Non-Invasive Early Molecular Detection of Gastric Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Epigenetic Alteration
3. MicroRNAs (miRNAs)
4. Epigenetic Field Defects and miR-34b/c Methylation
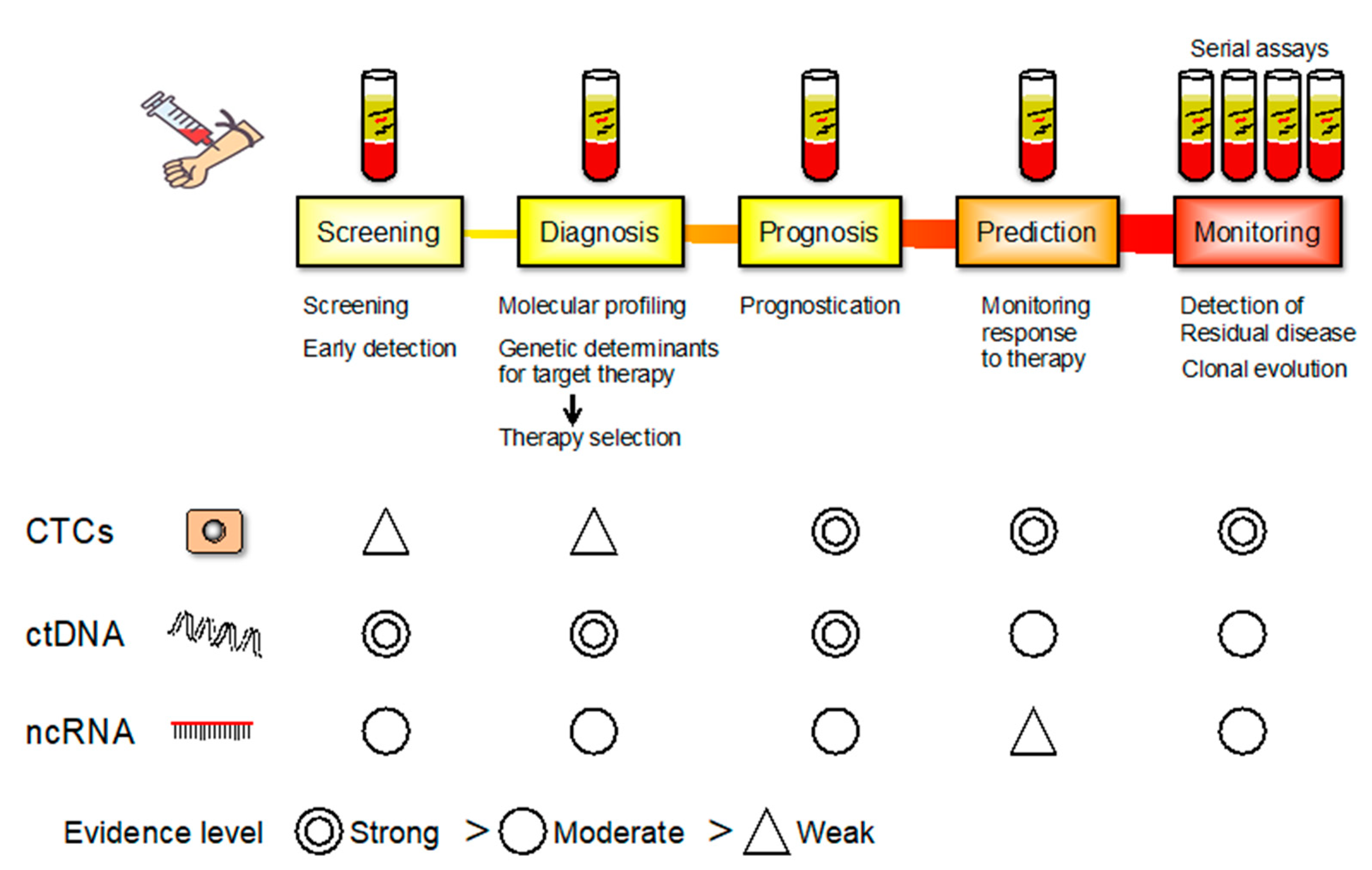
5. Circulating Tumor Cells (CTCs)
6. Cell-Free DNA (cfDNA)
7. Long Noncoding RNAs (LncRNAs)
8. Circular RNAs (CircRNAs)
9. Extracellular Vesicles (EVs)
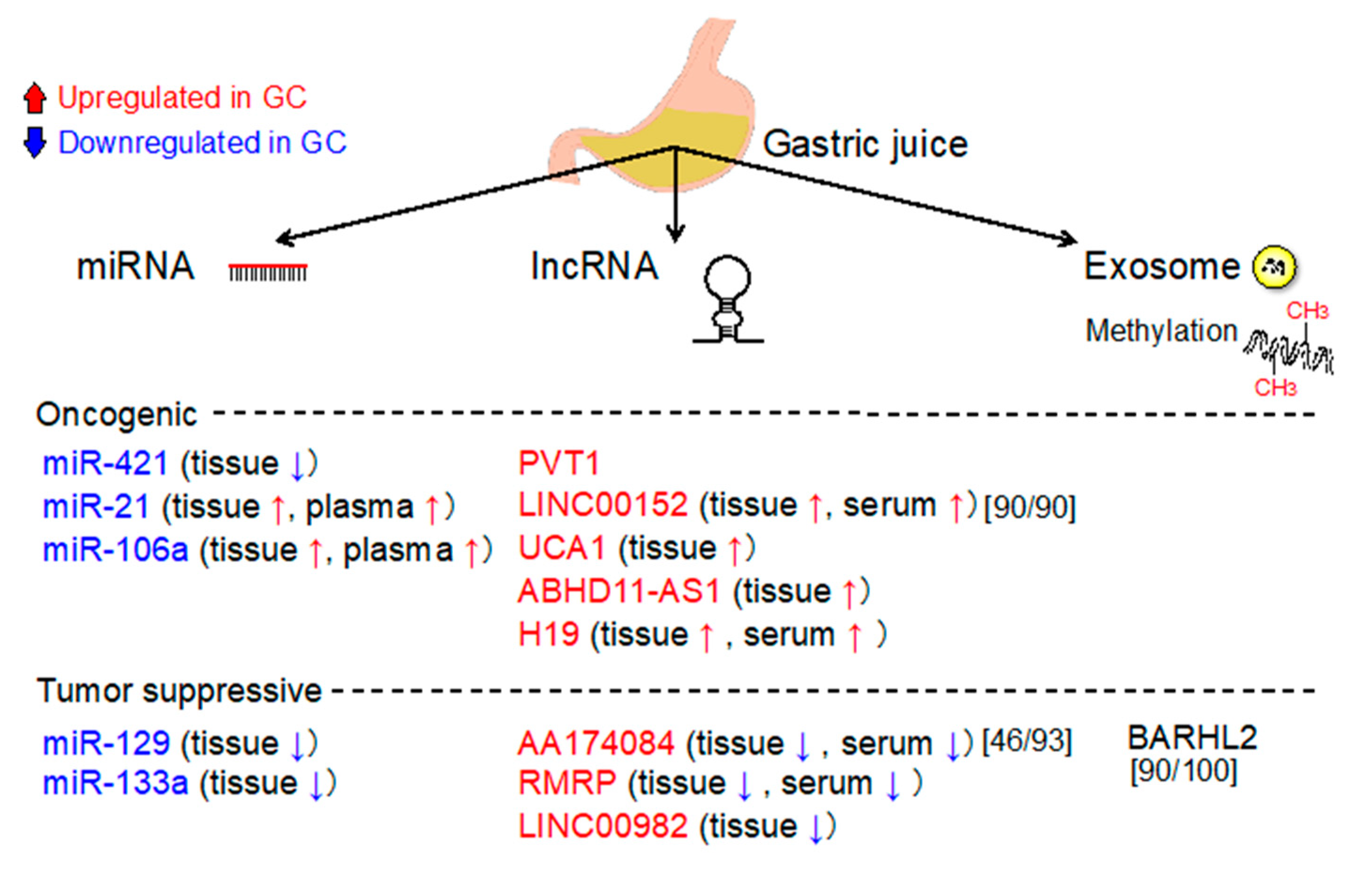
10. Analysis of Gastric Juices/Washes
10.1. Gastric Juice (GJ)-Based Biomarkers
10.2. DNA Methylation Analysis Using Gastric Washes (GWs)
10.3. Gastric Wash or Gastric Juice Exosomal DNA-Based Methylation Analysis of BARHL2
10.4. Analysis of Helicobacter pylori Genotypes Using GWs
11. Ongoing Studies Concerning the Early Molecular Detection of GC
12. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [Green Version]
- Wadhwa, R.; Song, S.; Lee, J.S.; Yao, Y.; Wei, Q.; Ajani, J.A. Gastric cancer-molecular and clinical dimensions. Nat. Rev. Clin. Oncol. 2013, 10, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Salati, M.; Orsi, G.; Smyth, E.; Aprile, G.; Beretta, G.; de Vita, F.; di Bartolomeo, M.; Fanotto, V.; Lonardi, S.; Morano, F.; et al. Gastric cancer: Translating novels concepts into clinical practice. Cancer Treat. Rev. 2019, 79, 101889. [Google Scholar] [CrossRef]
- Figueiredo, C.; Garcia-Gonzalez, M.A.; Machado, J.C. Molecular pathogenesis of gastric cancer. Helicobacter 2013, 18 (Suppl. 1), 28–33. [Google Scholar] [CrossRef] [PubMed]
- Conteduca, V.; Sansonno, D.; Lauletta, G.; Russi, S.; Ingravallo, G.; Dammacco, F.H. Pylori infection and gastric cancer: State of the art. Int. J. Oncol. 2013, 42, 5–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, I.; Gralnek, I.M. Complications of diagnostic colonoscopy, upper endoscopy, and enteroscopy. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, M.; Chen, S.; Hu, J.; Guo, Q.; Liu, R.; Zheng, H.; Jin, Z.; Yuan, Y.; Xi, Y.; et al. Endoscopic screening in Asian countries is associated with reduced gastric cancer mortality: A meta-analysis and systematic review. Gastroenterology 2018, 155, 347–354. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, E.; Suzuki, H.; Takamaru, H.; Yamamoto, H.; Toyota, M.; Shinomura, Y. Role of DNA methylation in the development of diffuse-type gastric cancer. Digestion 2011, 83, 241–249. [Google Scholar] [CrossRef]
- Baker, A.M.; Graham, T.A.; Wright, N.A. Pre-tumour clones, periodic selection and clonal interference in the origin and progression of gastrointestinal cancer: Potential for biomarker development. J. Pathol. 2013, 229, 502–514. [Google Scholar] [CrossRef]
- Yamamoto, H.; Watanabe, Y.; Maehata, T.; Morita, R.; Yoshida, Y.; Oikawa, R.; Ishigooka, S.; Ozawa, S.; Matsuo, Y.; Hosoya, K.; et al. An updated review of gastric cancer in the next-generation sequencing era: Insights from bench to bedside and vice versa. World J. Gastroenterol. 2014, 20, 3927–3937. [Google Scholar] [CrossRef]
