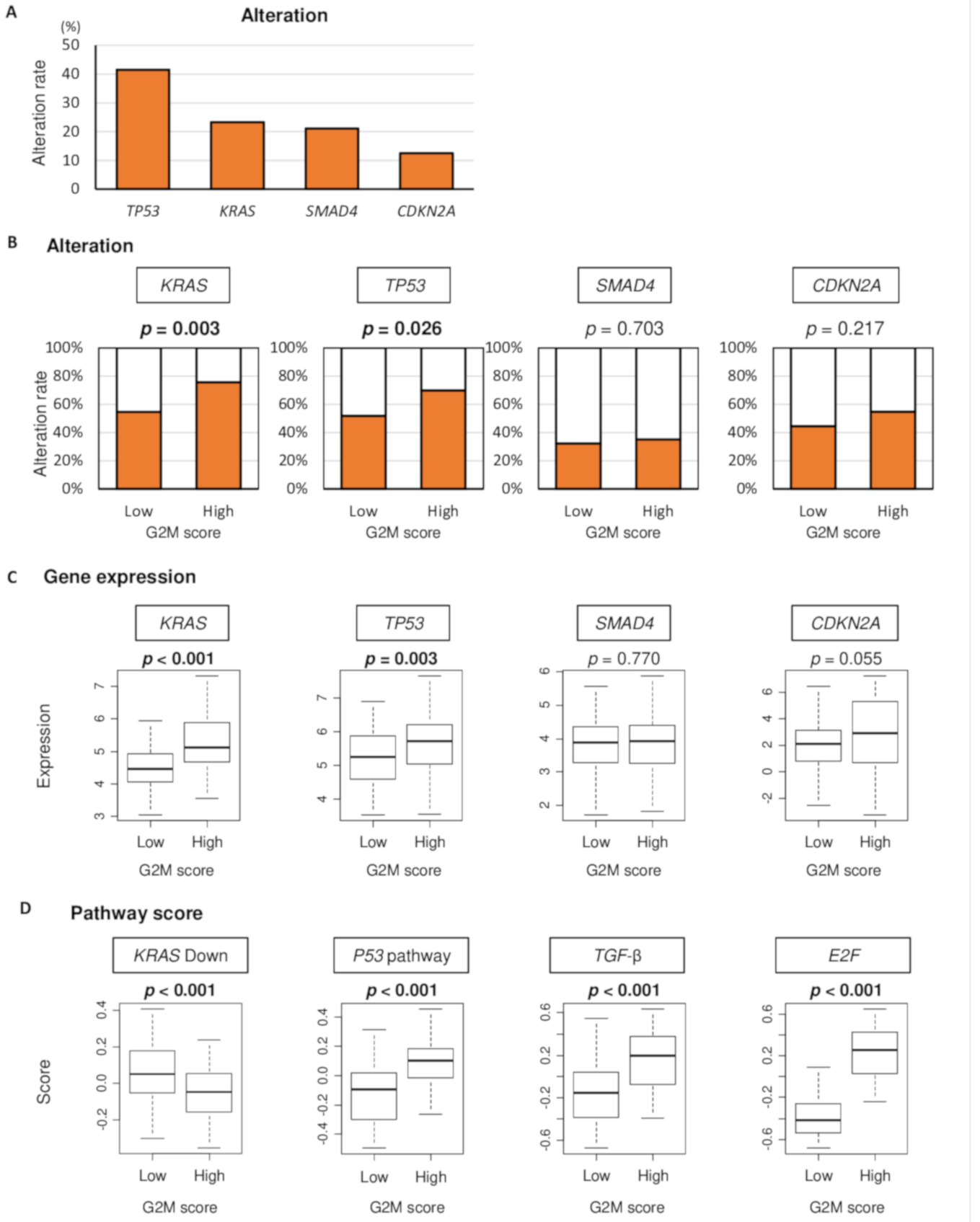
2.4. High G2M Pathway Score Tumor is Associated Not Only with KRAS and TP53 Gene Alteration but also with Their Signaling in Pancreatic Cancer
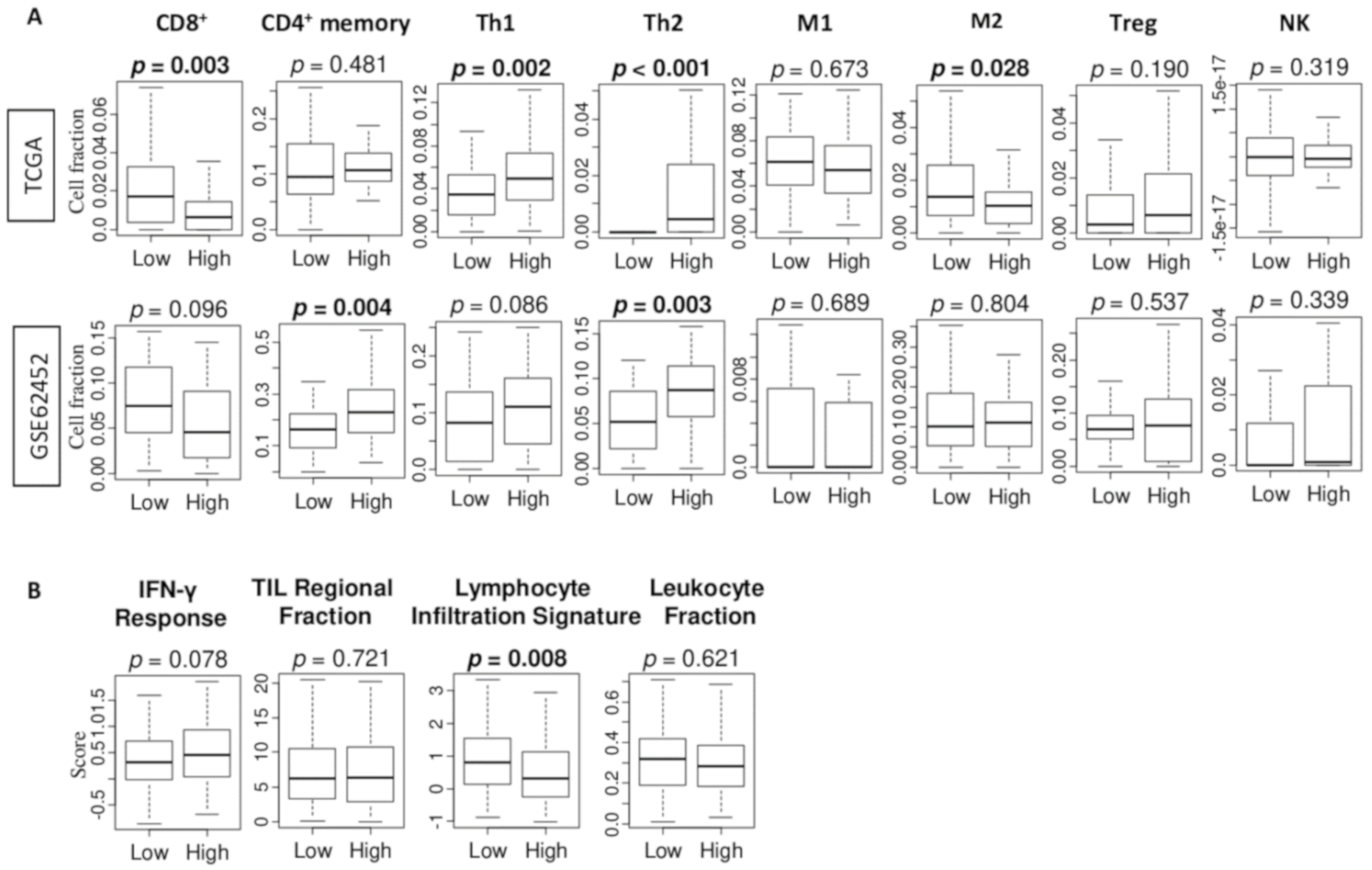
2.5. The High G2M Pathway Score is Significantly Associated with High Fraction of T Helper Type 2 (Th2) Immune Cells
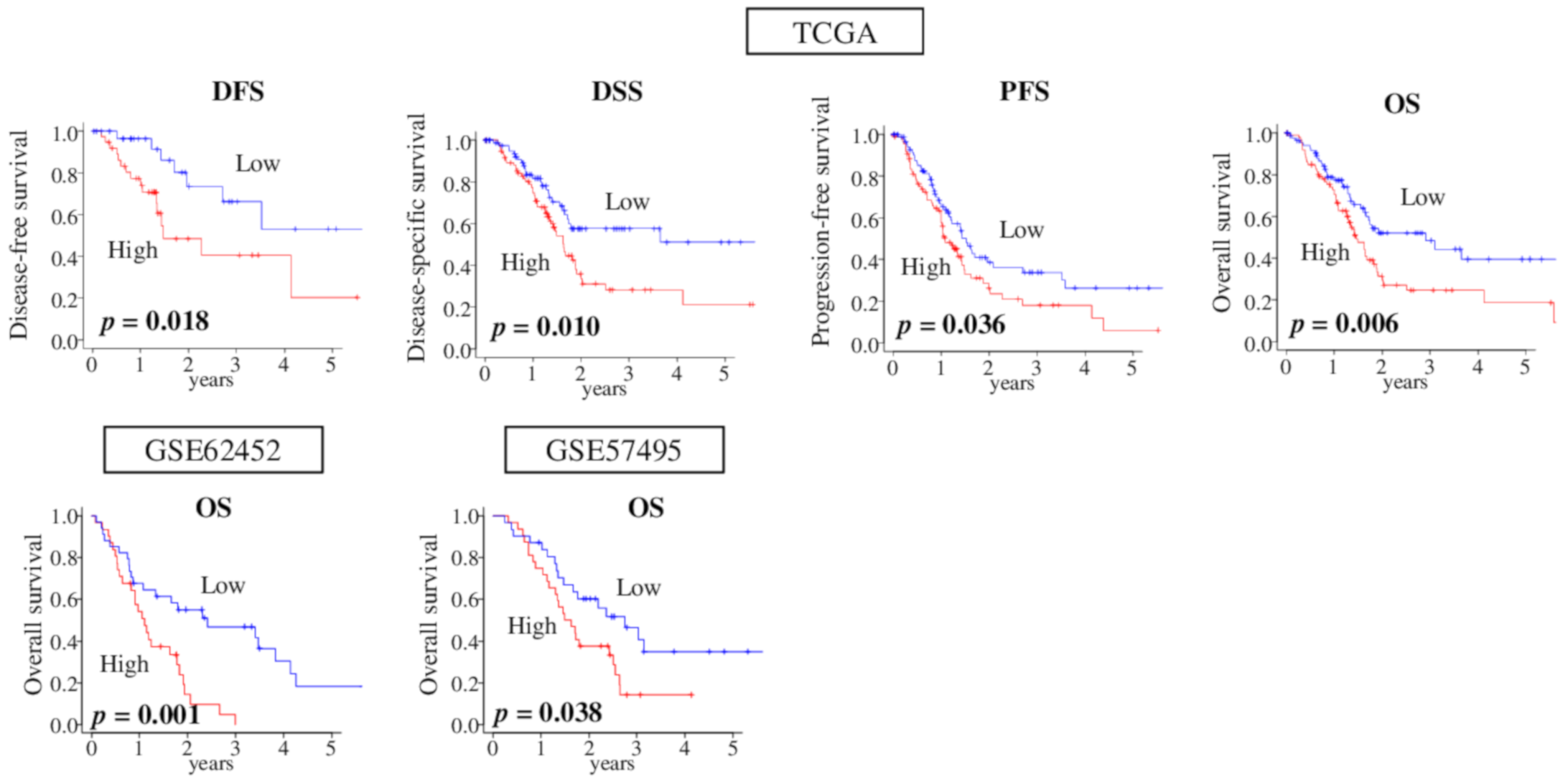
2.6. High-Score G2M Pathway Pancreatic Cancer was Significantly Associated with Worse Survival Consistently in Multiple Cohorts
2.7. High G2M Pathway Score Pancreatic Tumors were Significantly Associated with Worse Survival in Pathologically Margin-Positive Resection (R1 or R2), but not in Margin-Negative Resection (R0)
3. Discussion
4. Materials and Methods
4.1. Pancreatic Cancer Cohorts and Their Data
4.2. G2M Scoring Method
4.3. Gene Set Expression Analysis
4.4. Cell Composition Fraction Estimation
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Ansari, D.; Gustafsson, A.; Andersson, R. Update on the management of pancreatic cancer: Surgery is not enough. World J. Gastroenterol. 2015, 21, 3157–3165. [Google Scholar] [CrossRef]
- Katz, M.H.; Pisters, P.W.; Evans, D.B.; Sun, C.C.; Lee, J.E.; Fleming, J.B.; Vauthey, J.N.; Abdalla, E.K.; Crane, C.H.; Wolff, R.A.; et al. Borderline resectable pancreatic cancer: The importance of this emerging stage of disease. J. Am. Coll. Surg. 2008, 206, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Demir, I.E.; Jäger, C.; Schlitter, A.M.; Konukiewitz, B.; Stecher, L.; Schorn, S.; Tieftrunk, E.; Scheufele, F.; Calavrezos, L.; Schirren, R.; et al. R0 Versus R1 Resection Matters after Pancreaticoduodenectomy, and Less after Distal or Total Pancreatectomy for Pancreatic Cancer. Ann. Surg. 2018, 268, 1058–1068. [Google Scholar] [CrossRef]
- Rau, B.M.; Moritz, K.; Schuschan, S.; Alsfasser, G.; Prall, F.; Klar, E. R1 resection in pancreatic cancer has significant impact on long-term outcome in standardized pathology modified for routine use. Surgery 2012, 152, S103–S111. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Fibroblasts as a Biological Marker for Curative Resection in Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Duewell, P.; Steger, A.; Lohr, H.; Bourhis, H.; Hoelz, H.; Kirchleitner, S.V.; Stieg, M.R.; Grassmann, S.; Kobold, S.; Siveke, J.T.; et al. RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8(+) T cells. Cell Death Differ. 2014, 21, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.L.; Malgor, R.; Dickerson, E.; Weeraratna, A.T.; Slominski, A.; Wortsman, J.; Harii, N.; Kohn, A.D.; Moon, R.T.; Schwartz, F.L.; et al. Phenylmethimazole decreases Toll-like receptor 3 and noncanonical Wnt5a expression in pancreatic cancer and melanoma together with tumor cell growth and migration. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 4114–4122. [Google Scholar] [CrossRef]
