Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability
Abstract
:Simple Summary
Abstract
1. Introduction
2. Why Microsatellites Are Hot-Spots for Genomic Instability?
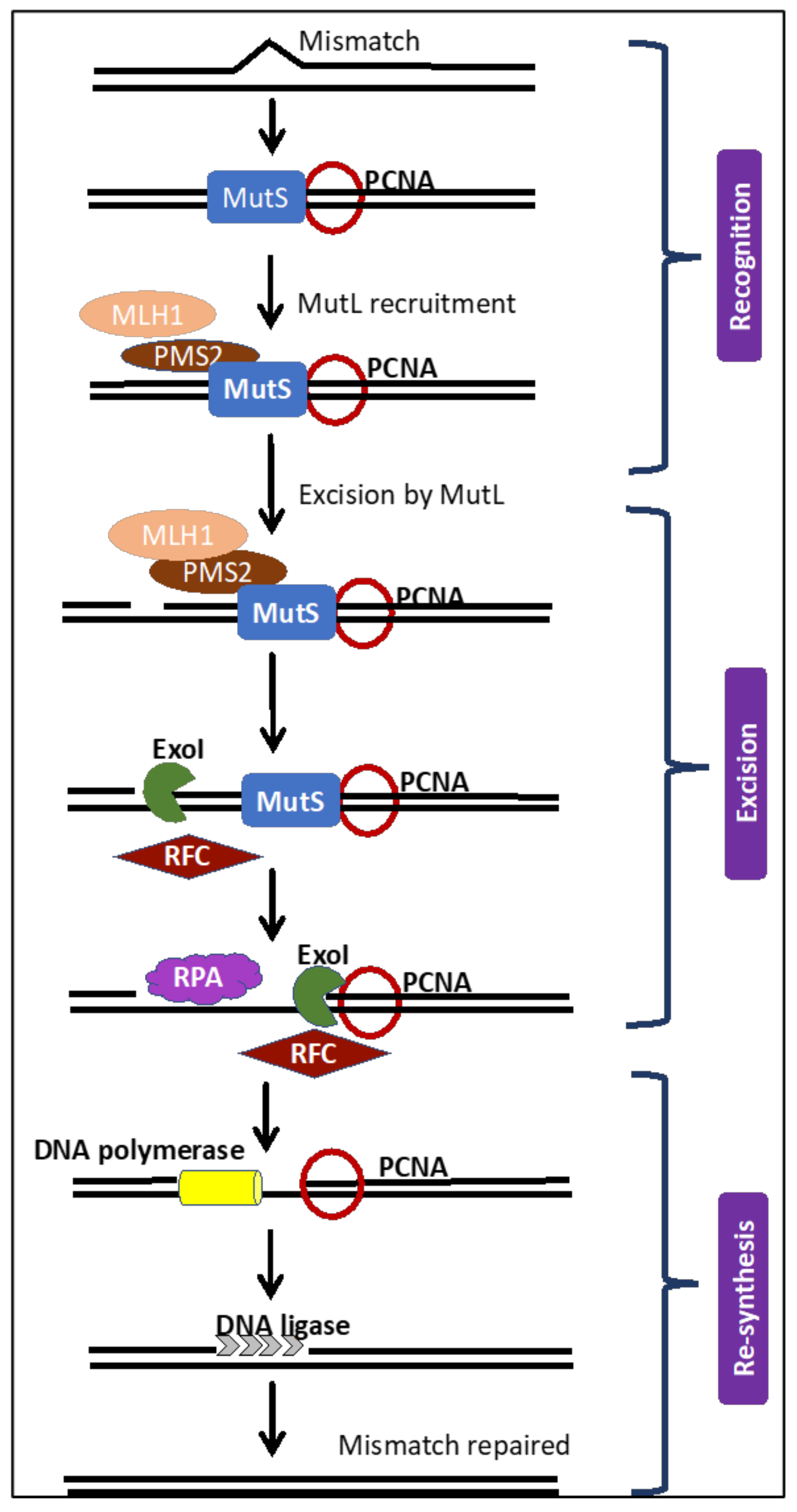
3. The MMR Repair Pathway and Its Kryptonite
4. Microsatellites Used as Markers for MSI/dMMR Cancer Diagnosis
| Repeat Type | Marker | Repeat Sequence | Gene | Studied for Detection of MSI | Reference |
|---|---|---|---|---|---|
| M O N O N U C L E O T I D E | BAT26 | (T)25 | MSH2 gene (MMR) | Ovarian cancer, Cervical cancer, Endometrial cancer | [63,64,65,66,67,68,69,70,71,72] |
| BAT25 | (A)26 | c-kit gene (oncogene) | Ovarian cancer, Endometrial cancer | [63,65,66,67,68,69,70,71] | |
| BAT34C4 | (T)3C(T)6C(T)17C(T)5C(T)3 | p53 | Endometrial cancer | [68] | |
| BAT40 | (A)40 | 3-beta-hydroxysteroid dehydrogenase gene | Endometrial cancer | [63,68] | |
| NR-21 | (A)21 | SLC7A8 | Ovarian cancer, Endometrial cancer | [63] | |
| NR-22 | (A)22 | Trans-membrane precursor B5 | Ovarian cancer, Endometrial cancer | [63,70] | |
| NR-24 | (T)24 | Zinc finger 2 | Endometrial cancer | [70] | |
| NR-27 | (T)27 | Inhibitor of apoptosis-Protein 1 | Ovarian cancer, Endometrial cancer | [63,70] | |
| TGFBR-II | (A)10 | TGF-beta receptor | Ovarian cancer Endometrial cancer | [73,74] | |
| D I N U C L E O T I D E | D2S123 | (CA)13(TA)(CA)15 | hMSH2 | Cervical cancer, Endometrial cancer, Ovarian cancer | [65,69,70,71] |
| D3S1260 | (AGAT)11 | XYLB gene | Cervical cancer, Endometrial cancer | [63] | |
| D3S1611 | (CA)11 | hMLH1 gene | Breast cancer | [71,75] | |
| D5S346 | (CA)26 | APC | Cervical cancer, Endometrial cancer, Ovarian cancer | [65,68,69,70,71] | |
| D10S197 | (CA)7…(CA)17 | GAD2 gene | Endometrial cancer, Ovarian cancer | [68,71] | |
| D11S1318 | (CA)15…(CA)5 | eIF3f gene | Ovarian cancer | [76] | |
| D11S904 | (CA)14(TA)5 | - | Ovarian cancer | [71,77] | |
| D17S807 | (CA)n | P53 gene | Breast cancer | [75] | |
| D17S796 | GT)n | P53 gene | Breast cancer | [75] | |
| D17S250 (Mfd15) | (TA)7….(CA)24 | BRCA1 gene | Cervical cancer, Endometrial cancer | [65,68,69,70,71] | |
| D18S55 | (GC)5GA(CA)17 | - | Endometrial cancer | [68] | |
| NME1 | Nucleoside diphosphate kinase1 | Ovarian cancer | [71] | ||
| T R I N U C L E O T I D E | AR | CAG | Androgen receptor | Breast cancer | [75] |
| DM1 | CAG | Myotonic dystrophy protein kinase | Ovarian cancer, Breast cancer | [78] | |
| T E T R A N U C L E O T I D E | D2S443 | (AAAG)n | - | Ovarian cancer | [72] |
| D8S321 | (AAAG)12 | - | Ovarian cancer | [72] | |
| D20S82 | (AAAG)10 | RM267 | Ovarian cancer | [72] | |
| DXS981 | TATC | Breast cancer, Ovarian cancer | [79] | ||
| DXS6800 | (TAGA)x-CA-(GATA)1-GAT-(GATA)y-GG-(TAGA)3-TC-(GATA)3 | X-chromosomal short tandem repeats | Ovarian cancer | [71] | |
