Therapeutic Efficacy of GC1118, a Novel Anti-EGFR Antibody, against Glioblastoma with High EGFR Amplification in Patient-Derived Xenografts
Simple Summary
Abstract
1. Introduction
2. Results
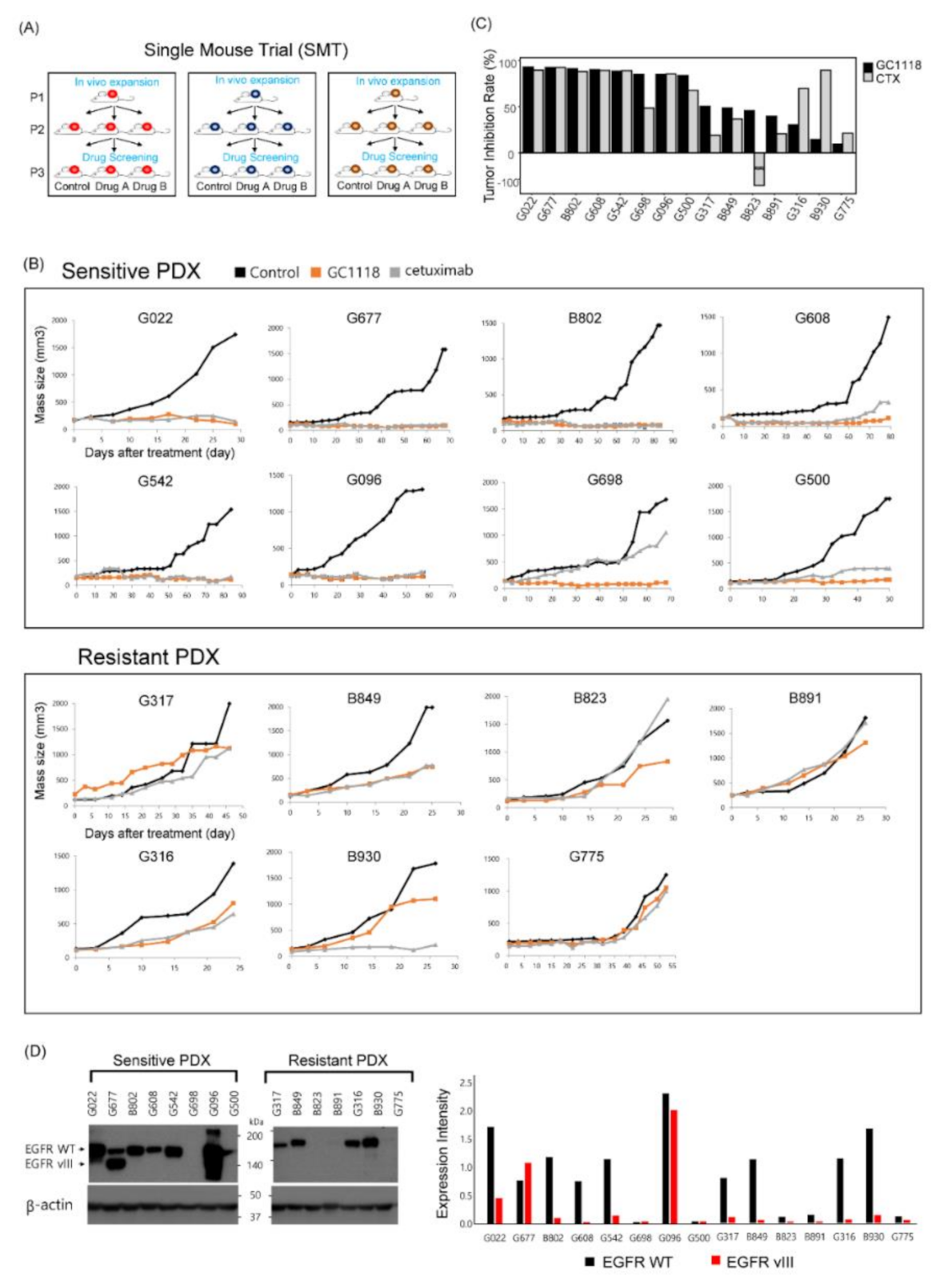
2.1. GC1118, a Novel Anti-EGFR Monoclonal Antibody, Exhibited Anti-Tumor Effects in Patient-Derived GBM Xenograft Models
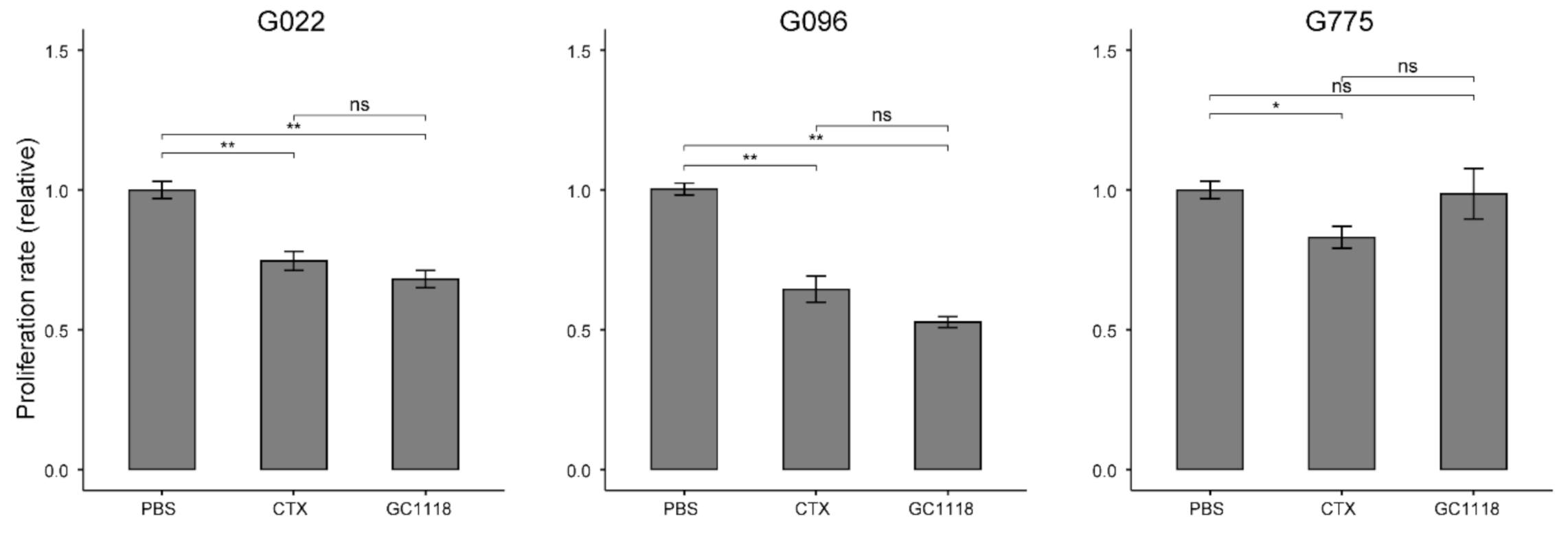
2.2. Anti-Proliferative Effect of GC1118 on GBM Patient-Derived Cells (PDCs)
2.3. EGFR Amplification as a Potent Biomarker to Predict the Therapeutic Efficacy of GC1118 in GBM Tumors
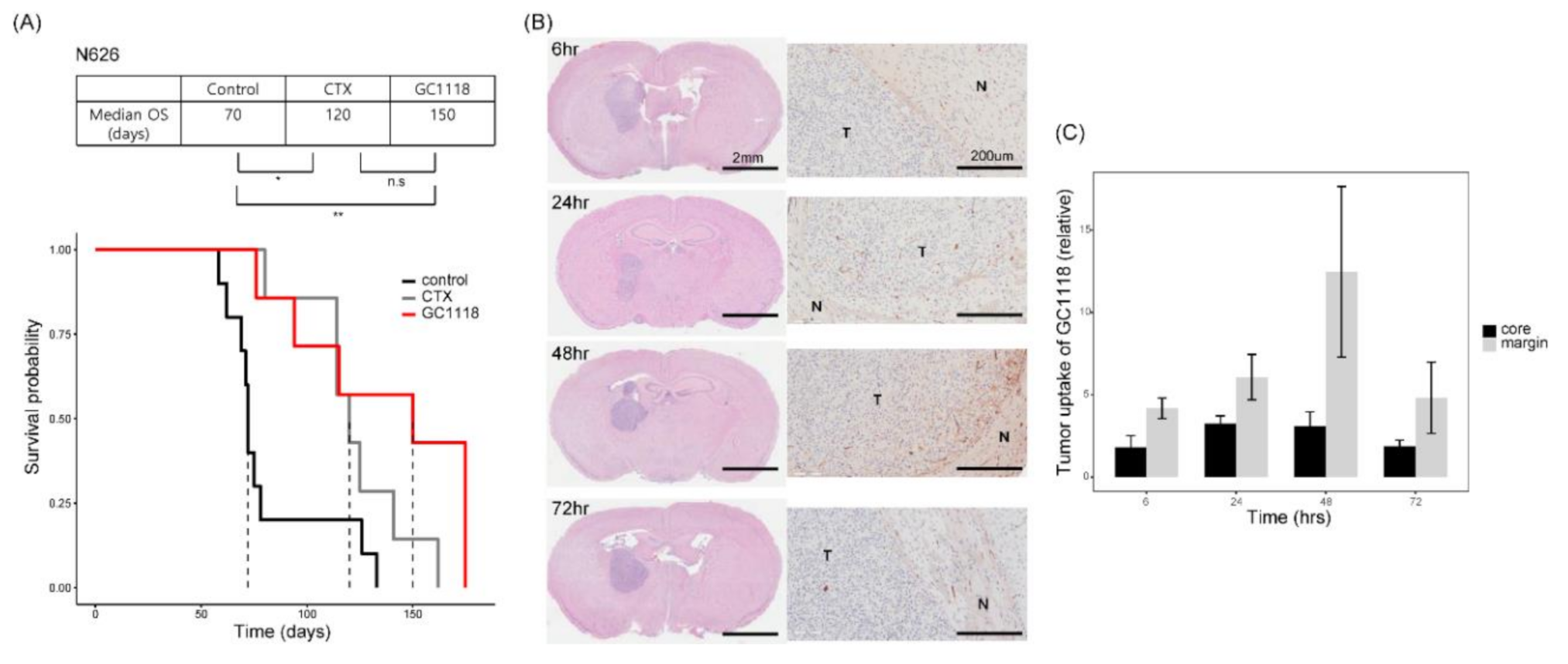
2.4. GC1118 Crossed the Blood–Brain Barrier, Thereby Conferring Survival Benefit in Orthotopic PDX Models
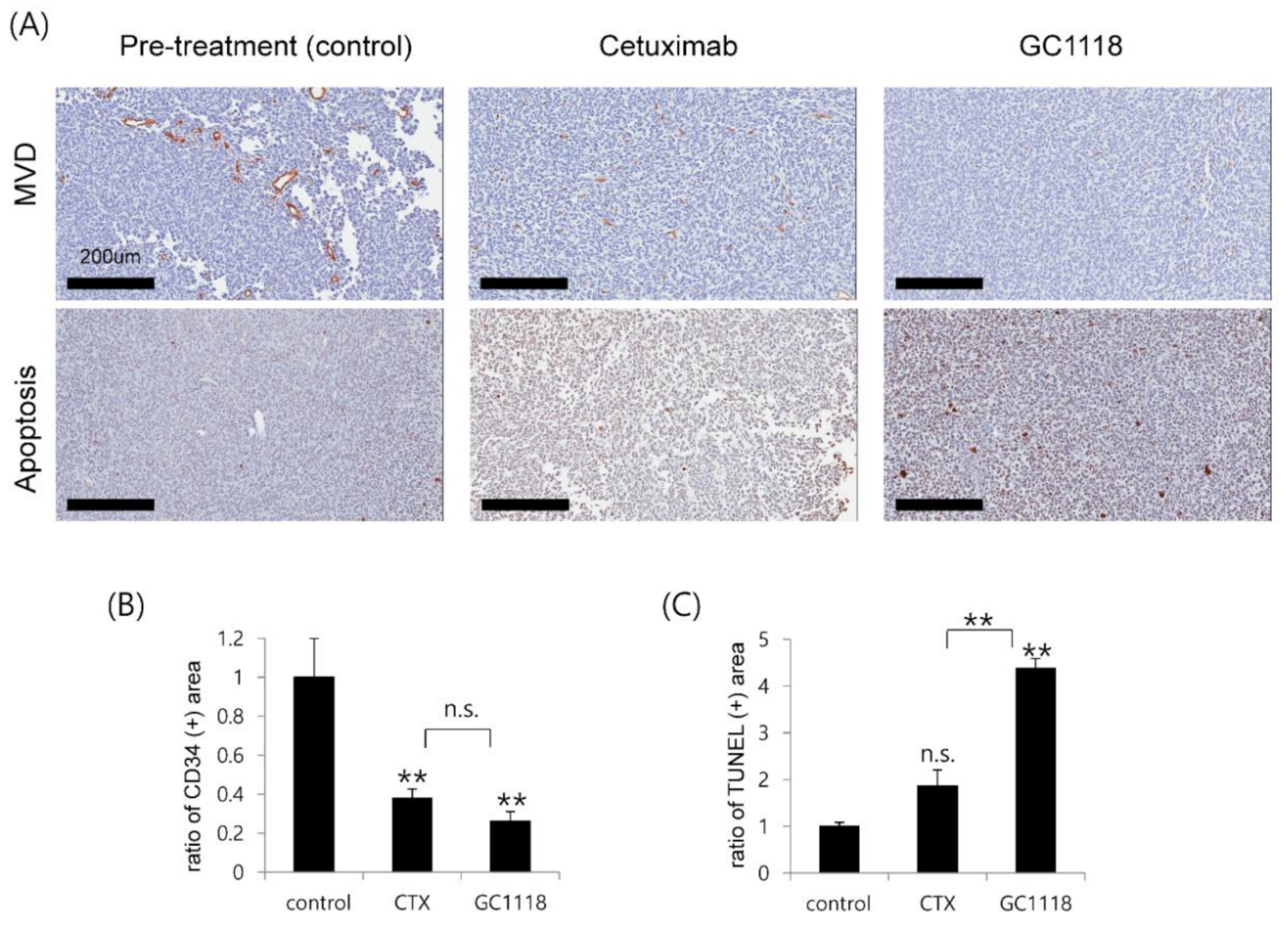
2.5. GC1118 Induced Tumor Cell Apoptosis and Exerted Anti-Angiogenic Effect in Orthotopic PDX Models
3. Discussion
4. Materials and Methods
4.1. Glioblastoma Specimens and Their Derivative Cells
4.2. Proliferation Assay
4.3. Single-Mouse Trial
4.4. Orthotopic Xenograft PDX Model
4.5. High-Throughput Drug Screening Experiment
4.6. Western Blot Analysis
4.7. In Vivo mAb Distribution Analysis
4.8. Immunohistochemistry (IHC)
4.9. Next Generation Sequencing (NGS)—Somatic Mutation
4.10. Target Sequencing
4.11. Copy Number Variation
4.12. Whole Transcriptome Sequencing and Analysis
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Vallentgoed, W.R.; French, P.J. Finding the Right Way to Target EGFR in Glioblastomas; Lessons from Lung Adenocarcinomas. Cancers 2018, 10, 489. [Google Scholar] [CrossRef] [PubMed]
