Immuno-Interface Score to Predict Outcome in Colorectal Cancer Independent of Microsatellite Instability Status
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patient Clinicopathological Characteristics
2.2. Summary Statistics of Immunogradient and Intratumoral Immune Cell Density Indicators
2.3. Associations of Clinicopathological Parameters, Immunogradient and Intratumoral Immune Cell Density Indicators
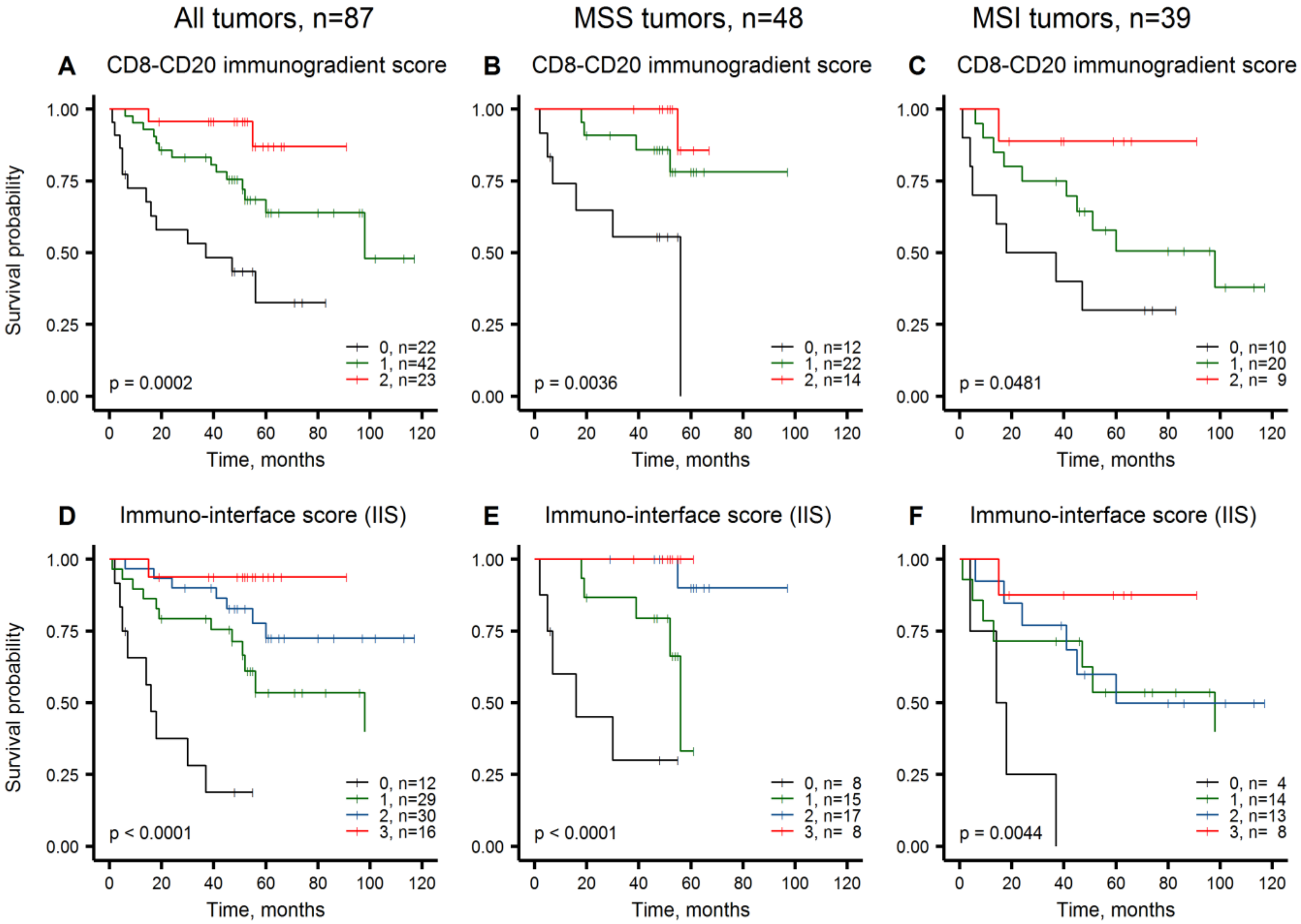
2.4. Immuno-Interface Score for Predicting Patient Overall Survival
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Ethics Statement
4.3. Digital Image Acquisition and Analysis
4.4. Extraction of Interface-Zone and Immunogradient Indicators
4.5. Statistic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Xie, Y.-H.; Chen, Y.X.; Fang, J. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 2020, 5, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Puppa, G.; Sonzogni, A.M.; Colombari, R.; Pelosi, G. TNM staging system of colorectal carcinoma: A critical appraisal of challenging issues. Arch. Pathol. Lab. Med. 2010, 134, 837. [Google Scholar] [PubMed]
- Turley, S.J.; Cremasco, V.; Astarita, J.L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 2015, 15, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Erratum: Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 268. [Google Scholar] [CrossRef] [PubMed]
- Angell, H.K.; Bruni, D.; Barrett, J.C.; Herbst, R.; Galon, J. The Immunoscore: Colon Cancer and Beyond. Clin. Cancer Res. 2019, 26, 332–339. [Google Scholar] [CrossRef]
- Garcia-Carbonero, N.; Martínez-Useros, J.; Li, W.; Orta, A.; Perez, N.; Caramés, C.; Hernandez, T.; Moreno, I.; Serrano, G.; García-Foncillas, J. KRAS and BRAF Mutations as Prognostic and Predictive Biomarkers for Standard Chemotherapy Response in Metastatic Colorectal Cancer: A Single Institutional Study. Cells 2020, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Giardiello, F.M.; Allen, J.I.; Axilbund, J.E.; Boland, R.C.; Burke, C.A.; Burt, R.W.; Church, J.M.; Dominitz, J.A.; Johnson, D.A.; Kaltenbach, T.; et al. Guidelines on Genetic Evaluation and Management of Lynch Syndrome: A Consensus Statement by the US Multi-Society Task Force on Colorectal Cancer. Am. J. Gastroenterol. 2014, 109, 1159–1179. [Google Scholar] [CrossRef]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.-J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair–Deficient/Microsatellite Instability–High Metastatic Colorectal Cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef]
- Le, D.T.; Kim, T.W.; Van Cutsem, E.; Geva, R.; Jäger, D.; Hara, H.; Burge, M.; O’Neil, B.; Kavan, P.; Yoshino, T.; et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J. Clin. Oncol. 2019, 38, 11–19. [Google Scholar] [CrossRef]
- Guastadisegni, C.; Colafranceschi, M.; Ottini, L.; Dogliotti, E. Microsatellite instability as a marker of prognosis and response to therapy: A meta-analysis of colorectal cancer survival data. Eur. J. Cancer 2010, 46, 2788–2798. [Google Scholar] [CrossRef] [PubMed]