| MYCL1 | (AAAG)21 | MYCL1 | Endometrial cancer | [68] | |
| UT5037 | (AAAG)19 | - | Ovarian cancer | [72] | |
| UT5320 | (AAAG)21 (AAAG)10 | 241A/241B | Ovarian cancer | [72] | |
| vWF-a | TCTA | Von Willebrand factor-alpha | Ovarian cancer, Breast cancer | [78] | |
| PENTA-NUCLEOTIDE | FMR2 | (CCAAA)6(CCAGA)2 | X chromosome | ||
| TP53Alu | (AAAAT)8 | p53 | Ovarian cancer | [80] |
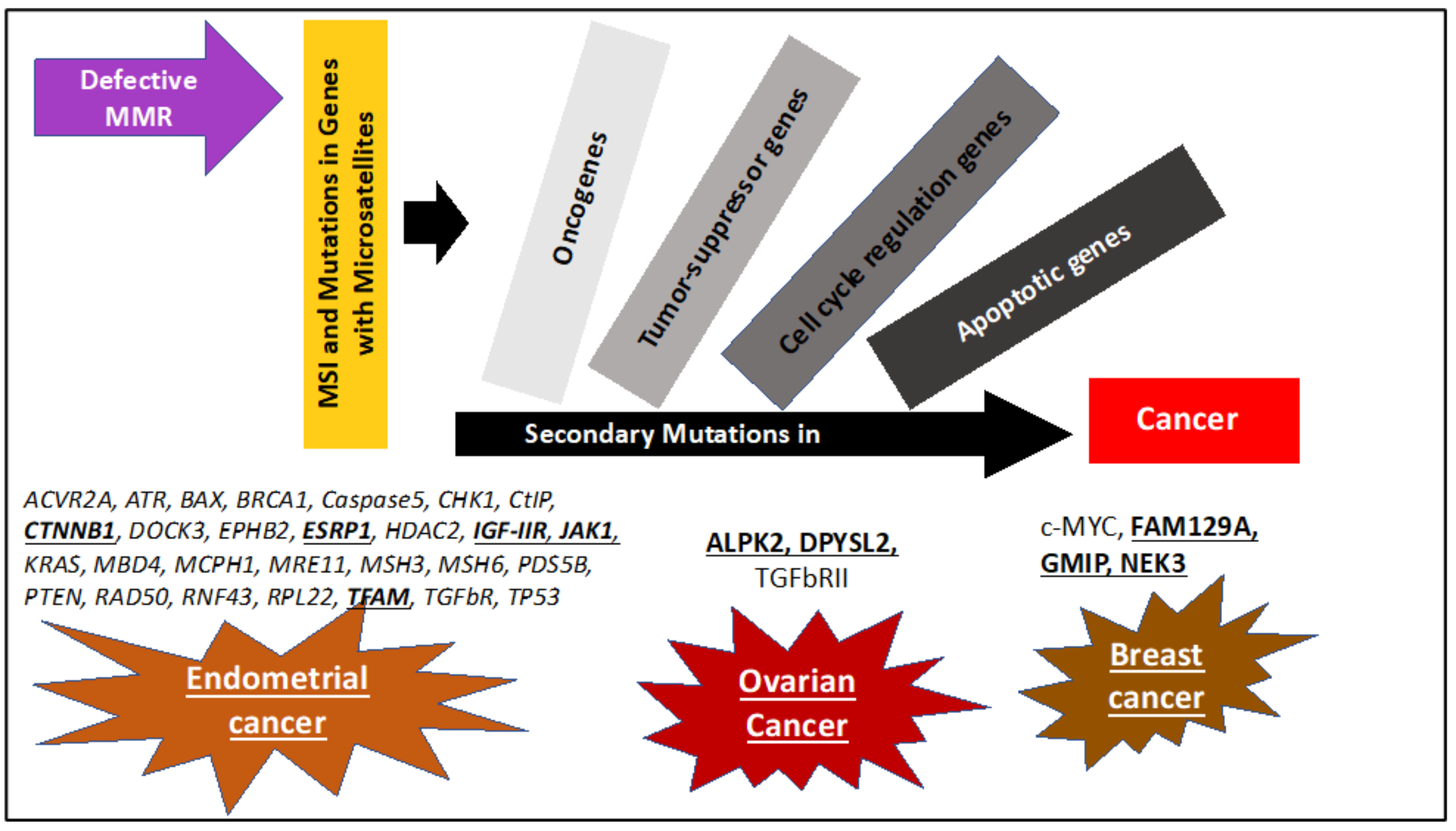
5. Sequential Steps Leading to MSI/dMMR Cancer Development
| Functional Group | Gene | Role | Repeat Sequence if Present | % Frequency of Mutation in MSI-H | |||
|---|---|---|---|---|---|---|---|
| Endometrial Cancers | Ovarian Cancers | Breast Cancer | Non-Gynecological Cancers | ||||
| Cell regulation/signaling | ACVR2A | Member of TGF-beta signaling pathway. Role in cell growth and tumor metastasis | 2(A)8 | 19% [97] | CRC 80% Stomach 75% [97] | ||
| CHK1 | DNA damage response | (A)9 | 29% [98] | ||||
| c-MYC | Cell division | (GT)n–(GC)n | 20% [44] | ||||
| DPYSL2 | Microtubule function. May play role in endocytosis | (CT)11 | 59% [54] | ||||
| ESRP1 | Protein-splicing regulator. May contribute to mesenchymal transition | (GGT)n | 20% [97] | ||||
| GMIP | Cell growth and survival. Ras pathway | 10% [54] | |||||
| HDAC2 | Histone deacetylase | (A)n | 11% [73] | ||||
| IGFRIIR | (G)8 | 14% [98] | |||||
| MBD4 | Methyl CpG | (A)10) | 31.8% [73] | ||||
| NEK3 | Mitotic regulator | (A)8 | 6% [54] | ||||
| PDS5B | DNA damage repair | (A)9 | 15% [69] | CRC 28% [69] | |||
| PTEN | DNA damage response | (A)6 | 15.8% [73], 88% [54] | CRC 28% [99] | |||
| RNF43 | Involved in controlling cell proliferation Negative regulator of WNT pathway. | (G)7 | 23% [97] | CRC 40% Stomach 35% [97] | |||
| RPL22 | Protein synthesis | (A)8 | 37% [54] 50% [93] 52% [100] | CRC 80% [100] | |||
| TGFBR | TGF-beta receptor | (A)10 | 36.3% [73] 5% [54] | CRC 90% [73] | |||
| Oncogenes | ARID1A | Tumor suppressor gene. Regulates transcription of certain genes by altering the chromatin structure around those genes | (AT)n | 37% [101] | |||
| JAK1 | Oncogene. Modulates IFN-gamma signaling pathway and enables tumor immune evasion Promotes tumor survival | (T)7, (T)8, (G)7 | 21% [97] 35% [102] | ||||
| KRAS | Oncogene | 35% [54] | CRC 31% [99] | ||||
| TP53 | Tumor suppressor | TP53 ALU (A)n (AAAAT)8 | 40% [93] | 21% | CRC 31% [99] | ||
| WNT pathway | CTNNB1 | Member of WNT pathway | (A)n | 30% [97] | CRC 6% [69] | ||
| DOCK3 | Protein dedicator of cytokinesis 3 Inhibits WNT pathway | 23% [97] | Stomach 40% [97] | ||||
| EPHB2 | Member of WNT pathway | (A)9 | 9% [73] 14% [103] | Gastric 39% [103] | |||
| Apoptosis pathway | ALPK2 | Apoptosis and DNA Repair | (T)3 | 17% [54] | |||