- Hou, J.; Wang, Z.; Li, H.; Zhang, H.; Luo, L. Gene Signature and Identification of Clinical Trait-Related m(6) A Regulators in Pancreatic Cancer. Front. Genet. 2020, 11, 522. [Google Scholar] [CrossRef]
- Chen, G.L.; Yang, L.; Rowe, T.C.; Halligan, B.D.; Tewey, K.M.; Liu, L.F. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J. Biol. Chem. 1984, 259, 13560–13566. [Google Scholar]
- Wheelden, M.; Yee, N.S. Clinical Evaluation of the Safety and Efficacy of Trifluridine/Tipiracil in the Treatment of Advanced Gastric/Gastroesophageal Junction Adenocarcinoma: Evidence to Date. Oncotargets Ther. 2020, 13, 7459–7465. [Google Scholar] [CrossRef] [PubMed]
- Long, K.; Suresh, K. Pulmonary toxicity of systemic lung cancer therapy. Respirology 2020. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Yost, S.E.; Yuan, Y. Neoadjuvant Treatment for Triple Negative Breast Cancer: Recent Progresses and Challenges. Cancers 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.K.; Collier, A.L.; Lee, D.; Hoefer, R.A.; Zheleva, V.; Siewertsz van Reesema, L.L.; Tang-Tan, A.M.; Guye, M.L.; Chang, D.Z.; Winston, J.S.; et al. Perspectives on Triple-Negative Breast Cancer: Current Treatment Strategies, Unmet Needs, and Potential Targets for Future Therapies. Cancers 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Dadi, N.; Stanley, M.; Shahda, S.; O’Neil, B.H.; Sehdev, A. Impact of Nab-Paclitaxel-based Second-line Chemotherapy in Metastatic Pancreatic Cancer. Anticancer Res. 2017, 37, 5533–5539. [Google Scholar] [CrossRef] [PubMed]
- Raufi, A.G.; Manji, G.A.; Chabot, J.A.; Bates, S.E. Neoadjuvant Treatment for Pancreatic Cancer. Semin. Oncol. 2019, 46, 19–27. [Google Scholar] [CrossRef]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. G2M Cell Cycle Pathway Score as a Prognostic Biomarker of Metastasis in Estrogen Receptor (ER)-Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2019, 51, 411–412. [Google Scholar] [CrossRef]
- Ohtsubo, K.; Watanabe, H.; Yamaguchi, Y.; Hu, Y.X.; Motoo, Y.; Okai, T.; Sawabu, N. Abnormalities of tumor suppressor gene p16 in pancreatic carcinoma: Immunohistochemical and genetic findings compared with clinicopathological parameters. J. Gastroenterol. 2003, 38, 663–671. [Google Scholar] [CrossRef][Green Version]