- Maby, P.; Tougeron, D.; Hamieh, M.; Mlecnik, B.; Kora, H.; Bindea, G.; Angell, H.K.; Fredriksen, T.; Elie, N.; Fauquembergue, E.; et al. Correlation between Density of CD8+ T-cell Infiltrate in Microsatellite Unstable Colorectal Cancers and Frameshift Mutations: A Rationale for Personalized Immunotherapy. Cancer Res. 2015, 75, 3446–3455. [Google Scholar] [CrossRef] [PubMed]
- Llosa, N.J.; Cruise, M.W.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2014, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Dieu-Nosjean, M.-C.; Sautès-Fridman, C.; Fridman, W.H. Cancer immune contexture and immunotherapy. Curr. Opin. Immunol. 2016, 39, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Van Der Woude, L.L.; Gorris, M.; Halilovic, A.; Figdor, C.G.; De Vries, I.J.M. Migrating into the Tumor: A Roadmap for T Cells. Trends Cancer 2017, 3, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Hendry, S.; Salgado, R.; Gevaert, T.; Russell, P.A.; John, T.; Thapa, B.; Christie, M.; Van De Vijver, K.; Estrada, M.V.; Gonzalez-Ericsson, P.I.; et al. Assessing tumor infiltrating lymphocytes in solid tumors: A practical review for pathologists and proposal for a standardized method from the International Immuno-Oncology Biomarkers Working Group: Part 1: Assessing the host immune response, TILs in invasive breast carcinoma and ductal carcinoma in situ, metastatic tumor deposits and areas for further research. Adv. Anat. Pathol. 2017, 24, 235–251. [Google Scholar] [CrossRef]
- Hendry, S.; Salgado, R.; Gevaert, T.; Russell, P.A.; John, T.; Thapa, B.; Christie, M.; Van De Vijver, K.; Estrada, M.V.; Gonzalez-Ericsson, P.I.; et al. Assessing tumor infiltrating lymphocytes in solid tumors: A practical review for pathologists and proposal for a standardized method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in melanoma, gastrointestinal tract carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv. Anat. Pathol. 2017, 24, 311–335. [Google Scholar] [CrossRef]
- Acs, B.; Ahmad, F.S.; Gupta, S.; Wong, P.F.; Gartrell, R.D.; Pradhan, J.S.; Rizk, E.M.; Rothberg, B.G.; Saenger, Y.M.; Rimm, D.L. An open source automated tumor infiltrating lymphocyte algorithm for prognosis in melanoma. Nat. Commun. 2019, 10, 1–7. [Google Scholar] [CrossRef]
- Abe, N.; Matsumoto, H.; Takamatsu, R.; Tamaki, K.; Takigami, N.; Uehara, K.; Kamada, Y.; Tamaki, N.; Motonari, T.; Unesoko, M.; et al. Quantitative digital image analysis of tumor-infiltrating lymphocytes in HER2-positive breast cancer. Virchows Archiv 2019, 476, 701–709. [Google Scholar] [CrossRef]