| BAX | Pro-apoptotic factor | (G)8 | 22.7% [73], 16% [96] 43% [98] | CRC 45% [73] | |||
| Caspase 5 | Pro-apoptotic factor | (A)10 | 4.5% [73], 5% [96] EC- 28% [104] | Stomach 44% CRC 62% [104] | |||
| FAM129A | Apoptosis regulator, Anti-apoptotic | - | 12% [54] | ||||
| TFAM | Apoptosis regulator, DNA damage repair | (A)10 | 20% [69] | ||||
| MMR genes | hMSH6 | Repair genes | (C)8 | 30% [73] | |||
| hMSH3 | Repair genes | (A)8 | 9% [73] | ||||
| DNA repair | ATR | DNA damage checkpoint | (A)10 | 15% [49] | |||
| BRCA1 | Tumor suppressor gene, DNA repair | (TA)7 (CA)24 Flanking sequences | 15% [49] | ||||
| CtIP | Promotes the resection of DNA double-strand breaks | (T)9 | 12% [49] | ||||
| MCPH1 | DNA damage response protein | (A)9 | 12% [49] | CRC 9.7% [105] | |||
| MRE11 | Double Strand Break Repair Nuclease | (T)11 | 15% [49] 50% [106] | CRC 83% [106] | |||
| RAD50 | Double Strand Break Repair Protein | (A)9 | 17% [49] | CRC 46% [107] | |||
| Other | PIK13CA | Role in protein kinase B signaling | - | 54% [101] | |||
| PIK3RI | Role in the metabolic actions of insulin | - | 40% [101] | ||||
6. Sporadic Malignancies Caused by MSI/dMMR
7. Inherited Malignancies Caused by MSI/dMMR-Lynch Syndrome
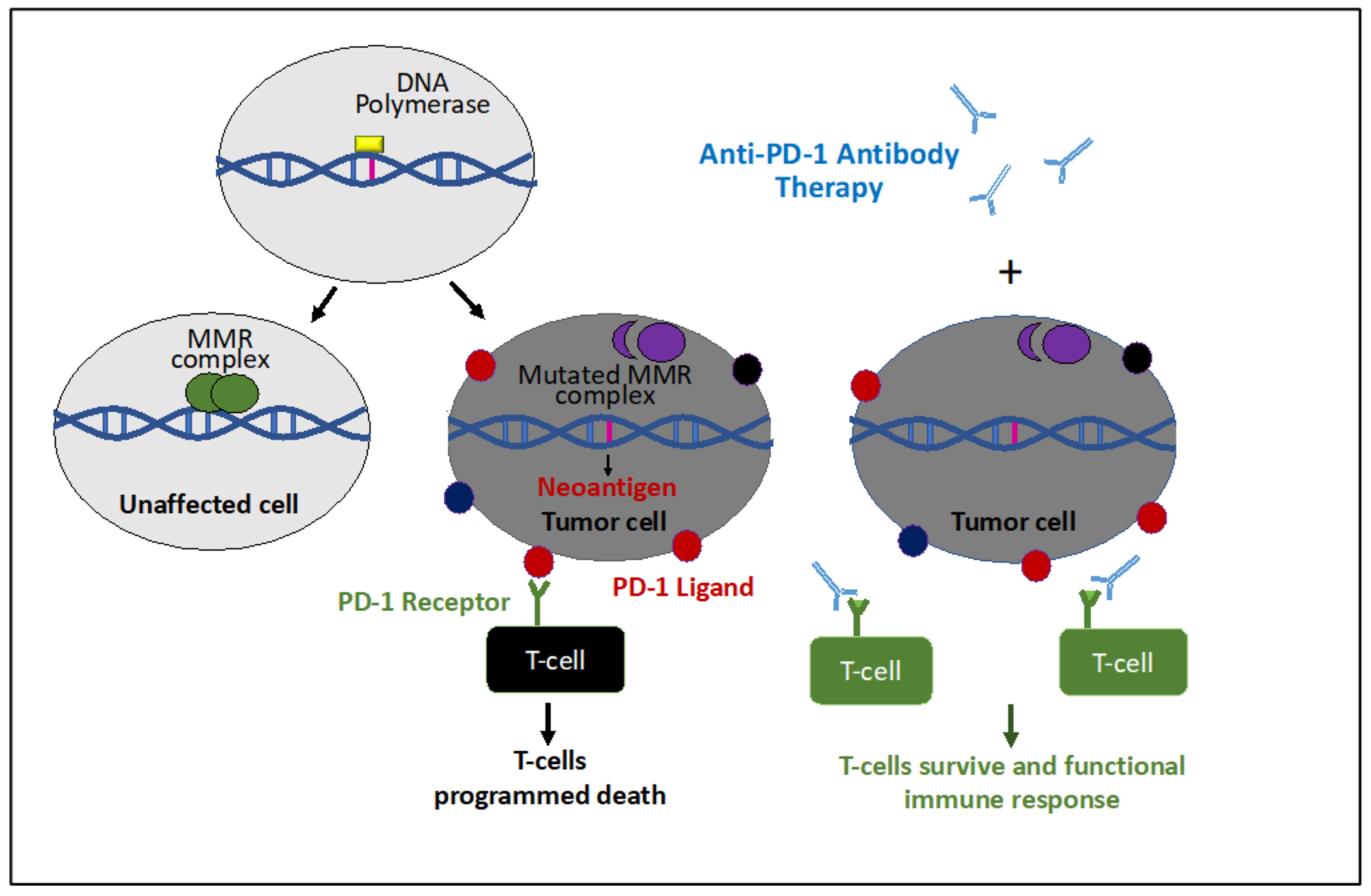
8. Immunotherapy for MSI/dMMR Gynecological Cancers
9. Conclusions
Funding
Conflicts of Interest
References
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.-Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.d.F. Microsatellite markers: What they mean and why they are so useful. Genet Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Schlötterer, C. Evolutionary dynamics of microsatellite DNA. Chromosoma 2000, 109, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008, 18, 85–98. [Google Scholar] [CrossRef] [Green Version]
- Moreira, L.; Balaguer, F.; Lindor, N.; de la Chapelle, A.; Hampel, H.; Aaltonen, L.A.; Hopper, J.L.; Marchand, L.L.; Gallinger, S.; Newcomb, P.A.; et al. Identification of Lynch Syndrome Among Patients With Colorectal Cancer. JAMA 2012, 308, 1555–1565. [Google Scholar] [CrossRef]
- Kane, M.F.; Loda, M.; Gaida, G.M.; Lipman, J.; Mishra, R.; Goldman, H.; Jessup, J.M.; Kolodner, R. Methylation of the hMLH1 Promoter Correlates with Lack of Expression of hMLH1 in Sporadic Colon Tumors and Mismatch Repair-defective Human Tumor Cell Lines. Cancer Res. 1997, 57, 808–811. [Google Scholar]
- Kakar, S.; Burgart, L.J.; Thibodeau, S.N.; Rabe, K.G.; Petersen, G.M.; Goldberg, R.M.; Lindor, N.M. Frequency of loss of hMLH1 expression in colorectal carcinoma increases with advancing age. Cancer 2003, 97, 1421–1427. [Google Scholar] [CrossRef]