- Oshima, M.; Okano, K.; Muraki, S.; Haba, R.; Maeba, T.; Suzuki, Y.; Yachida, S. Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer. Ann. Surg. 2013, 258, 336–346. [Google Scholar] [CrossRef]
- Masetti, M.; Acquaviva, G.; Visani, M.; Tallini, G.; Fornelli, A.; Ragazzi, M.; Vasuri, F.; Grifoni, D.; Di Giacomo, S.; Fiorino, S.; et al. Long-term survivors of pancreatic adenocarcinoma show low rates of genetic alterations in KRAS, TP53 and SMAD4. Cancer Biomark. Sect. A Dis. Markers 2018, 21, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Asaoka, M.; Yan, L.; Rashid, O.M.; Oshi, M.; Ishikawa, T.; Nagahashi, M.; Takabe, K. Biologically Aggressive Phenotype and Anti-cancer Immunity Counterbalance in Breast Cancer with High Mutation Rate. Sci. Rep. 2020, 10, 1852. [Google Scholar] [CrossRef] [PubMed]
- Notta, F.; Chan-Seng-Yue, M.; Lemire, M.; Li, Y.; Wilson, G.W.; Connor, A.A.; Denroche, R.E.; Liang, S.B.; Brown, A.M.; Kim, J.C.; et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 2016, 538, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, V.P.; Łuksza, M.; Zhao, J.N.; Makarov, V.; Moral, J.A.; Remark, R.; Herbst, B.; Askan, G.; Bhanot, U.; Senbabaoglu, Y.; et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017, 551, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef]
- Schlitter, A.M.; Segler, A.; Steiger, K.; Michalski, C.W.; Jäger, C.; Konukiewitz, B.; Pfarr, N.; Endris, V.; Bettstetter, M.; Kong, B.; et al. Molecular, morphological and survival analysis of 177 resected pancreatic ductal adenocarcinomas (PDACs): Identification of prognostic subtypes. Sci. Rep. 2017, 7, 41064. [Google Scholar] [CrossRef]
- Russo, A.A.; Tong, L.; Lee, J.O.; Jeffrey, P.D.; Pavletich, N.P. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 1998, 395, 237–243. [Google Scholar] [CrossRef]
- Blackford, A.; Serrano, O.K.; Wolfgang, C.L.; Parmigiani, G.; Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Eshleman, J.R.; et al. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 4674–4679. [Google Scholar] [CrossRef]
- Boone, B.A.; Sabbaghian, S.; Zenati, M.; Marsh, J.W.; Moser, A.J.; Zureikat, A.H.; Singhi, A.D.; Zeh, H.J., 3rd; Krasinskas, A.M. Loss of SMAD4 staining in pre-operative cell blocks is associated with distant metastases following pancreaticoduodenectomy with venous resection for pancreatic cancer. J. Surg. Oncol. 2014, 110, 171–175. [Google Scholar] [CrossRef]
- Winter, J.M.; Tang, L.H.; Klimstra, D.S.; Liu, W.; Linkov, I.; Brennan, M.F.; D’Angelica, M.I.; DeMatteo, R.P.; Fong, Y.; Jarnagin, W.R.; et al. Failure patterns in resected pancreas adenocarcinoma: Lack of predicted benefit to SMAD4 expression. Ann. Surg. 2013, 258, 331–335. [Google Scholar] [CrossRef]