- Matsuoka, T.; Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 2018, 24, 2818–2832. [Google Scholar] [CrossRef] [PubMed]
- Necula, L.; Matei, L.; Dragu, D.; Neagu, A.I.; Mambet, C.; Nedeianu, S.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 2019, 25, 2029–2044. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, T.; Yashiro, M. Precision medicine for gastrointestinal cancer: Recent progress and future perspective. World J. Gastrointest. Oncol. 2020, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Azad, N.; Zahnow, C.A.; Rudin, C.M.; Baylin, S.B. The future of epigenetic therapy in solid tumours—Lessons from the past. Nat. Rev. Clin. Oncol. 2013, 10, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Zouridis, H.; Deng, N.; Ivanova, T.; Zhu, Y.; Wong, B.; Huang, D.; Wu, Y.H.; Wu, Y.; Tan, I.B.; Liem, N. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci. Transl. Med. 2012, 4, 156ra140. [Google Scholar] [CrossRef]
- Gigek, C.O.; Chen, E.S.; Calcagno, D.Q.; Wisnieski, F.; Burbano, R.R.; Smith, M.A. Epigenetic mechanisms in gastric cancer. Epigenomics 2012, 4, 279–294. [Google Scholar] [CrossRef]
- Qu, Y.; Dang, S.; Hou, P. Gene methylation in gastric cancer. Clin. Chim. Acta 2013, 424, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Calcagno, D.Q.; Gigek, C.O.; Chen, E.S.; Burbano, R.R.; Smith, M.D.A.C. DNA and histone methylation in gastric carcinogenesis. World J. Gastroenterol. 2013, 19, 1182–1192. [Google Scholar] [CrossRef]
- Otani, K.; Li, X.; Arakawa, T.; Chan, F.K.; Yu, J. Epigenetic-mediated tumor suppressor genes as diagnostic or prognostic biomarkers in gastric cancer. Expert Rev. Mol. Diagn. 2013, 13, 445–455. [Google Scholar] [CrossRef]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo, S.A.; Esteller, M. Dysregulation of microRNAs in cancer: Playing with fire. FEBS Lett. 2011, 585, 2087–2099. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kouwenhove, M.; Kedde, M.; Agami, R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 2011, 11, 644–656. [Google Scholar] [CrossRef]
- Lopez-Serra, P.; Esteller, M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene 2012, 31, 1609–1622. [Google Scholar] [CrossRef] [Green Version]
- Ueda, T.; Volinia, S.; Okumura, H.; Shimizu, M.; Taccioli, C.; Rossi, S.; Alder, H.; Liu, C.G.; Oue, N.; Yasui, W. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010, 11, 136–146. [Google Scholar] [CrossRef] [Green Version]
- Albulescu, R.; Neagu, M.; Albulescu, L.; Tanase, C. Tissular and soluble miRNAs for diagnostic and therapy improvement in digestive tract cancers. Expert Rev. Mol. Diagn. 2011, 11, 101–120. [Google Scholar] [CrossRef]
- Pan, H.W.; Li, S.C.; Tsai, K.W. MicroRNA dysregulation in gastric cancer. Curr. Pharm. Des. 2013, 19, 1273–1284. [Google Scholar]
- Tong, F.; Cao, P.; Yin, Y.; Xia, S.; Lai, R.; Liu, S. MicroRNAs in gastric cancer: From benchtop to bedside. Dig. Dis. Sci. 2014, 59, 24–30. [Google Scholar] [CrossRef]