- Thorne, A.H.; Zanca, C.; Furnari, F. Epidermal growth factor receptor targeting and challenges in glioblastoma. Neuro Oncol. 2016, 18, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Roth, P.; Weller, M. Challenges to targeting epidermal growth factor receptor in glioblastoma: Escape mechanisms and combinatorial treatment strategies. Neuro Oncol. 2014, 16, viii14–viii19. [Google Scholar] [CrossRef]
- Humphrey, P.A.; Wong, A.J.; Vogelstein, B.; Friedman, H.S.; Werner, M.H.; Bigner, D.D.; Bigner, S.H. Amplification and expression of the epidermal growth factor receptor gene in human glioma xenografts. Cancer Res. 1988, 48, 2231–2238. [Google Scholar]
- Nathanson, D.A.; Gini, B.; Mottahedeh, J.; Visnyei, K.; Koga, T.; Gomez, G.; Eskin, A.; Hwang, K.; Wang, J.; Masui, K.; et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 2014, 343, 72–76. [Google Scholar] [CrossRef]
- Westphal, M.; Maire, C.L.; Lamszus, K. EGFR as a Target for Glioblastoma Treatment: An Unfulfilled Promise. CNS Drugs 2017, 31, 723–735. [Google Scholar] [CrossRef]
- Combs, S.E.; Heeger, S.; Haselmann, R.; Edler, L.; Debus, J.; Schulz-Ertner, D. Treatment of primary glioblastoma multiforme with cetuximab, radiotherapy and temozolomide (GERT)--phase I/II trial: Study protocol. BMC Cancer 2006, 6, 133. [Google Scholar] [CrossRef]
- Neyns, B.; Sadones, J.; Joosens, E.; Bouttens, F.; Verbeke, L.; Baurain, J.F.; D’Hondt, L.; Strauven, T.; Chaskis, C.; In’t Veld, P.; et al. Stratified phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann. Oncol. 2009, 20, 1596–1603. [Google Scholar] [CrossRef]