- Orhan, A.; Vogelsang, R.P.; Andersen, M.B.; Madsen, M.T.; Hölmich, E.R.; Raskov, H.; Gögenur, I. The prognostic value of tumour-infiltrating lymphocytes in pancreatic cancer: A systematic review and meta-analysis. Eur. J. Cancer 2020, 132, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Steele, K.E.; Tan, T.H.; Korn, R.; Dacosta, K.; Brown, C.; Kuziora, M.; Zimmermann, J.; Laffin, B.; Widmaier, M.; Rognoni, L.; et al. Measuring multiple parameters of CD8+ tumor-infiltrating lymphocytes in human cancers by image analysis. J. Immunother. Cancer 2018, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Koelzer, V.; Sirinukunwattana, K.; Rittscher, J.; Mertz, K.D. Precision immunoprofiling by image analysis and artificial intelligence. Virchows Archiv 2018, 474, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Pagès, F.; Marincola, F.M.; Angell, H.K.; Thurin, M.; Lugli, A.; Zlobec, I.; Berger, A.; Bifulco, C.B.; Botti, G.; et al. Cancer classification using the Immunoscore: A worldwide task force. J. Transl. Med. 2012, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.-S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Marliot, F.; Chen, X.; Kirilovsky, A.; Sbarrato, T.; El Sissy, C.; Batista, L.; Eynde, M.V.D.; Haicheur-Adjouri, N.; Anitei, M.-G.; Musina, A.-M.; et al. Analytical validation of the Immunoscore and its associated prognostic value in patients with colon cancer. J. Immunother. Cancer 2020, 8, e000272. [Google Scholar] [CrossRef]
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Maby, P.; Angelova, M.; Tougeron, D.; Church, S.E.; Lafontaine, L.; Fischer, M.; Fredriksen, T.; et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.S.; Mouradov, D.; Newman, M.R.; Amini, E.; Nickless, D.K.; Fang, C.G.; Palmieri, M.; Sakthianandeswaren, A.; Li, S.; Ward, R.L.; et al. Tumour infiltrating lymphocyte status is superior to histological grade, DNA mismatch repair and BRAF mutation for prognosis of colorectal adenocarcinomas with mucinous differentiation. Mod. Pathol. 2020, 33, 1420–1432. [Google Scholar] [CrossRef]
- Rozek, L.S.; Schmit, S.L.; Greenson, J.K.; Tomsho, L.P.; Rennert, H.S.; Rennert, G.; Gruber, S.B. Tumor-Infiltrating Lymphocytes, Crohn’s-Like Lymphoid Reaction, and Survival From Colorectal Cancer. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Bosman, F.; Carneiro, F.; Hruban, R.; Theise, N. Digestive System Tumours. WHO Classification of Tumours, 5th ed.; World Health Organization: Geneva, Switzerland, 2019; Volume 1, ISBN 13-978-92-832-4499-8. (Print Book). [Google Scholar]
- Nearchou, I.P.; Lillard, K.; Gavriel, C.G.; Ueno, H.; Harrison, D.J.; Caie, P.D. Automated Analysis of Lymphocytic Infiltration, Tumor Budding, and Their Spatial Relationship Improves Prognostic Accuracy in Colorectal Cancer. Cancer Immunol. Res. 2019, 7, 609–620. [Google Scholar] [CrossRef]
- Nearchou, I.P.; Gwyther, B.M.; Georgiakakis, E.C.T.; Gavriel, C.G.; Lillard, K.; Kajiwara, Y.; Ueno, H.; Harrison, D.J.; Caie, P.D. Spatial immune profiling of the colorectal tumor microenvironment predicts good outcome in stage II patients. NPJ Digit. Med. 2020, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rasmusson, A.; Zilenaite, D.; Nestarenkaite, A.; Augulis, R.; Laurinaviciene, A.; Ostapenko, V.; Poskus, T.; Laurinavicius, A. Immunogradient Indicators for Antitumor Response Assessment by Automated Tumor-Stroma Interface Zone Detection. Am. J. Pathol. 