Quantification of Receptor Occupancy by Ligand—An Understudied Class of Potential Biomarkers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Soluble Ligand–Receptor Complexes
3. Signaling Mechanisms of Soluble Ligand–Receptor Complexes
4. Agonistic Activity
4.1. Initiation
4.2. Sensitization
5. Antagonistic Activity
6. Quantification of Ligand–Receptor Complexes and Fractional Occupancy
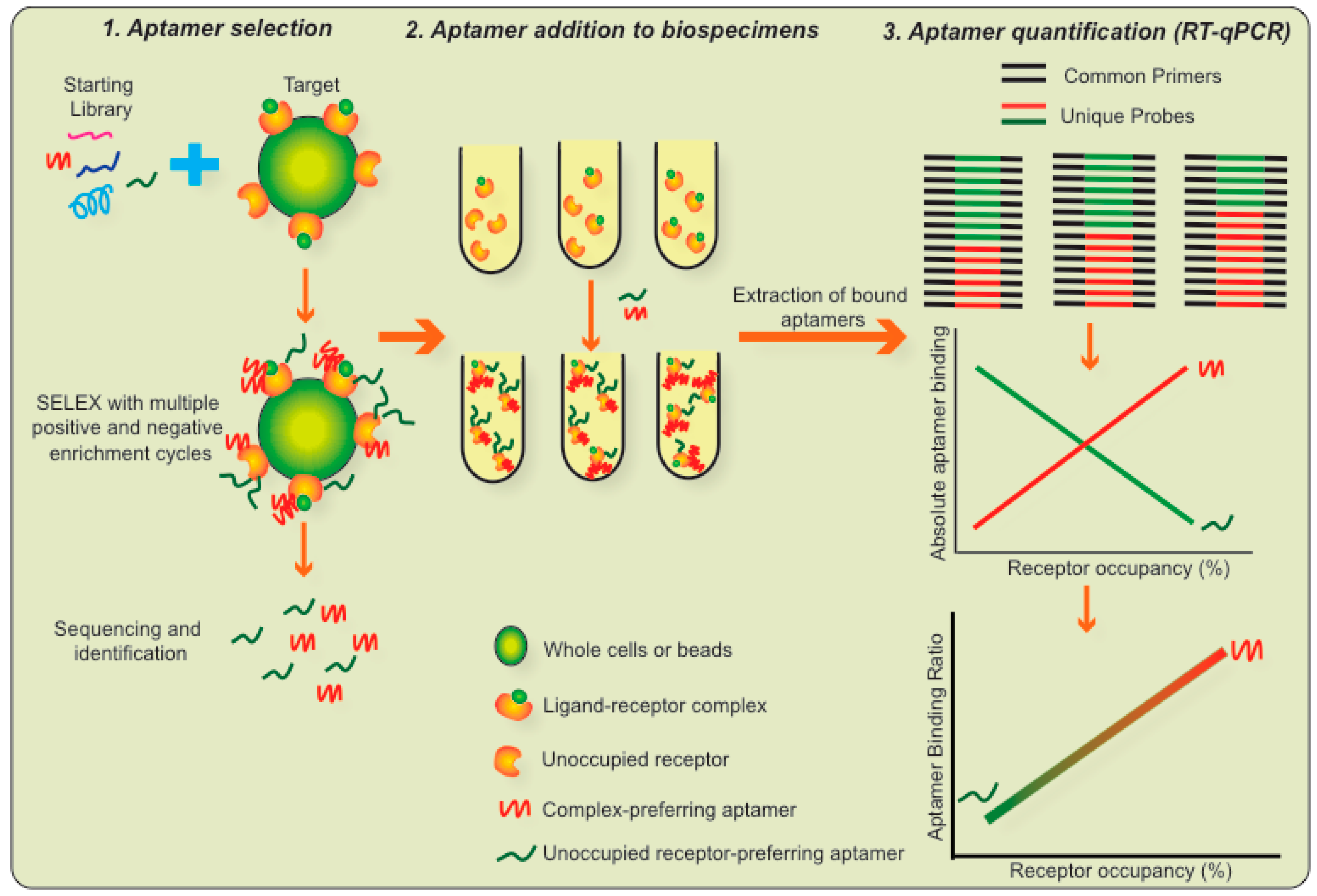
7. A Novel Assay Platform Based on LIRECAP
- Aptamer selection: A pair of RNA aptamers are identified, where one aptamer binds preferentially to the ligand–receptor complex and the second aptamer binds preferentially to the unoccupied receptor. Differences in their variable regions are responsible for this distinct binding. For the LIRECAP assay to be effective, such aptamers should be of the same length and have the same 5′ and 3′ primer-binding regions. The selected aptamers should not cross block each other, nor interfere with the binding of the ligand to the receptor.