- Ligtenberg, M.J.L.; Kuiper, R.P.; Chan, T.L.; Goossens, M.; Hebeda, K.M.; Voorendt, M.; Lee, T.Y.H.; Bodmer, D.; Hoenselaar, E.; Hendriks-Cornelissen, S.J.B.; et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009, 41, 112–117. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Simpkins, S.B.; Bocker, T.; Swisher, E.M.; Mutch, D.G.; Gersell, D.J.; Kovatich, A.J.; Palazzo, J.P.; Fishel, R.; Goodfellow, P.J. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum. Mol. Genet. 1999, 8, 661–666. [Google Scholar] [CrossRef] [Green Version]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Macaubas, C.; Jin, L.; Hallmayer, J.; Kimura, A.; Mignot, E. The Complex Mutation Pattern of a Microsatellite. Genome Res. 1997, 7, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhargava, A.; Fuentes, F.F. Mutational Dynamics of Microsatellites. Mol. Biotechnol. 2010, 44, 250–266. [Google Scholar] [CrossRef] [PubMed]
- López Castel, A.; Cleary, J.D.; Pearson, C.E. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 2010, 11, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, S.M. Expandable DNA repeats and human disease. Nature 2007, 447, 932–940. [Google Scholar] [CrossRef]
- Richard, G.-F.; Pâques, F. Mini- and microsatellite expansions: The recombination connection. EMBO Rep. 2000, 1, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, Y.D.; Tyekucheva, S.; Chiaromonte, F.; Makova, K.D. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008, 18, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Strassmann, J.E.; Queller, D.C. Insertions, substitutions, and the origin of microsatellites. Genet. Res. 2000, 76, 227–236. [Google Scholar] [CrossRef]
- Kim, J.C.; Harris, S.T.; Dinter, T.; Shah, K.A.; Mirkin, S.M. The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat. Struct. Mol. Biol. 2017, 24, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Jackson, A.; Okely, E.A.; Leach, D.R.F. Expansion of CAG Repeats in Escherichia coli Is Controlled by Single-Strand DNA Exonucleases of Both Polarities. Genetics 2014, 198, 509–517. [Google Scholar] [CrossRef] [Green Version]
- Cleary, J.D.; Nichol, K.; Wang, Y.-H.; Pearson, C.E. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 2002, 31, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Freudenreich, C.H.; Stavenhagen, J.B.; Zakian, V.A. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell Biol. 1997, 17, 2090–2098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.; Jaworski, A.; Ohshima, K.; Wells, R.D. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 1995, 10, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chen, X.; Bissler, J.J.; Sinden, R.R.; Leffak, M. Replication-dependent instability at (CTG)•(CAG) repeat hairpins in human cells. Nat. Chem. Biol. 2010, 6, 652–659. [Google Scholar] [CrossRef] [Green Version]