- Hasselbalch, B.; Lassen, U.; Hansen, S.; Holmberg, M.; Sorensen, M.; Kosteljanetz, M.; Broholm, H.; Stockhausen, M.T.; Poulsen, H.S. Cetuximab, bevacizumab, and irinotecan for patients with primary glioblastoma and progression after radiation therapy and temozolomide: A phase II trial. Neuro Oncol. 2010, 12, 508–516. [Google Scholar] [CrossRef]
- Westphal, M.; Heese, O.; Steinbach, J.P.; Schnell, O.; Schackert, G.; Mehdorn, M.; Schulz, D.; Simon, M.; Schlegel, U.; Senft, C.; et al. A randomised, open label phase III trial with nimotuzumab, an anti-epidermal growth factor receptor monoclonal antibody in the treatment of newly diagnosed adult glioblastoma. Eur. J. Cancer 2015, 51, 522–532. [Google Scholar] [CrossRef]
- Lee, H.W.; Son, E.; Lee, K.; Lee, Y.; Kim, Y.; Lee, J.C.; Lim, Y.; Hur, M.; Kim, D.; Nam, D.H. Promising Therapeutic Efficacy of GC1118, an Anti-EGFR Antibody, against KRAS Mutation-Driven Colorectal Cancer Patient-Derived Xenografts. Int. J. Mol. Sci. 2019, 20, 5894. [Google Scholar] [CrossRef]
- Park, J.E.; Jin, M.H.; Hur, M.; Nam, A.R.; Bang, J.H.; Won, J.; Oh, D.Y.; Bang, Y.J. GC1118, a novel anti-EGFR antibody, has potent KRAS mutation-independent antitumor activity compared with cetuximab in gastric cancer. Gastric Cancer 2019, 22, 932–940. [Google Scholar] [CrossRef]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Bady, P.; Kamoshima, Y.; Kouwenhoven, M.C.; Delorenzi, M.; Lambiv, W.L.; Hamou, M.F.; Matter, M.S.; Koch, A.; et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial. Mol. Cancer Ther. 2011, 10, 1102–1112. [Google Scholar] [CrossRef]