2020, 190, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Kakar, S.; Burgart, L.J.; Thibodeau, S.N.; Rabe, K.G.; Petersen, G.M.; Goldberg, R.M.; Lindor, N.M. Frequency of loss ofhMLH1 expression in colorectal carcinoma increases with advancing age. Cancer 2003, 97, 1421–1427. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite Instability in Colorectal Cancer. Gastroenterology 2010, 138, 2073–2087. [Google Scholar] [CrossRef]
- Weisenberger, D.J.; Siegmund, K.D.; Campan, M.; Young, J.P.; Long, T.I.; Faasse, M.A.; Kang, G.H.; Widschwendter, M.; Weener, D.; Buchanan, D.; et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006, 38, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, M.A.; Hayashi, S.; O’Shea, A.-M.; Burgart, L.J.; Smyrk, T.C.; Shimizu, D.; Waring, P.M.; Ruszkiewicz, A.R.; Pollett, A.; Redston, M.; et al. Pathology Features in Bethesda Guidelines Predict Colorectal Cancer Microsatellite Instability: A Population-Based Study. Gastroenterology 2007, 133, 48–56. [Google Scholar] [CrossRef]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Győrffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A Comprehensive and Straightforward Web Application Enabling Rapid Biomarker Cutoff Optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef]
- Waniczek, D.; Lorenc, Z.; Śnietura, M.; Wesecki, M.; Kopec, A.; Muc-Wierzgoń, M. Tumor-Associated Macrophages and Regulatory T Cells Infiltration and the Clinical Outcome in Colorectal Cancer. Arch. Immunol. Ther. Exp. 2017, 65, 445–454. [Google Scholar] [CrossRef]
- Kim, Y.; Wen, X.; Bae, J.M.; Kim, J.H.; Cho, N.-Y.; Kang, G.H. The distribution of intratumoral macrophages correlates with molecular phenotypes and impacts prognosis in colorectal carcinoma. Histopathology 2018, 73, 663–671. [Google Scholar] [CrossRef]
- Wouters, M.C.A.; Nelson, B.H. Prognostic Significance of Tumor-Infiltrating B Cells and Plasma Cells in Human Cancer. Clin. Cancer Res. 2018, 24, 6125–6135. [Google Scholar] [CrossRef]
- Meshcheryakova, A.; Tamandl, D.; Bajna, E.; Stift, J.; Mittlboeck, M.; Svoboda, M.; Heiden, D.; Stremitzer, S.; Jensen-Jarolim, E.; Gruenberger, T.; et al. B Cells and Ectopic Follicular Structures: Novel Players in Anti-Tumor Programming with Prognostic Power for Patients with Metastatic Colorectal Cancer. PLoS ONE 2014, 9, e99008. [Google Scholar] [CrossRef] [PubMed]
- Berntsson, J.; Nodin, B.; Eberhard, J.; Micke, P.; Jirström, K. Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer. Int. J. Cancer 2016, 139, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.S.; Sahota, R.A.; Milne, K.; Kost, S.E.; Nesslinger, N.J.; Watson, P.H.; Nelson, B.H. CD20+ Tumor-Infiltrating Lymphocytes Have an Atypical CD27- Memory Phenotype and Together with CD8+ T Cells Promote Favorable Prognosis in Ovarian Cancer. Clin. Cancer Res. 2012, 18, 3281–3292. [Google Scholar] [CrossRef] [PubMed]