- Miret, J.J.; Pessoa-Brandão, L.; Lahue, R.S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1998, 95, 12438–12443. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, R.; Krasilnikova, M.M.; Samadashwily, G.M.; Lahue, R.; Mirkin, S.M. Replication and Expansion of Trinucleotide Repeats in Yeast. Mol. Cell Biol. 2003, 23, 1349–1357. [Google Scholar] [CrossRef] [Green Version]
- Samadashwily, G.M.; Raca, G.; Mirkin, S.M. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 1997, 17, 298–304. [Google Scholar] [CrossRef]
- Leffak, M.; Gadgil, R.; Barthelemy, J.; Lewis, T. Replication stalling and DNA microsatellite instability. Biophys. Chem. 2017, 225, 38–48. [Google Scholar] [CrossRef]
- Leffak, M. Break-induced replication links microsatellite expansion to complex genome rearrangements. Bioessays 2017, 39. [Google Scholar] [CrossRef]
- Mirkin, S.M. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biology 2006, 16, 351–358. [Google Scholar] [CrossRef]
- Frank-Kamenetskii, M.D.; Mirkin, S.M. Triplex DNA structures. Annu. Rev. Biochem. 1995, 64, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003, 4, R13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brazda, V.; Fojta, M.; Bowater, R.P. Structures and stability of simple DNA repeats from bacteria. Biochem. J. 2020, 477, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bacolla, A.; Wang, G.; Vasquez, K.M. Non-B DNA structure-induced genetic instability and evolution. Cell Mol. Life Sci. 2010, 67, 43–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckert, K.A.; Hile, S.E. Every Microsatellite is Different: Intrinsic DNA Features Dictate Mutagenesis of Common Microsatellites Present in the Human Genome. Mol. Carcinog. 2009, 48, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Hile, S.E.; Eckert, K.A. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 2008, 36, 688–696. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Vasquez, K.M. Impact of Alternative DNA Structures on DNA Damage, DNA Repair, and Genetic Instability. DNA Repair (Amst.) 2014, 19, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Gerhardt, J.; Bhalla, A.D.; Butler, J.S.; Puckett, J.W.; Dervan, P.B.; Rosenwaks, Z.; Napierala, M. Stalled DNA Replication Forks at the Endogenous GAA Repeats Drive Repeat Expansion in Friedreich’s Ataxia Cells. Cell Rep. 2016, 16, 1218–1227. [Google Scholar] [CrossRef] [Green Version]
- Krasilnikova, M.M.; Kireeva, M.L.; Petrovic, V.; Knijnikova, N.; Kashlev, M.; Mirkin, S.M. Effects of Friedreich’s ataxia (GAA)n*(TTC)n repeats on RNA synthesis and stability. Nucleic Acids Res. 2007, 35, 1075–1084. [Google Scholar] [CrossRef]