- Stojanovic, J.; Tognetto, A.; Tiziano, D.F.; Leoncini, E.; Posteraro, B.; Pastorino, R.; Boccia, S. MicroRNAs expression profiles as diagnostic biomarkers of gastric cancer: A systematic literature review. Biomarkers 2019, 24, 110–119. [Google Scholar] [CrossRef]
- Telonis, A.G.; Magee, R.; Loher, P.; Chervoneva, I.; Londin, E.; Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017, 45, 2973–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Link, A.; Kupcinskas, J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J. Gastroenterol. 2018, 24, 3313–3329. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, G.; Wang, Z.; Yao, Y.; Chen, R.; Pu, X.; Wang, J. Circulating microRNA-21 is a potential diagnostic biomarker in gastric cancer. Dis. Markers 2015, 2015, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.M.; Wang, C.S.; Tsai, C.Y.; Huang, C.G.; Lee, K.F.; Huang, H.W.; Lin, Y.H.; Chi, H.C.; Kuo, L.M.; Lu, P.H.; et al. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur. J. Cancer 2016, 64, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.S.; Chen, C.Y.; Chen, W.T.; Kuo, C.Y.; Fang, W.L.; Huang, K.H.; Chiu, P.C.; Lo, S.S. miR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS ONE 2017, 12, e0177346. [Google Scholar] [CrossRef]
- Hanke, M.; Hoefig, K.; Merz, H.; Feller, A.C.; Kausch, I.; Jocham, D.; Warnecke, J.M.; Sczakiel, G. A robust methodology to study urine microRNA as tumor marker: MicroRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 2010, 28, 655–661. [Google Scholar] [CrossRef]
- Zhou, J.; Gong, G.; Tan, H.; Dai, F.; Zhu, X.; Chen, Y.; Wang, J.; Liu, Y.; Chen, P.; Wu, X.; et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015, 33, 2915–2923. [Google Scholar] [CrossRef]
- Zhu, C.; Ren, C.; Han, J.; Ding, Y.; Du, J.; Dai, N.; Dai, J.; Ma, H.; Hu, Z.; Shen, H.; et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br. J. Cancer 2014, 110, 2291–2299. [Google Scholar] [CrossRef]
- Huang, Z.; Zhu, D.; Wu, L.; He, M.; Zhou, X.; Zhang, L.; Zhang, H.; Wang, W.; Zhu, J.; Cheng, W.; et al. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol. Biomark. Prev. 2017, 26, 188–196. [Google Scholar] [CrossRef] [Green Version]
- Nik Mohamed Kamal, N.N.; Shahidan, W.N. Non-exosomal and exosomal circulatory microRNAs: Which are more valid as biomarkers? Front. Pharmacol. 2020, 10, 1500. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, H.; Zhou, X.; Wang, T.; Zhang, J.Y.; Zhu, W.; Zhu, H.; Cheng, W. Five serum-based miRNAs were identified as potential diagnostic biomarkers in gastric cardia adenocarcinoma. Cancer Biomark. 2018, 23, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, L.; Yang, Y.; Gong, L.; Xiao, B.; Liu, X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem. Biophys. Res. Commun. 2017, 493, 1322–1328. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Hesari, A.; Ghasemi, F.; Sahebkar, A. Expression of microRNAs and IRAK1 pathway genes are altered in gastric cancer patients with Helicobacter pylori infection. J. Cell. Biochem. 2018, 119, 7570–7576. [Google Scholar] [CrossRef] [PubMed]