- Yachida, S.; White, C.M.; Naito, Y.; Zhong, Y.; Brosnan, J.A.; Macgregor-Das, A.M.; Morgan, R.A.; Saunders, T.; Laheru, D.A.; Herman, J.M.; et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 6339–6347. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Yan, L.; Asaoka, M.; Rashid, O.; Takabe, K. Late recurrence of breast cancer is associated with pro-cancerous immune microenvironment in the primary tumor. Sci. Rep. 2019, 9, 16942. [Google Scholar] [CrossRef] [PubMed]
- McDonald, K.A.; Kawaguchi, T.; Qi, Q.; Peng, X.; Asaoka, M.; Young, J.; Opyrchal, M.; Yan, L.; Patnaik, S.; Otsuji, E.; et al. Tumor Heterogeneity Correlates with Less Immune Response and Worse Survival in Breast Cancer Patients. Ann. Surg. Oncol. 2019, 26, 2191–2199. [Google Scholar] [CrossRef]
- Narayanan, S.; Kawaguchi, T.; Peng, X.; Qi, Q.; Liu, S.; Yan, L.; Takabe, K. Tumor Infiltrating Lymphocytes and Macrophages Improve Survival in Microsatellite Unstable Colorectal Cancer. Sci. Rep. 2019, 9, 13455. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Clinical relevance of tumor microenvironment: Immune cells, vessels, and mouse models. Hum. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Nagahashi, M.; Matsuyama, R.; Endo, I.; Takabe, K. The E2F Pathway Score as a Predictive Biomarker of Response to Neoadjuvant Therapy in ER+/HER2- Breast Cancer. Cells 2020, 9. [Google Scholar] [CrossRef]
- Oshi, M.; Katsuta, E.; Yan, L.; Ebos, J.M.L.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. A Novel 4-Gene Score to Predict Survival, Distant Metastasis and Response to Neoadjuvant Therapy in Breast Cancer. Cancers 2020, 12. [Google Scholar] [CrossRef]
- Okano, M.; Oshi, M.; Butash, A.L.; Katsuta, E.; Tachibana, K.; Saito, K.; Okayama, H.; Peng, X.; Yan, L.; Kono, K.; et al. Triple-Negative Breast Cancer with High Levels of Annexin A1 Expression Is Associated with Mast Cell Infiltration, Inflammation, and Angiogenesis. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Okano, M.; Oshi, M.; Butash, A.L.; Asaoka, M.; Katsuta, E.; Peng, X.; Qi, Q.; Yan, L.; Takabe, K. Estrogen Receptor Positive Breast Cancer with High Expression of Androgen Receptor has Less Cytolytic Activity and Worse Response to Neoadjuvant Chemotherapy but Better Survival. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Joyce, D.; Takabe, K. Current status and limitations of immunotherapy for breast cancer. Surgery 2020, 167, 628–630. [Google Scholar] [CrossRef]
- Lesinski, G.B. Braking the cell’s cycle and invigorating T-cell immunity against pancreatic cancer. Gut 2020. [Google Scholar] [CrossRef]