- Chacko, A.M.; Li, C.; Pryma, D.A.; Brem, S.; Coukos, G.; Muzykantov, V. Targeted delivery of antibody-based therapeutic and imaging agents to CNS tumors: Crossing the blood-brain barrier divide. Expert Opin. Drug Deliv. 2013, 10, 907–926. [Google Scholar] [CrossRef] [PubMed]
- Lassman, A.B.; Rossi, M.R.; Raizer, J.J.; Abrey, L.E.; Lieberman, F.S.; Grefe, C.N.; Lamborn, K.; Pao, W.; Shih, A.H.; Kuhn, J.G.; et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: Tissue analysis from North American Brain Tumor Consortium Trials 01-03 and 00-01. Clin. Cancer Res. 2005, 11, 7841–7850. [Google Scholar] [CrossRef]
- Tabernero, J. The role of VEGF and EGFR inhibition: Implications for combining anti-VEGF and anti-EGFR agents. Mol. Cancer Res. 2007, 5, 203–220. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Yoo, J.; Kim, M.S.; Hur, M.; Lee, E.H.; Hur, H.S.; Lee, J.C.; Lee, S.N.; Park, T.W.; Lee, K.; et al. GC1118, an Anti-EGFR Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity against High-Affinity EGFR Ligands. Mol. Cancer Ther. 2016, 15, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Fukai, J.; Nishio, K.; Itakura, T.; Koizumi, F. Antitumor activity of cetuximab against malignant glioma cells overexpressing EGFR deletion mutant variant III. Cancer Sci. 2008, 99, 2062–2069. [Google Scholar] [CrossRef]