- Ruggieri, V.; Russi, S.; Zoppoli, P.; La Rocca, F.; Angrisano, T.; Falco, G.; Calice, G.; Laurino, S. The role of microRNAs in the regulation of gastric cancer stem cells: A meta-analysis of the current status. J. Clin. Med. 2019, 8, 639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.; Yamamoto, E.; Nojima, M.; Kai, M.; Yamano, H.O.; Yoshikawa, K.; Kimura, T.; Kudo, T.; Harada, E.; Sugai, T.; et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis 2010, 31, 2066–2073. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Yamamoto, E.; Nojima, M.; Maruyama, R.; Yamano, H.O.; Yoshikawa, K.; Kimura, T.; Harada, T.; Ashida, M.; Niinuma, T.; et al. Aberrant methylation of microRNA-34b/c is a predictive marker of metachronous gastric cancer risk. J. Gastroenterol. 2014, 49, 1135–1144. [Google Scholar] [CrossRef] [Green Version]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy in 2016: Circulating tumour cells and cell-free DNA in gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 73–74. [Google Scholar] [CrossRef]
- Guimarães, C.T.U.; Martins, N.N.F.; Oliveira, K.C.D.S.; Almeida, C.M.; Pinheiro, T.M.; Gigek, C.O.; Cavalléro, S.R.D.A.; Assumpção, P.P.; Smith, M.A.C.; Burbano, R.R.; et al. Liquid biopsy provides new insights into gastric cancer. Oncotarget 2018, 9, 15144–15156. [Google Scholar]
- Russi, S.; Calice, G.; Ruggieri, V.; Laurino, S.; La Rocca, F.; Amendola, E.; Lapadula, C.; Compare, D.; Nardone, G.; Musto, P.; et al. Gastric normal adjacent mucosa versus healthy and cancer tissues: Distinctive transcriptomic profiles and biological features. Cancers 2019, 11, 1248. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Okumura, T.; Hirano, K.; Yamaguchi, T.; Sekine, S.; Nagata, T.; Tsukada, K. Circulating tumor cells expressing cancer stem cell marker CD44 as a diagnostic biomarker in patients with gastric cancer. Oncol. Lett. 2017, 13, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Huong, P.T.; Gurshaney, S.; Binh, N.T.; Anh, P.G.; Nguyen, H.H.; Nguyen, X.T.; Pham-The, H.; Tran, P.-T.; Vu, K.T.; Duong, N.X.; et al. Emerging role of circulating tumor cells in gastric cancer. Cancers 2020, 12, 695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, L.; Zhao, S.; Liu, W.; Parchim, N.F.; Huang, J.; Tang, Y.; Gan, P.; Zhong, M. Diagnostic accuracy of circulating tumor cells detection in gastric cancer: Systematic review and meta-analysis. BMC Cancer 2013, 13, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. ctDNA and CTCs in liquid biopsy—Current status and where we need to progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Koldby, K.M.; Mortensen, M.B.; Detlefsen, S.; Pfeiffer, P.; Thomassen, M.; Kruse, T.A. Tumor-specific genetic aberrations in cell-free DNA of gastroesophageal cancer patients. J. Gastroenterol. 2019, 54, 108–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alix-Panabières, C.; Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [Green Version]