- Wang, B.; Sun, L.; Yuan, Z.; Tao, Z. Wee1 kinase inhibitor AZD1775 potentiates CD8+ T cell-dependent antitumour activity via dendritic cell activation following a single high dose of irradiation. Med Oncol. 2020, 37, 66. [Google Scholar] [CrossRef]
- Isaji, S.; Mizuno, S.; Windsor, J.A.; Bassi, C.; Fernández-Del Castillo, C.; Hackert, T.; Hayasaki, A.; Katz, M.H.G.; Kim, S.W.; Kishiwada, M.; et al. International consensus on definition and criteria of borderline resectable pancreatic ductal adenocarcinoma 2017. Pancreatology 2018, 18, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, I.T.; Warshaw, A.L.; Allen, J.N.; Blaszkowsky, L.S.; Castillo, C.F.; Deshpande, V.; Hong, T.S.; Kwak, E.L.; Lauwers, G.Y.; Ryan, D.P.; et al. Pancreatic ductal adenocarcinoma: Is there a survival difference for R1 resections versus locally advanced unresectable tumors? What is a “true” R0 resection? Ann. Surg. 2013, 257, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Kim, J.H.; Joo, I.; Chang, W.; Han, J.K. MDCT findings predicting post-operative residual tumor and survival in patients with pancreatic cancer. Eur. Radiol. 2019, 29, 3714–3724. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, E.S.; O’Reilly, E.M.; Brody, J.R.; Witkiewicz, A.K. Genetic Diversity of Pancreatic Ductal Adenocarcinoma and Opportunities for Precision Medicine. Gastroenterology 2016, 150, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Witkiewicz, A.K.; Borja, N.A.; Franco, J.; Brody, J.R.; Yeo, C.J.; Mansour, J.; Choti, M.A.; McCue, P.; Knudsen, E.S. Selective impact of CDK4/6 suppression on patient-derived models of pancreatic cancer. Oncotarget 2015, 6, 15788–15801. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Tang, J.; Tudur-Smith, C.; Neoptolemos, J.P.; Ghaneh, P. Meta-analysis of immunohistochemical prognostic markers in resected pancreatic cancer. Br. J. Cancer 2011, 104, 1440–1451. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Chou, A.; Froio, D.; Nagrial, A.M.; Parkin, A.; Murphy, K.J.; Chin, V.T.; Wohl, D.; Steinmann, A.; Stark, R.; Drury, A.; et al. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut 2018, 67, 2142–2155. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nicolay, B.N.; Chick, J.M.; Gao, X.; Geng, Y.; Ren, H.; Gao, H.; Yang, G.; Williams, J.A.; Suski, J.M.; et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 2017, 546, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Barbero, B.; Álvarez-Fernández, M.; Zapatero-Solana, E.; El Bakkali, A.; Menéndez, M.D.C.; López-Casas, P.P.; Di Domenico, T.; Xie, T.; VanArsdale, T.; Shields, D.J.; et al. CDK4/6 Inhibitors Impair Recovery from Cytotoxic Chemotherapy in Pancreatic Adenocarcinoma. Cancer Cell 2020, 37, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Yan, L.; Qi, Q.; Peng, X.; Edge, S.B.; Young, J.; Yao, S.; Liu, S.; Otsuji, E.; Takabe, K. Novel MicroRNA-Based Risk Score Identified by Integrated Analyses to Predict Metastasis and Poor Prognosis in Breast Cancer. Ann. Surg. Oncol. 