- Baptiste, B.A.; Jacob, K.D.; Eckert, K.A. Genetic Evidence That Both dNTP-Stabilized and Strand Slippage Mechanisms May Dictate DNA Polymerase Errors Within Mononucleotide Microsatellites. DNA Repair (Amst.) 2015, 29, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Sood, A.K.; Skilling, J.S.; Buller, R.E. Ovarian Cancer Genomic Instability Correlates with p53 Frameshift Mutations. Cancer Res. 1997, 57, 1047–1049. [Google Scholar] [PubMed]
- Rimokh, R.; Rouault, J.P.; Wahbi, K.; Gadoux, M.; Lafage, M.; Archimbaud, E.; Charrin, C.; Gentilhomme, O.; Germain, D.; Samarut, J. A chromosome 12 coding region is juxtaposed to the MYC protooncogene locus in a t(8;12)(q24;q22) translocation in a case of B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer 1991, 3, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, N.; Singh, R.K.; Mondal, S.; Roy, A.; Mondal, R.; Roychowdhury, S.; Panda, C.K. Analysis of molecular alterations in chromosome 8 associated with the development of uterine cervical carcinoma of Indian patients. Gynecol. Oncol. 2004, 95, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Toyama, T.; Iwase, H.; Yamashita, H.; Iwata, H.; Yamashita, T.; Ito, K.; Hara, Y.; Suchi, M.; Kato, T.; Nakamura, T.; et al. Microsatellite instability in sporadic human breast cancers. Int. J. Cancer 1996, 68, 447–451. [Google Scholar] [CrossRef]
- Schildkraut, J.M.; Murphy, S.K.; Palmieri, R.T.; Iversen, E.; Moorman, P.G.; Huang, Z.; Halabi, S.; Calingaert, B.; Gusberg, A.; Marks, J.R.; et al. Trinucleotide repeat polymorphisms in the androgen receptor gene and risk of ovarian cancer. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 473–480. [Google Scholar] [CrossRef] [Green Version]
- Catasús, L.; Matias-Guiu, X.; Machin, P.; Muñoz, J.; Prat, J. BAX somatic frameshift mutations in endometrioid adenocarcinomas of the endometrium: Evidence for a tumor progression role in endometrial carcinomas with microsatellite instability. Lab. Invest. 1998, 78, 1439–1444. [Google Scholar]
- Markowitz, S.; Wang, J.; Myeroff, L.; Parsons, R.; Sun, L.; Lutterbaugh, J.; Fan, R.S.; Zborowska, E.; Kinzler, K.W.; Vogelstein, B. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 1995, 268, 1336–1338. [Google Scholar] [CrossRef]
- Bilbao, C.; Rodríguez, G.; Ramírez, R.; Falcón, O.; León, L.; Chirino, R.; Rivero, J.F.; Falcón, O.; Díaz-Chico, B.N.; Díaz-Chico, J.C.; et al. The relationship between microsatellite instability and PTEN gene mutations in endometrial cancer. Int. J. Cancer 2006, 119, 563–570. [Google Scholar] [CrossRef]
- Bilbao, C.; Ramírez, R.; Rodríguez, G.; Falcón, O.; León, L.; Díaz-Chico, N.; Perucho, M.; Díaz-Chico, J.C. Double strand break repair components are frequent targets of microsatellite instability in endometrial cancer. Eur. J. Cancer 2010, 46, 2821–2827. [Google Scholar] [CrossRef]