- Sumbal, S.; Javed, A.; Afroze, B.; Zulfiqar, H.F.; Javed, F.; Noreen, S.; Ijaz, B. Circulating tumor DNA in blood: Future genomic biomarkers for cancer detection. Exp. Hematol. 2018, 65, 17–28. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [Green Version]
- Saini, A.; Pershad, Y.; Albadawi, H.; Kuo, M.; Alzubaidi, S.; Naidu, S.; Knuttinen, M.G.; Oklu, R. Liquid biopsy in gastrointestinal cancers. Diagnostics 2018, 8, 75. [Google Scholar] [CrossRef] [Green Version]
- Shoda, K.; Ichikawa, D.; Fujita, Y.; Masuda, K.; Hiramoto, H.; Hamada, J.; Arita, T.; Konishi, H.; Kosuga, T.; Komatsu, S.; et al. Clinical utility of circulating cell-free Epstein-Barr virus DNA in patients with gastric cancer. Oncotarget 2017, 8, 28796–28804. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, Y.-X.; Wang, Z. Non-coding RNAs in gastric cancer. Gene 2015, 560, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Gao, G.; Cao, Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis. Markers 2016, 2016, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Long noncoding RNAs as biomarkers in cancer. Dis. Markers 2017, 2017, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.; Sun, W.; Ye, G.; Ding, X.; Liu, Z.; Zhang, S.; Xia, T.; Xiao, B.-X.; Xi, Y.; Guo, J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J. Transl. Med. 2013, 11, 225. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.-Y.; Pan, H.-F.; Leng, R.-X.; Ye, D.-Q. Long noncoding RNAs: Novel insights into gastric cancer. Cancer Lett. 2015, 356, 357–366. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Wang, J.; Song, Y.; Gao, P.; Shi, J.; Chen, P.; Wang, Z. Long noncoding RNAs in gastric cancer: Functions and clinical applications. Onco Targets Ther. 2016, 9, 681–697. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.L.; Li, H.; Zhu, L.; Liu, Z.; Zhou, J.; Shu, Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma 2016, 63, 442–449. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Guo, Q.; Chen, J.; Hu, J.; Wang, S.; Sun, Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: A clinical and in vitro investigation. Oncol. Rep. 2014, 31, 358–364. [Google Scholar] [CrossRef] [Green Version]
- Xian, H.P.; Zhuo, Z.L.; Sun, Y.J.; Liang, B.; Zhao, X.T. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol. Lett. 2018, 16, 4689–4698. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Ye, M.; Jiang, X.; Sun, W.; Ding, X.; Liu, Z.; Ye, G.; Zhang, X.; Xiao, B.; Guo, J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 2014, 120, 3320–3328. [Google Scholar] [CrossRef] [PubMed]
- Arita, T.; Ichikawa, D.; Konishi, H.; Komatsu, S.; Shiozaki, A.; Shoda, K.; Kawaguchi, T.; Hirajima, S.; Nagata, H.; Kubota, T.; et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013, 33, 3185–3193. [Google Scholar] [PubMed]