2018, 25, 4037–4046. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Katsuta, E.; Oshi, M.; Sporn, J.C.; Yan, L.; Le, L.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K.; et al. High Expression of miR-34a Associated with Less Aggressive Cancer Biology but Not with Survival in Breast Cancer. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Tokumaru, Y.; Asaoka, M.; Oshi, M.; Katsuta, E.; Yan, L.; Narayanan, S.; Sugito, N.; Matsuhashi, N.; Futamura, M.; Akao, Y.; et al. High Expression of microRNA-143 is Associated with Favorable Tumor Immune Microenvironment and Better Survival in Estrogen Receptor Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Kawaguchi, T.; Yan, L.; Peng, X.; Qi, Q.; Morris, L.G.T.; Chan, T.A.; Tsung, A.; Otsuji, E.; Takabe, K. Immune Cytolytic Activity for Comprehensive Understanding of Immune Landscape in Hepatocellular Carcinoma. Cancers 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Elkhanany, A.; Oshi, M.; Dai, T.; Opyrchal, M.; Mohammadpour, H.; Repasky, E.A.; Takabe, K. Contribution of Immune Cells to Glucocorticoid Receptor Expression in Breast Cancer. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Katsuta, E.; Yan, L.; Dasgupta, S.; Takabe, K. High expression of Annexin A2 is associated with DNA repair, metabolic alteration, and worse survival in pancreatic ductal adenocarcinoma. Surgery 2019, 166, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Qi, Q.; Peng, X.; Hochwald, S.N.; Yan, L.; Takabe, K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci. Rep. 2019, 9, 1310. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Katsuta, E.; Yan, L.; Tokumaru, Y.; Katz, M.H.G.; Takabe, K. Transcriptomic Profile of Lymphovascular Invasion, a Known Risk Factor of Pancreatic Ductal Adenocarcinoma Metastasis. Cancers 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, M.; Patnaik, S.K.; Zhang, F.; Ishikawa, T.; Takabe, K. Lymphovascular invasion in breast cancer is associated with gene expression signatures of cell proliferation but not lymphangiogenesis or immune response. Breast Cancer Res. Treat. 2020, 181, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Oshi, M.; Asaoka, M.; Yan, L.; Endo, I.; Takabe, K. Molecular Biological Features of Nottingham Histological Grade 3 Breast Cancers. Ann. Surg. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Oshi, M.; Asaoka, M.; Ishikawa, T.; Endo, I.; Takabe, K. ASO Author Reflections: Transitioning From Morphology to Transcriptomics in Capturing Tumor Biology. Ann. Surg. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Yan, L.; Takeshita, T.; McDonald, K.A.; Dasgupta, S.; Opyrchal, M.; Takabe, K. High MYC mRNA Expression Is More Clinically Relevant than MYC DNA Amplification in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2019, 21. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Oshi, M.; Katsuta, E.; Yan, L.; Satyananda, V.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K.; Takabe, K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am. J. Cancer Res. 2020, 10, 897–907. [Google Scholar] [PubMed]
- Yang, S.; He, P.; Wang, J.; Schetter, A.; Tang, W.; Funamizu, N.; Yanaga, K.; Uwagawa, T.; Satoskar, A.R.; Gaedcke, J.; et al. A Novel MIF Signaling Pathway Drives the Malignant Character of Pancreatic Cancer by Targeting NR3C2. Cancer Res. 2016, 76, 3838–3850. [Google Scholar] [CrossRef]
- Chen, D.T.; Davis-Yadley, A.H.; Huang, P.Y.; Husain, K.; Centeno, B.A.; Permuth-Wey, J.; Pimiento, J.M.; Malafa, M. Prognostic Fifteen-Gene Signature for Early Stage Pancreatic Ductal Adenocarcinoma. PLoS ONE 2015, 10, e0133562. [Google Scholar] [CrossRef]
- Bocklandt, S.; Lin, W.; Sehl, M.E.; Sánchez, F.J.; Sinsheimer, J.S.; Horvath, S.; Vilain, E. Epigenetic predictor of age. PLoS ONE 2011, 6, e14821. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
| Clinical Variables | G2M-Low (n = 88) | G2M-High (n = 88) | p-Value |
|---|---|---|---|
| Age at diagnosis | 0.950 | ||
| Median | 65.0 | 65.0 | |
| IQR | 57–74 | 58–71 | |
| Gender | 0.650 | ||
| Male | 50 (56.8%) | 46 (52.3%) | |
| Female | 38 (43.2%) | 42 (47.7%) | |
| Race | 1.00 | ||
| White | 78 (88.6%) | 77 (87.5%) | |
| Other | 10 (11.4%) | 11 (12.5%) | |
| Primary site | 0.680 | ||
| Head | 66 (75.0%) | 71 (80.7%) | |
| Body/Tail | 15 (17.0%) | 13 (14.8%) | |
| Unknown | 7 (8.0%) | 4 (4.5%) | |
| Histology | 0.429 | ||
| PDAC | 70 | 75 | |
| Others | 18 | 13 | |
| Grade | 0.047 | ||
| G1 | 21 (23.9%) | 10 (11.4%) | |
| G2 | 47 (53.4%) | 46 (52.3%) | |
| G3 | 18 (20.5%) | 30 (34.1%) | |
| G4 | 1 (1.1%) | 1 (1.1%) | |
| Unknown | 1 (1.1%) | 1 (1.1%) | |
| AJCC T-category | |||
| T1 | 4 (4.5%) | 3 (3.4%) | 0.524 |
| T2 | 14 (15.9%) | 9 (10.2%) | |
| T3 | 66 (75.0%) | 75 (85.3%) | |
| T4 | 2 (2.3%) | 1 (1.1%) | |
| Unknown | 2 (2.3%) | 0 (0.0%) | |
| AJCC N-category | |||
| N− | 26 (29.5%) | 23 (26.1%) | 0.612 |