- Maletzki, C.; Jahnke, A.; Ostwald, C.; Klar, E.; Prall, F.; Linnebacher, M. Ex-vivo Clonally Expanded B Lymphocytes Infiltrating Colorectal Carcinoma Are of Mature Immunophenotype and Produce Functional IgG. PLoS ONE 2012, 7, e32639. [Google Scholar] [CrossRef]
- Edin, S.; Kaprio, T.; Hagström, J.; Larsson, P.; Mustonen, H.; Böckelman, C.; Strigård, K.; Gunnarsson, U.; Haglund, C.; Palmqvist, R. The Prognostic Importance of CD20+ B lymphocytes in Colorectal Cancer and the Relation to Other Immune Cell subsets. Sci. Rep. 2019, 9, 19997–19999. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Mlecnik, B.; Eynde, M.V.D.; Bindea, G.; Church, S.E.; Vasaturo, A.; Fredriksen, T.; Lafontaine, L.; Haicheur, N.; Marliot, F.; Debetancourt, D.; et al. Comprehensive Intrametastatic Immune Quantification and Major Impact of Immunoscore on Survival. J. Natl. Cancer Inst. 2017, 110, 97–108. [Google Scholar] [CrossRef]
- Zlobec, I.; Baker, K.; Minoo, P.; Hayashi, S.; Terracciano, L.; Lugli, A. Tumor border configuration added to TNM staging better stratifies stage II colorectal cancer patients into prognostic subgroups. Cancer 2009, 115, 4021–4029. [Google Scholar] [CrossRef]
- Morikawa, T.; Kuchiba, A.; Qian, Z.R.; Mino-Kenudson, M.; Hornick, J.L.; Yamauchi, M.; Imamura, Y.; Liao, X.; Nishihara, R.; Meyerhardt, J.A.; et al. Prognostic significance and molecular associations of tumor growth pattern in colorectal cancer. Ann. Surg. Oncol. 2011, 19, 1944–1953. [Google Scholar] [CrossRef]
- Pinheiro, R.S.; Herman, P.; Lupinacci, R.M.; Lai, Q.; Mello, E.S.; Coelho, F.F.; Perini, M.V.; Pugliese, V.; Andraus, W.; Cecconello, I.; et al. Tumor growth pattern as predictor of colorectal liver metastasis recurrence. Am. J. Surg. 2014, 207, 493–498. [Google Scholar] [CrossRef]
- Zlobec, I.; Lugli, A. Invasive front of colorectal cancer: Dynamic interface ofpro-/anti-tumor factors. World J. Gastroenterol. 2009, 15, 5898–5906. [Google Scholar] [CrossRef] [PubMed]
- Halvorsen, T.B.; Seim, E. Association between invasiveness, inflammatory reaction, desmoplasia and survival in colorectal cancer. J. Clin. Pathol. 1989, 42, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Weis, C.-A.; Kather, J.N.; Melchers, S.; Al-Ahmdi, H.; Pollheimer, M.J.; Langner, C.; Gaiser, T. Automatic evaluation of tumor budding in immunohistochemically stained colorectal carcinomas and correlation to clinical outcome. Diagn. Pathol. 2018, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Zlobec, I.; Terracciano, L.M.; Lugli, A. Local Recurrence in Mismatch Repair-Proficient Colon Cancer Predicted by an Infiltrative Tumor Border and Lack of CD8+ Tumor-Infiltrating Lymphocytes. Clin. Cancer Res. 2008, 14, 3792–3797. [Google Scholar] [CrossRef]
- Lang-Schwarz, C.; Melcher, B.; Haumaier, F.; Schneider-Fuchs, A.; Lang-Schwarz, K.; Krugmann, J.; Vieth, M.; Sterlacci, W. Budding, tumor-infiltrating lymphocytes, gland formation: Scoring leads to new prognostic groups in World Health Organization low-grade colorectal cancer with impact on survival. Hum. Pathol. 2019, 89, 81–89. [Google Scholar] [CrossRef]