- Aptamer addition to biospecimens: The pair of aptamers identified above is added in equimolar concentrations to a biospecimen and unbound aptamers are removed by washing.
- Aptamer quantification: Bound aptamers are extracted using standard molecular biology procedures and quantified using a standard TaqMan RT-qPCR reaction. The TaqMan amplification utilizes a set of PCR primers that bind to the 5′ and 3′ ends of both aptamers, thus amplifying both aptamers proportionately. TaqMan probes that bind to the individual variable central regions of the aptamers, each labeled with a different color, allow for quantification of each of the aptamers individually and for calculation of their ratio of binding to the samples.
- Use of a standard curve to calculate fractional occupancy: A standard curve is constructed based on the fractional occupancy of receptor by ligand using control samples with known fractional occupancy. Results of the test samples are compared to the standard curve. Using the binding ratio provides an internal control so the specificity of the two aptamers (for ligand–receptor complex and unoccupied receptor, respectively) does not have to be absolute, only relative. Thus, by using two aptamers and a standard curve, the relative binding of the two aptamers precisely reflects the fractional occupancy of the receptor by ligand.
8. Measuring Soluble IL2-IL2Ra Complexes in Cancer Biospecimens Using the LIRECAP Assay
9. Potential of the LIRECAP Assay with Other Ligand Receptor Pairs
10. Future Directions and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Waldmann, T.A. The biology of interleukin-2 and interleukin-15: Implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 2006, 6, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Zhao, N.; Zeng, Z.; Zhou, X.; Chang, C.; Zu, Y. Interleukin-2 Functions in Anaplastic Large Cell Lymphoma Cells through Augmentation of Extracellular Signal-Regulated Kinases 1/2 Activation. Int. J. Biomed. Sci. 2011, 7, 181–190. [Google Scholar] [PubMed]
- Malek, T.R.; Castro, I. Interleukin-2 Receptor Signaling: At the Interface between Tolerance and Immunity. Immunity 2010, 33, 153–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peggs, K.S.; Quezada, S.A.; Chambers, C.A.; Korman, A.J.; Allison, J.P. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti–CTLA-4 antibodies. J. Exp. Med. 2009, 206, 1717–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whiteside, T.L.; Schuler, P.; Schilling, B. Induced and natural regulatory T cells in human cancer. Expert Opin. Biol. Ther. 2012, 12, 1383–1397. [Google Scholar] [CrossRef] [Green Version]
- Binder, M.; O ’Byrne, M.M.; Maurer, M.J.; Ansell, S.; Feldman, A.L.; Cerhan, J.; Novak, A.; Porrata, L.F.; Markovic, S.; Link, B.K.; et al. Associations between elevated pre-treatment serum cytokines and peripheral blood cellular markers of immunosuppression in patients with lymphoma. Am. J. Hematol. 2017, 92, 752–758. [Google Scholar] [CrossRef] [Green Version]
- Mir, M.A.; Maurer, M.J.; Ziesmer, S.C.; Slager, S.L.; Habermann, T.; Macon, W.R.; Link, B.K.; Syrbu, S.; Witzig, T.; Friedberg, J.W.; et al. Elevated serum levels of IL-2R, IL-1RA, and CXCL9 are associated with a poor prognosis in follicular lymphoma. Blood 2015, 125, 992–998. [Google Scholar] [CrossRef] [Green Version]
- Van Der Merwe, P.A.; Davis, S.J. Molecular interactions mediating T cell antigen recognition. Annu. Rev. Immunol. 2003, 21, 659–684. [Google Scholar] [CrossRef]
- Syedbasha, M.; Linnik, J.; Santer, D.; O’Shea, D.; Barakat, K.; Joyce, M.; Khanna, N.; Tyrrell, D.L.; Houghton, M.; Egli, A. An ELISA Based Binding and Competition Method to Rapidly Determine Ligand-receptor Interactions. J. Vis. Exp. 2016, 2016. [Google Scholar] [CrossRef] [Green Version]