| N+ | 58 (65.9%) | 64 (72.7%) | |
| Unknown | 4 (4.6%) | 1 (1.1%) | |
| AJCC M-category | |||
| M− | 37 (42.0%) | 41 (46.6%) | 1.00 |
| M+ | 2 (2.3%) | 2 (2.3%) | |
| Unknown | 49 (55.7%) | 45 (51.1%) | |
| AJCC Stage at diagnosis | 0.540 | ||
| I | 13 (14.8%) | 8 (9.1%) | |
| II | 69 (78.4%) | 76 (86.4%) | |
| III | 2 (2.3%) | 1 (1.1%) | |
| IV | 2 (2.3%) | 2 (2.3%) | |
| Unknown | 2 (2.3%) | 1 (1.1%) | |
| Resection status | 0.102 | ||
| R0 | 57 (64.8%) | 48 (54.6%) | |
| R1/2 | 23 (26.1%) | 34 (38.6%) | |
| Unknown | 8 (9.1%) | 6 (6.8%) |
| TCGA (DSS) | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Age | 1.02 (1.00–1.04) | 0.109 | ||
| Gender (Male vs. Female) | 0.77 (0.48–1.22) | 0.267 | ||
| Race (Caucasian vs. other) | 1.35 (0.45–2.83) | 0.420 | ||
| Primary tumor site (Head vs. Body/Tail) | 1.69 (0.86–3.31) | 0.126 | ||
| Grade (G3/4 vs. G1/2) | 3.28 (1.19–9.07) | 0.022 * | 0.71 (0.12–4.27) | 0.706 |
| AJCC-T category (pT3/4 vs. pT1/2) | 3.08 (1.33–7.14) | 0.009 * | 2.46 (0.59–10.19) | 0.215 |
| AJCC-N category (N+ vs. N-) | 2.58 (1.39–4.81) | 0.003 * | 1.93 (0.96–3.86) | 0.064 |
| AJCC-M category (M+ vs. M-) | 1.21 (0.29–5.09) | 0.799 | ||
| Resection (R1/2 vs. R0) | 1.82 (1.11–2.98) | 0.017 * | 1.58 (0.96–2.61) | 0.072 |
| G2M score | 2.65 (1.36–5.16) | 0.004 * | 2.08 (1.02–4.24) | 0.045 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oshi, M.; Newman, S.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Katz, M.H.G.; Takabe, K. High G2M Pathway Score Pancreatic Cancer is Associated with Worse Survival, Particularly after Margin-Positive (R1 or R2) Resection. Cancers 2020, 12, 2871. https://doi.org/10.3390/cancers12102871
Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. High G2M Pathway Score Pancreatic Cancer is Associated with Worse Survival, Particularly after Margin-Positive (R1 or R2) Resection. Cancers. 2020; 12(10):2871. https://doi.org/10.3390/cancers12102871
Chicago/Turabian StyleOshi, Masanori, Stephanie Newman, Yoshihisa Tokumaru, Li Yan, Ryusei Matsuyama, Itaru Endo, Matthew H. G. Katz, and Kazuaki Takabe. 2020. "High G2M Pathway Score Pancreatic Cancer is Associated with Worse Survival, Particularly after Margin-Positive (R1 or R2) Resection" Cancers 12, no. 10: 2871. https://doi.org/10.3390/cancers12102871
APA StyleOshi, M., Newman, S., Tokumaru, Y., Yan, L., Matsuyama, R., Endo, I., Katz, M. H. G., & Takabe, K. (2020). High G2M Pathway Score Pancreatic Cancer is Associated with Worse Survival, Particularly after Margin-Positive (R1 or R2) Resection. Cancers, 12(10), 2871. https://doi.org/10.3390/cancers12102871