- Susanti, S.; Fadhil, W.; Ebili, H.O.; Asiri, A.; Nestarenkaite, A.; Hadjimichael, E.; Ham-Karim, H.A.; Field, J.; Stafford, K.; Matharoo-Ball, B.; et al. N_LyST: A simple and rapid screening test for Lynch syndrome. J. Clin. Pathol. 2018, 71, 713–720. [Google Scholar] [CrossRef]
- Fadhil, W.; Ibrahem, S.; Seth, R.; Ilyas, M. Quick-multiplex-consensus (QMC)-PCR followed by high-resolution melting: A simple and robust method for mutation detection in formalin-fixed paraffin-embedded tissue. J. Clin. Pathol. 2010, 63, 134–140. [Google Scholar] [CrossRef]
- Plancoulaine, B.; Laurinaviciene, A.; Herlin, P.; Besusparis, J.; Meškauskas, R.; Baltrušaitytė, I.; Iqbal, Y.; Laurinavicius, A. A methodology for comprehensive breast cancer Ki67 labeling index with intra-tumor heterogeneity appraisal based on hexagonal tiling of digital image analysis data. Virchows Archiv 2015, 467, 711–722. [Google Scholar] [CrossRef]
- Laurinavicius, A.; Plancoulaine, B.; Herlin, P.; Laurinaviciene, A. Comprehensive Immunohistochemistry: Digital, Analytical and Integrated. Pathobiology 2016, 83, 156–163. [Google Scholar] [CrossRef]
- Rushing, C.; Bulusu, A.; Hurwitz, H.I.; Nixon, A.B.; Pang, H. A leave-one-out cross-validation SAS macro for the identification of markers associated with survival. Comput. Biol. Med. 2014, 57, 123–129. [Google Scholar] [CrossRef]


| Clinicopathological Parameters | MSS CRC, n (%) | MSI CRC, n (%) | p-Value * | |
|---|---|---|---|---|
| Total | 48 (100) | 39 (100) | - | |
| OS follow-up, months | Median | 52 | 46 | - |
| Range | 2–97 | 1–117 | ||
| Deceased | 5-year follow-up | 11 (12.6) | 17 (19.5) | - |
| 10-year follow-up | 11 (12.6) | 18 (20.7) | ||
| Age groups by median | ≤71 years | 32 (66.7) | 13 (33.3) | 0.0026 * |
| >71 years | 16 (33.3) | 26 (66.7) | ||
| Sex | Female | 23 (47.9) | 26 (66.7) | 0.0878 |
| Male | 25 (52.1) | 13 (33.3) | ||
| TNM stage | I | 0 | 1 (2.6) | 0.9999 |
| II | 31 (64.5) | 23 (58.9) | ||
| III | 16 (33.3) | 13 (33.3) | ||
| IV | 1 (2.1) | 2 (5.1) | ||
| Histological grade (G) | G2 | 44 (91.7) | 20 (51.3) | <0.0001 * |
| G3 | 4 (8.3) | 19 (48.72) | ||
| Tumor invasion (pT) | pT2 | 1 (2.1) | 1 (2.6) | 0.8115 |
| pT3 | 36 (75) | 27 (69.2) | ||
| pT4 | 11 (22.9) | 11 (28.2) | ||
| Lymph node metastasis (pN) | pN0 | 32 (66.7) | 25 (64.1) | 0.9027 |
| pN1 | 8 (16.7) | 8 (20.5) | ||
| pN2 | 8 (16.7) | 6 (15.4) | ||
| Distant metastasis (M) | M0 | 47 (97.9) | 37 (94.9) | 0.5850 |
| M1 | 1 (2.1) | 2 (5.1) | ||
| Lymphovascular invasion (LVI) | LVI0 | 28 (58.3) | 24 (61.5) | 0.8279 |
| LVI1 | 20 (41.7) | 15 (38.5) | ||
| Perineural invasion (Pne) | Pne0 | 42 (87.5) | 32 (82.1) | 0.5529 |
| Pne1 | 6 (12.5) | 7 (18.9) | ||
| Tumor location | Left | 28 (58.3) | 3 (7.7) | <0.0001 * |
| Transverse | 0 | 1 (2.56) | ||