- Hulme, E.C.; Trevethick, M.A. Ligand binding assays at equilibrium: Validation and interpretation. Br. J. Pharmacol. 2010, 161, 1219–1237. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.; German, N.; Decker, A.M.; Li, J.X.; Wiley, J.L.; Thomas, B.F.; Kenakin, T.P.; Zhang, Y. Structure–activity relationships of substituted 1H-indole-2-carboxamides as CB1 receptor allosteric modulators. Bioorg. Med. Chem. 2015, 23, 2195–2203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu-Farha, M.; Elisma, F.; Figeys, D. Identification of Protein–Protein Interactions by Mass Spectrometry Coupled Techniques. Adv. Biochem. Eng. Biotechnol. 2008, 110, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Markham, K.; Bai, Y.; Eschmitt-Ulms, G. Co-immunoprecipitations revisited: An update on experimental concepts and their implementation for sensitive interactome investigations of endogenous proteins. Anal. Bioanal. Chem. 2007, 389, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Nohe, A.; Petersen, N.O. Analyzing for co-localization of proteins at a cell membrane. Curr. Pharm. Biotechnol. 2004, 5, 213–220. [Google Scholar] [CrossRef]
- Li, S. Mechanisms of Cellular Signal Transduction. Int. J. Biol. Sci. 2005, 1, 152. [Google Scholar] [CrossRef] [Green Version]
- Rose-John, S. IL-6 Trans-Signaling via the Soluble IL-6 Receptor: Importance for the Pro-Inflammatory Activities of IL-6. Int. J. Biol. Sci. 2012, 8, 1237–1247. [Google Scholar] [CrossRef]
- Goyonlo, V.M.; Elnour, H.; Nordlind, K. Interleukin-2 Expression in Lupoid and Usual Types of Old World Cutaneous Leishmaniasis. Iran. Red. Crescent Med. J. 2014, 16, 5410. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.; Ibrahim, S.; Patel, K.; Luthra, R.; Duvic, M.; Medeiros, L.J. Degree of CD25 Expression in T-Cell Lymphoma Is Dependent on Tissue Site: Implications for Targeted Therapy. Clin. Cancer Res. 2004, 10, 5587–5594. [Google Scholar] [CrossRef] [Green Version]
- Kasprzak, A.; Spachacz, R.; Wachowiak, J.; Stefańska, K.; Kaczmarek, E.; Zabel, M. Tissue Expression of Interleukin 2 (IL-2) and IL-2 Receptor (IL-2Rα/CD25) in non-Hodgkin B-cell Lymphomas in Children: Correlations with clinical data. J. Pediatr. Hematol. 2010, 32, 462–471. [Google Scholar] [CrossRef]
- Miyamoto, C.; Neto, R.B.M.; Di Cesare, S.; Junior, R.B.; Belfort, R. Use of CD25 as an immunohistochemical marker for acquired ocular toxoplasmosis. Arq. Bras. Oftalmol. 2010, 73, 443–446. [Google Scholar] [CrossRef] [Green Version]
- Wargo, J.; Reddy, S.M.; Reuben, A.; Seliger, B. Monitoring immune responses in the tumor microenvironment. Curr. Opin. Immunol. 2016, 41, 23–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.Z.; Grote, D.M.; Ziesmer, S.C.; Manske, M.K.; Witzig, T.E.; Novak, A.J.; Ansell, S.M. Soluble IL-2Rα facilitates IL-2–mediated immune responses and predicts reduced survival in follicular B-cell non-Hodgkin lymphoma. Blood 2011, 118, 2809–2820. [Google Scholar] [CrossRef] [PubMed]
- Nixon, A.B.; Schalper, K.A.; Jacobs, I.; Potluri, S.; Wang, I.-M.; Fleener, C. Peripheral immune-based biomarkers in cancer immunotherapy: Can we realize their predictive potential? J. Immunother. Cancer 2019, 7, 325. [Google Scholar] [CrossRef] [PubMed]
- Briso, E.M.; Dienz, O.; Rincon, M. Soluble IL-6R is produced by IL-6R ectodomain shedding in activated CD4 T cells. J. Immunol. 2008, 180, 7102–7106. [Google Scholar] [CrossRef] [Green Version]
- De Paiva, C.S.; Yoon, K.-C.; Pangelinan, S.B.; Pham, S.; Puthenparambil, L.M.; Chuang, E.Y.; Farley, W.J.; E Stern, M.; Li, D.Q.; Pflugfelder, S.C. Cleavage of functional IL-2 receptor alpha chain (CD25) from murine corneal and conjunctival epithelia by MMP-9. J. Inflamm. Lond. 2009, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Levine, S.J. Molecular Mechanisms of Soluble Cytokine Receptor Generation. J. Biol. Chem. 2008, 283, 14177–14181. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, N.; Oda, M.; Kuroda, Y.; Katayama, Y.; Okikawa, Y.; Masunari, T.; Fujiwara, M.; Nishisaka, T.; Sasaki, N.; Sadahira, Y.; et al. Clinical Significance of sIL-2R Levels in B-Cell Lymphomas. PLoS ONE 2013, 8, e78730. [Google Scholar] [CrossRef]