- Shah, S.N.; Hile, S.E.; Eckert, K.A. Defective Mismatch Repair, Microsatellite Mutation Bias, and Variability in Clinical Cancer Phenotypes. Cancer Res. 2010, 70, 431–435. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, P.; Yamane, K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 2008, 129, 391–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sameer, A.S.; Nissar, S.; Fatima, K. Mismatch repair pathway: Molecules, functions, and role in colorectal carcinogenesis. Eur. J. Cancer Prev. 2014, 23, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.; Zhang, Y. The Devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. USA 2017, 114, 3552–3554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortes-Ciriano, I.; Lee, S.; Park, W.-Y.; Kim, T.-M.; Park, P.J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shilpa, V.; Rahul, B.; Premalata, C.S.; Pallavi, V.R.; Lakshmi, K. Microsatellite instability, promoter methylation and protein expression of the DNA mismatch repair genes in epithelial ovarian cancer. Genomics 2014, 104, 257–263. [Google Scholar] [CrossRef]
- Cederquist, K.; Emanuelsson, M.; Wiklund, F.; Golovleva, I.; Palmqvist, R.; Grönberg, H. Two Swedish founder MSH6 mutations, one nonsense and one missense, conferring high cumulative risk of Lynch syndrome. Clin. Genet. 2005, 68, 533–541. [Google Scholar] [CrossRef]
- Pal, T.; Permuth-Wey, J.; Sellers, T.A. A review of the clinical relevance of mismatch-repair deficiency in ovarian cancer. Cancer 2008, 113, 733–742. [Google Scholar] [CrossRef] [Green Version]
- Espenschied, C.R.; LaDuca, H.; Li, S.; McFarland, R.; Gau, C.-L.; Hampel, H. Multigene Panel Testing Provides a New Perspective on Lynch Syndrome. JCO 2017, 35, 2568–2575. [Google Scholar] [CrossRef]
- Roberts, M.E.; Jackson, S.A.; Susswein, L.R.; Zeinomar, N.; Ma, X.; Marshall, M.L.; Stettner, A.R.; Milewski, B.; Xu, Z.; Solomon, B.D.; et al. MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet. Med. 2018, 20, 1167–1174. [Google Scholar] [CrossRef] [Green Version]
- Boland, C.R.; Thibodeau, S.N.; Hamilton, S.R.; Sidransky, D.; Eshleman, J.R.; Burt, R.W.; Meltzer, S.J.; Rodriguez-Bigas, M.A.; Fodde, R.; Ranzani, G.N.; et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998, 58, 5248–5257. [Google Scholar]
- Murphy, M.A.; Wentzensen, N. Frequency of mismatch repair deficiency in ovarian cancer: A systematic review. Int. J. Cancer 2011, 129, 1914–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hempelmann, J.A.; Lockwood, C.M.; Konnick, E.Q.; Schweizer, M.T.; Antonarakis, E.S.; Lotan, T.L.; Montgomery, B.; Nelson, P.S.; Klemfuss, N.; Salipante, S.J.; et al. Microsatellite instability in prostate cancer by PCR or next-generation sequencing. J. Immunother. Cancer 2018, 6, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagin, A.; Zerimech, F.; Leclerc, J.; Wacrenier, A.; Lejeune, S.; Descarpentries, C.; Escande, F.; Porchet, N.; Buisine, M.