| Right | 19 (39.6) | 33 (84.6) | ||
| Multiple sites | 1 (2.1) | 2 (5.1) | ||
| Tumor growth pattern | Pushing margin | 23 (47.9) | 26 (66.7) | 0.0878 |
| Infiltrative margin | 25 (52.1) | 13 (33.3) | ||
| Tumor budding | Low | 33 (68.8) | 25 (64.1) | 0.6557 |
| High | 15 (31.2) | 14 (35.9) | ||
| Peritumoral lymphocytes | Inconspicuous | 35 (72.9) | 20 (52.6) | 0.0707 |
| Conspicuous | 13 (27.1) | 18 (47.4) | ||
| BRAF mutation status | Wild-type | 44 (91.7) | 18 (46.2) | <0.0001 * |
| Mutant | 4 (8.3) | 21 (53.8) | ||
| KRAS mutation status | Wild-type | 25 (52.1) | 32 (82.2) | 0.0060 * |
| Mutant | 23 (47.9) | 7 (17.9) | ||
| PIK3CA mutation status | Wild-type | 40 (83.3) | 31 (79.5) | 0.7822 |
| Mutant | 8 (16.7) | 8 (20.5) |
| Immunogradient and Intratumoral Cell Density (Cells/mm2) Indicators | MSS CRC, n = 48 | MSI CRC, n = 39 | |||||
|---|---|---|---|---|---|---|---|
| Mean | Median | sd | Mean | Median | sd | p-Value * | |
| CD8_CM | −0.35 | −0.35 | 0.17 | −0.20 | −0.18 | 0.21 | 0.0006 * |
| CD8_d_S | 193.78 | 147.06 | 147.73 | 370.76 | 294.91 | 404.69 | 0.0024 * |
| CD8_d_TE | 141.82 | 90.03 | 128.49 | 339.94 | 208.15 | 400.52 | 0.0004 * |
| CD8_d_T | 76.47 | 49.24 | 92.49 | 262.40 | 140.22 | 342.64 | 0.0001 * |
| INT_CD8 | 65.37 | 37.59 | 81.99 | 238.90 | 133.46 | 311.26 | <0.0001 * |
| CD20_CM | −0.49 | −0.54 | 0.23 | −0.59 | −0.63 | 0.14 | 0.0141 * |
| CD20_d_S | 54.26 | 32.81 | 68.44 | 71.37 | 36.78 | 83.35 | 0.3650 |
| CD20_d_TE | 31.61 | 14.01 | 59.39 | 30.56 | 18.93 | 33.44 | 0.7857 |
| CD20_d_T | 12.20 | 4.66 | 30.68 | 5.40 | 3.87 | 6.12 | 0.0899 |
| INT_CD20 | 13.75 | 4.19 | 31.21 | 9.70 | 5.88 | 12.90 | 0.6003 |
| CD68_CM | −0.26 | −0.28 | 0.14 | −0.11 | −0.08 | 0.14 | <0.0001 * |
| CD68_d_S | 173.95 | 158.15 | 118.19 | 182.45 | 173.88 | 104.31 | 0.5616 |
| CD68_d_TE | 145.14 | 120.25 | 99.73 | 190.06 | 175.17 | 106.06 | 0.0281 * |
| CD68_d_T | 72.49 | 55.29 | 73.41 | 126.52 | 100.39 | 82.40 | <0.0001 * |
| INT_CD68 | 60.04 | 48.90 | 55.89 | 112.15 | 95.33 | 71.48 | <0.0001 * |
| Clinicopathological Parameters, Immunogradient and Intratumoral Cell Density Indicators | CRC, n = 87 | ||
|---|---|---|---|
| HR | 95% CI | p-Value * | |
| Age group (>median vs. ≤median) | 1.33 | 0.64–2.77 | 0.4480 |
| Sex (male vs. female) | 0.84 | 0.40–1.77 | 0.6481 |
| TNM stage (III-IV vs. I-II) | 1.06 | 0.49–2.30 | 0.8825 |
| pT status (pT4 vs. pT2-3) | 1.05 | 0.45–2.46 | 0.9151 |
| pN status (pN1-2 vs. pN0) | 0.98 | 0.45–2.18 | 0.9683 |
| M status (M1 vs. M0) | 3.41 | 0.80–14.60 | 0.0978 |
| G stage (G3 vs. G2) | 1.60 | 0.74–3.46 | 0.2312 |
| LVI status (LVI1 vs. LVI0) | 1.77 | 0.56–2.43 | 0.6737 |
| Pne status (Pne1 vs. Pne0) | 1.67 | 0.68–4.12 | 0.2648 |
| Tumor location (right/transverse/multiple vs. left) | 2.00 | 0.85–4.68 | 0.1128 |
| Tumor growth pattern (infiltrative vs. pushing margin) | 2.81 | 1.32–5.98 | 0.0075 * |
| Tumor budding (high vs. low) | 2.05 | 0.98–4.29 | 0.0556 |