- Zhou, X.; Yin, C.; Dang, Y.; Ye, F.; Zhang, G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 2015, 5, 11516. [Google Scholar] [CrossRef] [PubMed]
- Hashad, D.; Elbanna, A.; Ibrahim, A.; Khedr, G. Evaluation of the role of circulating long non-coding RNA H19 as a promising novel biomarker in plasma of patients with gastric cancer. J. Clin. Lab. Anal. 2016, 30, 1100–1105. [Google Scholar] [CrossRef]
- Yörüker, E.E.; Keskin, M.; Kulle, C.B.; Holdenrieder, S.; Gezer, U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed. Rep. 2018, 9, 181–186. [Google Scholar]
- Li, Q.; Shao, Y.; Zhang, X.; Zheng, T.; Miao, M.; Qin, L.; Wang, B.; Ye, G.; Xiao, B.; Guo, J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol. 2015, 36, 2007–2012. [Google Scholar] [CrossRef]
- Tan, L.; Yang, Y.; Shao, Y.; Zhang, H.; Guo, J. Plasma lncRNA-GACAT2 is a valuable marker for the screening of gastric cancer. Oncol. Lett. 2016, 12, 4845–4849. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Cao, R.; Mu, H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 12936–12942. [Google Scholar]
- Elsayed, E.T.; Salem, P.E.; Darwish, A.M.; Fayed, H.M. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer. Int. J. Biol. Markers 2018, 33, 528–533. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Shi, H.; Xi, H.; Wu, X.; Cui, J.; Gao, Y.; Liang, W.; Hu, C.; Liu, Y.; Li, J.; et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer. Theranostics 2017, 7, 213–227. [Google Scholar] [CrossRef]
- Dong, L.; Qi, P.; Xu, M.-D.; Ni, S.-J.; Huang, D.; Xu, Q.-H.; Weng, W.; Tan, C.; Sheng, W.; Zhou, X.-Y.; et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int. J. Cancer 2015, 137, 1128–1135. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shen, Y.; Li, Z.; Ruan, Y.; Li, T.; Xiao, B.; Sun, W. The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J. Clin. Lab. Anal. 2020, 34, e23049. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell. Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, S.; Chen, H.; Mo, X.; Li, T.; Shao, Y.; Xiao, B.; Guo, J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta. 2015, 444, 132–136. [Google Scholar] [CrossRef]
- Vidal, A.F.; Ribeiro-Dos-Santos, A.M.; Sandoval, T.V.; De Magalhães, L.L.; Pinto, P.; Anaissi, A.K.M.; Demachki, S.; De Assumpção, P.P.; Dos Santos, S.E.B.; Ribeiro-Dos-Santos, A. The comprehensive expression analysis of circular RNAs in gastric cancer and its association with field cancerization. Sci. Rep. 2017, 7, 14551. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Hong, F.; Shah, M.W.; Shen, X. Circular RNAs as diagnostic biomarkers in gastric cancer: A meta-analysis review. Pathol. Res. Pract. 2019, 215, 152419. [Google Scholar] [CrossRef]
- Sun, H.; Tang, W.; Rong, D.; Jin, H.; Fu, K.; Zhang, W.; Liu, Z.; Cao, H.; Cao, X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018, 21, 299–306. [Google Scholar] [CrossRef]