- Eller, J.L.; Longo, S.L.; Kyle, M.M.; Bassano, D.; Hicklin, D.J.; Canute, G.W. Anti-epidermal growth factor receptor monoclonal antibody cetuximab augments radiation effects in glioblastoma multiforme in vitro and in vivo. Neurosurgery 2005, 56, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Combs, S.E.; Schulz-Ertner, D.; Roth, W.; Herold-Mende, C.; Debus, J.; Weber, K.J. In vitro responsiveness of glioma cell lines to multimodality treatment with radiotherapy, temozolomide, and epidermal growth factor receptor inhibition with cetuximab. Int. J. Radiat Oncol. Biol. Phys. 2007, 68, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bent, M.; Eoli, M.; Sepulveda, J.M.; Smits, M.; Walenkamp, A.; Frenel, J.S.; Franceschi, E.; Clement, P.M.; Chinot, O.; De Vos, F.; et al. INTELLANCE 2/EORTC 1410 randomized phase II study of Depatux-M alone and with temozolomide vs temozolomide or lomustine in recurrent EGFR amplified glioblastoma. Neuro Oncol. 2020, 22, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Soderstrom, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- Wang, W.; Erbe, A.K.; Hank, J.A.; Morris, Z.S.; Sondel, P.M. NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity in Cancer Immunotherapy. Front. Immunol. 2015, 6, 368. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Banerji, S.; Cibulskis, K.; Rangel-Escareno, C.; Brown, K.K.; Carter, S.L.; Frederick, A.M.; Lawrence, M.S.; Sivachenko, A.Y.; Sougnez, C.; Zou, L.; et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012, 486, 405–409. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Shin, H.; Sa, J.K.; Bae, J.S.; Koo, H.; Jin, S.; Cho, H.J.; Choi, S.W.; Kyoung, J.M.; Kim, J.Y.; Seo, Y.J.; et al. Clinical Targeted Next-Generation sequencing Panels for Detection of Somatic Variants in Gliomas. Cancer Res. Treat. 