- Levine, S.J. Mechanisms of Soluble Cytokine Receptor Generation. J. Immunol. 2004, 173, 5343–5348. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.E.; Hanna, R.; Della, F.; Moore, H.; Chen, H.; Farese, A.M.; MacVittie, T.J.; Virca, G.D.; Sims, J.E. The Soluble Form of IL-1 Receptor Accessory Protein Enhances the Ability of Soluble Type II IL-1 Receptor to Inhibit IL-1 Action. Immunity 2003, 18, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Arondel, J.; Singer, M.; Matsukawa, A.; Zychlinsky, A.; Sansonetti, P.J. Increased Interleukin-1 (IL-1) and Imbalance between IL-1 and IL-1 Receptor Antagonist during Acute Inflammation in Experimental Shigellosis. Infect. Immun. 1999, 67, 6056–6066. [Google Scholar] [CrossRef] [Green Version]
- Brou, C.; Logeat, F.; Gupta, N.; Bessia, C.; LeBail, O.; Doedens, J.R.; Cumano, A.; Roux, P.; A Black, R.; Israël, A. A Novel Proteolytic Cleavage Involved in Notch Signaling: The role of the disintegrin-metalloprotease TACE. Mol. Cell 2000, 5, 207–216. [Google Scholar] [CrossRef]
- Jones, S.A.; Rose-John, S. The role of soluble receptors in cytokine biology: The agonistic properties of the sIL-6R/IL-6 complex. Biochim. Biophys. Acta 2002, 1592, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Taga, T.; Hibi, M.; Hirata, Y.; Yamasaki, K.; Yasukawa, K.; Matsuda, T.; Hirano, T.; Kishimoto, T. Interleukin-6 triggers the association of its receptor with a possible signal transducer, gp130. Cell 1989, 58, 573–581. [Google Scholar] [CrossRef]
- Peters, M.; Schirmacher, P.; Goldschmitt, J.; Odenthal, M.; Peschel, C.; Fattori, E.; Ciliberto, G.; Dienes, H.P.; Büschenfelde, K.-H.M.Z.; Rose-John, S. Extramedullary Expansion of Hematopoietic Progenitor Cells in Interleukin (IL)-6–sIL-6R Double Transgenic Mice. J. Exp. Med. 1997, 185, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Caruso, C.; Candore, G.; Cigna, D.; Colucci, A.T.; Modica, M.A. Biological significance of soluble IL-2 receptor. Mediat. Inflamm. 1993, 2, 3–21. [Google Scholar] [CrossRef]
- Veeramani, S.; Blackwell, S.E.; Thiel, W.H.; Yang, Z.-Z.; Ansell, S.M.; Giangrande, P.H.; Weiner, G.J. An RNA Aptamer-Based Biomarker Platform Demonstrates High Soluble CD25 Occupancy by IL2 in the Serum of Follicular Lymphoma Patients. Cancer Immunol. Res. 2019, 7, 1511–1522. [Google Scholar] [CrossRef]
- Rosenzweig, J.M.; Elei, J.; Burd, I. Interleukin-1 Receptor Blockade in Perinatal Brain Injury. Front. Pediatr. 2014, 2, 108. [Google Scholar] [CrossRef]
- Bresnihan, B.; Alvaro-Garcia, J.M.; Cobby, M.; Doherty, M.; Domljan, Z.; Emery, P.; Nuki, G.; Pavelka, K.; Rau, R.; Rozman, B.; et al. Treatment of rheumatoid arthritis with recombinant human interleukin-1 receptor antagonist. Arthritis Rheum. 1998, 41, 2196–2204. [Google Scholar] [CrossRef]
- Campion, G.V.; Lebsack, M.E.; Lookabaugh, J.; Gordon, G.; Catalano, M. Dose-range and dose-frequency study of recombinant human interleukin-1 receptor antagonist in patients with rheumatoid arthritis. The IL-1Ra Arthritis Study Group. Arthritis Rheum. 1996, 39, 1092–1101. [Google Scholar] [CrossRef]
- Nuki, G.; Bresnihan, B.; Bear, M.B.; McCabe, D. Long-term safety and maintenance of clinical improvement following treatment with anakinra (recombinant human interleukin-1 receptor antagonist) in patients with rheumatoid arthritis: Extension phase of a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2002, 46, 2838–2846. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Li, X.; Xi, D.; Mao, R.; Wu, X.; Cheng, S.; Sun, X.; Yi, C.; Ling, Z.; et al. Potential contribution of increased soluble IL-2R to lymphopenia in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 878–880. [Google Scholar] [CrossRef] [PubMed]