- Chen, S.; Li, T.; Zhao, Q.; Xiao, B.; Guo, J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta 2017, 466, 167–171. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, S.; Li, T.; Xiao, B.; Zhang, X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J. Clin. Lab. Anal. 2018, 32, e22333. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Shao, Y.; Fu, L.; Xie, Y.; Zhu, L.; Sun, W.; Yu, R.; Xiao, B.; Guo, J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J. Mol. Med. 2018, 96, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Gu, J.; Jiang, P.; Qian, H.; Xu, W.; Zhang, X. Exosomes in gastric cancer: Roles, mechanisms, and applications. Mol. Cancer 2019, 18, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.; Song, C.; Zheng, L.; Xia, L.; Li, Y.; Zhou, Y. The roles of extracellular vesicles in gastric cancer development, microenvironment, anti-cancer drug resistance, and therapy. Mol. Cancer 2019, 18, 62. [Google Scholar] [CrossRef] [PubMed]
- Kahroba, H.; Hejazi, M.S.; Samadi, N. Exosomes: From carcinogenesis and metastasis to diagnosis and treatment of gastric cancer. Cell. Mol. Life Sci. 2019, 76, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; Van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; De Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 3. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ridder, K.; Keller, S.; Dams, M.; Rupp, A.K.; Schlaudraff, J.; del Turco, D.; Starmann, J.; Jadranka Macas, J.; Karpova, D.; Devraj, K.; et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014, 12, e1001874. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas, F.A.S.; Allenson, K.; Bernard, V.; Castillo, J.; Kim, D.U.; Ellis, K.; Ehli, E.A.; Davies, G.E.; Petersen, J.L.; Li, D.; et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann. Oncol. 2016, 27, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allenson, K.; Castillo, J.; Lucas, F.A.S.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Möhrmann, L.; Huang, H.J.; Hong, D.S.; Tsimberidou, A.M.; Fu, S.; Piha-Paul, S.A.; Subbiah, V.; Karp, D.D.; Naing, A.; Krug, A.; et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin. Cancer Res. 2018, 24, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Virgilio, E.; Giarnieri, E.; Giovagnoli, M.R.; Montagnini, M.; Proietti, A.; D’Urso, R.; Mercantini, P.; Balducci, G.; Cavallini, M. Gastric juice microRNAs as potential biomarkers for screening gastric cancer: A systematic review. Anticancer. Res. 2018, 38, 613–616. [Google Scholar]
- Yu, X.; Le, Y.; Guo, J. Gastric juice MicroRNAs as potential biomarkers for the screening of gastric cancer. Cancer 2013, 119, 1618–1626. [Google Scholar]
- Pang, Q.; Ge, J.; Shao, Y.; Sun, W.; Song, H.; Xia, T.; Xiao, B.; Guo, J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumor Biol. 2014, 35, 5441–5447. [Google Scholar] [CrossRef]
- Yang, Y.; Shao, Y.; Zhu, M.; Li, Q.; Yang, F.; Lu, X.; Xu, C.; Xiao, B.; Sun, Y.; Guo, J. Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumor Biol. 2016, 37, 1183–1188. [Google Scholar] [CrossRef]