| Peritumoral lymphocytes (inconspicuous vs. conspicuous) | 1.28 | 0.61–2.69 | 0.5234 |
| MSI status (MSI vs. MSS) | 2.07 | 0.97–4.43 | 0.0614 |
| BRAF status (mutant vs. wild-type) | 0.98 | 0.44–2.18 | 0.9501 |
| KRAS status (mutant vs. wild-type) | 0.78 | 0.36–1.72 | 0.5369 |
| PIK3CA status (mutant vs. wild-type) | 0.59 | 0.21–1.70 | 0.3264 |
| CD8_CM (high vs. low) | 0.31 | 0.15–0.66 | 0.0013 * |
| CD8_d_S (high vs. low) | 1.46 | 0.64–3.31 | 0.3600 |
| CD8_d_TE (high vs. low) | 0.64 | 0.31–1.35 | 0.2400 |
| CD8_d_T (high vs. low) | 0.53 | 0.25–1.10 | 0.0850 |
| INT_CD8 (high vs. low) | 2.13 | 0.93–4.88 | 0.0670 |
| CD20_CM (high vs. low) | 0.39 | 0.16–0.91 | 0.0230 * |
| CD20_d_S (high vs. low) | 0.30 | 0.12–0.75 | 0.0061 * |
| CD20_d_TE (high vs. low) | 0.33 | 0.10–1.08 | 0.0530 |
| CD20_d_T (high vs. low) | 0.43 | 0.20–0.90 | 0.0210 * |
| INT_CD20 (high vs. low) | 0.41 | 0.18–0.90 | 0.0230 * |
| CD68_CM (high vs. low) | 1.77 | 0.84–3.74 | 0.1300 |
| CD68_d_S (high vs. low) | 0.59 | 0.28–1.23 | 0.1500 |
| CD68_d_TE (high vs. low) | 0.65 | 0.30–1.43 | 0.2800 |
| CD68_d_T (high vs. low) | 1.82 | 0.77–4.26 | 0.1600 |
| INT_CD68 (high vs. low) | 1.73 | 0.79–3.81 | 0.1700 |
| Clinicopathological and Immunogradient Indicators | CRC, n = 87 | ||
|---|---|---|---|
| Model#1, LR: 23.03; p < 0.0001 | HR | 95% CI | p-Value |
| CD8_CM (high) | 0.31 | 1.42–0.67 | 0.0029 |
| CD20_CM (high) | 0.33 | 0.14–0.78 | 0.0113 |
| Tumor growth pattern (infiltrative) | 2.90 | 1.34–6.29 | 0.0071 |
| Model#2, LR: 15.50; p = 0.0004 | HR | 95% CI | p-Value |
| CD8_CM (high) | 0.30 | 0.14–0.64 | 0.0019 |
| CD20_CM (high) | 0.37 | 0.16–0.87 | 0.0228 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nestarenkaite, A.; Fadhil, W.; Rasmusson, A.; Susanti, S.; Hadjimichael, E.; Laurinaviciene, A.; Ilyas, M.; Laurinavicius, A. Immuno-Interface Score to Predict Outcome in Colorectal Cancer Independent of Microsatellite Instability Status. Cancers 2020, 12, 2902. https://doi.org/10.3390/cancers12102902
Nestarenkaite A, Fadhil W, Rasmusson A, Susanti S, Hadjimichael E, Laurinaviciene A, Ilyas M, Laurinavicius A. Immuno-Interface Score to Predict Outcome in Colorectal Cancer Independent of Microsatellite Instability Status. Cancers. 2020; 12(10):2902. https://doi.org/10.3390/cancers12102902
Chicago/Turabian StyleNestarenkaite, Ausrine, Wakkas Fadhil, Allan Rasmusson, Susanti Susanti, Efthymios Hadjimichael, Aida Laurinaviciene, Mohammad Ilyas, and Arvydas Laurinavicius. 2020. "Immuno-Interface Score to Predict Outcome in Colorectal Cancer Independent of Microsatellite Instability Status" Cancers 12, no. 10: 2902. https://doi.org/10.3390/cancers12102902
APA StyleNestarenkaite, A., Fadhil, W., Rasmusson, A., Susanti, S., Hadjimichael, E., Laurinaviciene, A., Ilyas, M., & Laurinavicius, A. (2020). Immuno-Interface Score to Predict Outcome in Colorectal Cancer Independent of Microsatellite Instability Status. Cancers, 12(10), 2902. https://doi.org/10.3390/cancers12102902