- Rizk, S.S.; Paduch, M.; Heithaus, J.H.; Duguid, E.M.; Sandstrom, A.; Kossiakoff, A.A. Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins. Nat. Struct. Mol. Biol. 2011, 18, 437–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saylor, K.; Gillam, F.; Lohneis, T.; Zhang, C. Designs of Antigen Structure and Composition for Improved Protein-Based Vaccine Efficacy. Front. Immunol. 2020, 11, 283. [Google Scholar] [CrossRef]
- Paduch, M.; Koide, A.; Uysal, S.; Rizk, S.S.; Koide, S.; Kossiakoff, A.A. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods 2013, 60, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Zu, Y. A Highlight of Recent Advances in Aptamer Technology and Its Application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Rossi, J. Erratum: Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 440. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Ao, X.; Yang, Y.; Chen, Z.; Xu, X. Soluble immune checkpoints in cancer: Production, function and biological significance. J. Immunother. Cancer 2018, 6, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heaney, M.L.; Golde, D.W. Soluble receptors in human disease. J. Leukoc. Biol. 1998, 64, 135–146. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Grote, D.M.; Ziesmer, S.C.; Xiu, B.; Yates, N.R.; Secreto, F.J.; Hodge, L.S.; Witzig, T.E.; Novak, A.J.; Ansell, S.M. Soluble and Membrane-Bound TGF-β-Mediated Regulation of Intratumoral T Cell Differentiation and Function in B-Cell Non-Hodgkin Lymphoma. PLoS ONE 2013, 8, e59456. [Google Scholar] [CrossRef] [Green Version]


| Primary Application | Technique(s) | Limitations | Citations |
|---|---|---|---|
| Quantify receptors or ligands | RIA and ELISAs using labeled antibodies | Usually used to measure individual receptors or ligands; ELISA-based assays that combine both anti-receptor and anti-ligand antibodies to detect complexes are designed to quantify complexes and not assess fractional occupancy of a receptor by a ligand | [9] |
| Determine the ligand–receptor interaction kinetics | Labeled ligands (e.g., radioactively labeled), Surface Plasmon Resonance (SPR) and Fluorescence Resonance Energy Transfer (FRET) | Limited use in clinical diagnosis due to challenges in handling large number of samples simultaneously | [10,11] |
| Identify interacting molecules | Co-immunoprecipitation; mass spectrometry | Data is qualitative to semi-quantitative; high-throughput assay is not feasible | [12,13] |
| Determine co-localization in the cellular environment | Confocal microscopy | Data is qualitative | [14] |
| Measure biological function | Assessment of signal transduction (e.g., arrays, such as ZeptoMARK), cell proliferation or differentiation induced as a result of ligand–receptor binding | Provides indirect analysis of ligand–receptor interaction | [2,15,16] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veeramani, S.; Weiner, G.J. Quantification of Receptor Occupancy by Ligand—An Understudied Class of Potential Biomarkers. Cancers 2020, 12, 2956. https://doi.org/10.3390/cancers12102956
Veeramani S, Weiner GJ. Quantification of Receptor Occupancy by Ligand—An Understudied Class of Potential Biomarkers. Cancers. 2020; 12(10):2956. https://doi.org/10.3390/cancers12102956
Chicago/Turabian StyleVeeramani, Suresh, and George J. Weiner. 2020. "Quantification of Receptor Occupancy by Ligand—An Understudied Class of Potential Biomarkers" Cancers 12, no. 10: 2956. https://doi.org/10.3390/cancers12102956
APA StyleVeeramani, S., & Weiner, G. J. (2020). Quantification of Receptor Occupancy by Ligand—An Understudied Class of Potential Biomarkers. Cancers, 12(10), 2956. https://doi.org/10.3390/cancers12102956