- Watanabe, Y.; Kim, H.S.; Castoro, R.J.; Chung, W.; Estecio, M.R.; Kondo, K.; Guo, Y.; Ahmed, S.S.; Toyota, M.; Itoh, F. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology 2009, 136, 2149–2158. [Google Scholar] [CrossRef] [Green Version]
- Oishi, Y.; Watanabe, Y.; Yoshida, Y.; Sato, Y.; Hiraishi, T.; Oikawa, R.; Maehata, T.; Suzuki, H.; Toyota, M.; Niwa, H.; et al. Hypermethylation of Sox17 gene is useful as a molecular diagnostic application in early gastric cancer. Tumor Biol. 2012, 33, 383–393. [Google Scholar] [CrossRef]
- Yamamoto, H.; Watanabe, Y.; Oikawa, R.; Morita, R.; Yoshida, Y.; Maehata, T.; Yasuda, H.; Itoh, F. BARHL2 methylation using gastric wash DNA or gastric juice exosomal DNA is a useful marker for early detection of gastric cancer in an H. pylori-independent manner. Clin. Transl. Gastroenterol. 2016, 7, e184. [Google Scholar] [CrossRef] [PubMed]
- Baba, S.; Oishi, Y.; Watanabe, Y.; Oikawa, R.; Morita, R.; Yoshida, Y.; Hiraishi, T.; Maehata, T.; Nagase, Y.; Fukuda, Y.; et al. Gastric wash-based molecular testing for antibiotic resistance in Helicobacter pylori. Digestion 2011, 84, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Watanabe, Y.; Oikawa, R.; Ono, S.; Mabe, K.; Kudo, T.; Yamamoto, H.; Itoh, F.; Kato, M.; Sakamoto, N. Analysis of Helicobacter pylori genotypes in clinical gastric wash samples. Tumor Biol. 2016, 37, 10123–10132. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, R.; Watanabe, Y.; Miyamoto, S.; Sato, Y.; Ono, S.; Mabe, K.; Yamamoto, H.; Kato, M.; Itoh, F. Enrichment of Helicobacter pylori mutant strains after eradication therapy analyzed by gastric wash-based quantitative pyrosequencing. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, Y.; Oikawa, R.; Kodaka, Y.; Sato, Y.; Ono, S.; Kenmochi, T.; Suzuki, H.; Futagami, S.; Kato, M.; Yamamoto, H.; et al. Cancer-related genetic variants of Helicobacter pylori strains determined using gastric wash-based whole genome analysis with single-molecule real-time technology. Int. J. Cancer 2020. [Google Scholar] [CrossRef]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, H.; Watanabe, Y.; Sato, Y.; Maehata, T.; Itoh, F. Non-Invasive Early Molecular Detection of Gastric Cancers. Cancers 2020, 12, 2880. https://doi.org/10.3390/cancers12102880
Yamamoto H, Watanabe Y, Sato Y, Maehata T, Itoh F. Non-Invasive Early Molecular Detection of Gastric Cancers. Cancers. 2020; 12(10):2880. https://doi.org/10.3390/cancers12102880
Chicago/Turabian StyleYamamoto, Hiroyuki, Yoshiyuki Watanabe, Yoshinori Sato, Tadateru Maehata, and Fumio Itoh. 2020. "Non-Invasive Early Molecular Detection of Gastric Cancers" Cancers 12, no. 10: 2880. https://doi.org/10.3390/cancers12102880
APA StyleYamamoto, H., Watanabe, Y., Sato, Y., Maehata, T., & Itoh, F. (2020). Non-Invasive Early Molecular Detection of Gastric Cancers. Cancers, 12(10), 2880. https://doi.org/10.3390/cancers12102880