2020, 52, 41–50. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 30 October 2020).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Koo, H.; Kim, Y.; Kim, D.; Son, E.; Yang, H.; Lim, Y.; Hur, M.; Lee, H.W.; Choi, S.W.; et al. Therapeutic Efficacy of GC1118, a Novel Anti-EGFR Antibody, against Glioblastoma with High EGFR Amplification in Patient-Derived Xenografts. Cancers 2020, 12, 3210. https://doi.org/10.3390/cancers12113210
Lee K, Koo H, Kim Y, Kim D, Son E, Yang H, Lim Y, Hur M, Lee HW, Choi SW, et al. Therapeutic Efficacy of GC1118, a Novel Anti-EGFR Antibody, against Glioblastoma with High EGFR Amplification in Patient-Derived Xenografts. Cancers. 2020; 12(11):3210. https://doi.org/10.3390/cancers12113210
Chicago/Turabian StyleLee, Kyoungmin, Harim Koo, Yejin Kim, Donggeon Kim, Eunju Son, Heekyoung Yang, Yangmi Lim, Minkyu Hur, Hye Won Lee, Seung Won Choi, and et al. 2020. "Therapeutic Efficacy of GC1118, a Novel Anti-EGFR Antibody, against Glioblastoma with High EGFR Amplification in Patient-Derived Xenografts" Cancers 12, no. 11: 3210. https://doi.org/10.3390/cancers12113210
APA StyleLee, K., Koo, H., Kim, Y., Kim, D., Son, E., Yang, H., Lim, Y., Hur, M., Lee, H. W., Choi, S. W., & Nam, D.-H. (2020). Therapeutic Efficacy of GC1118, a Novel Anti-EGFR Antibody, against Glioblastoma with High EGFR Amplification in Patient-Derived Xenografts. Cancers, 12(11), 3210. https://doi.org/10.3390/cancers12113210