-P. Evaluation of a new panel of six mononucleotide repeat markers for the detection of DNA mismatch repair-deficient tumours. Br. J. Cancer 2013, 108, 2079–2087. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.-F.; Cheung, T.-H.; Poon, K.-Y.; Wang, V.W.; Li, J.C.B.; Lo, K.W.-K.; Yim, S.-F.; Yu, M.-Y.; Lahr, G.; Chung, T.K.-H. The role of microsatellite instability in cervical intraepithelial neoplasia and squamous cell carcinoma of the cervix. Gynecol. Oncol. 2003, 89, 434–439. [Google Scholar] [CrossRef]
- Chung, T.K.; Cheung, T.H.; Wang, V.W.; Yu, M.Y.; Wong, Y.F. Microsatellite instability, expression of hMSH2 and hMLH1 and HPV infection in cervical cancer and their clinico-pathological association. Gynecol. Obstet. Invest. 2001, 52, 98–103. [Google Scholar] [CrossRef]
- Gurin, C.C.; Federici, M.G.; Kang, L.; Boyd, J. Causes and Consequences of Microsatellite Instability in Endometrial Carcinoma. Cancer Res. 1999, 59, 462–466. [Google Scholar]
- Helleman, J.; van Staveren, I.L.; Dinjens, W.N.; van Kuijk, P.F.; Ritstier, K.; Ewing, P.C.; van der Burg, M.E.; Stoter, G.; Berns, E.M. Mismatch repair and treatment resistance in ovarian cancer. BMC Cancer 2006, 6, 201. [Google Scholar] [CrossRef] [Green Version]
- Walsh, M.D.; Cummings, M.C.; Buchanan, D.D.; Dambacher, W.M.; Arnold, S.; McKeone, D.; Byrnes, R.; Barker, M.A.; Leggett, B.A.; Gattas, M.; et al. Molecular, Pathologic, and Clinical Features of Early-Onset Endometrial Cancer: Identifying Presumptive Lynch Syndrome Patients. Clin. Cancer Res. 2008, 14, 1692–1700. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.-M.; Laird, P.W.; Park, P.J. The Landscape of Microsatellite Instability in Colorectal and Endometrial Cancer Genomes. Cell 2013, 155, 858–868. [Google Scholar] [CrossRef] [Green Version]
- Wong, Y.F.; Cheung, T.H.; Lo, K.W.K.; Yim, S.F.; Chan, L.K.Y.; Buhard, O.; Duval, A.; Chung, T.K.H.; Hamelin, R. Detection of microsatellite instability in endometrial cancer: Advantages of a panel of five mononucleotide repeats over the National Cancer Institute panel of markers. Carcinogenesis 2006, 27, 951–955. [Google Scholar] [CrossRef] [Green Version]
- Yoon, B.-S.; Kim, Y.-T.; Kim, J.-H.; Kim, S.-W.; Nam, E.-J.; Cho, N.-H.; Kim, J.-W.; Kim, S. Clinical Significance of Microsatellite Instability in Sporadic Epithelial Ovarian Tumors. Yonsei Med. J. 2008, 49, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Singer, G.; Kallinowski, T.; Hartmann, A.; Dietmaier, W.; Wild, P.J.; Schraml, P.; Sauter, G.; Mihatsch, M.J.; Moch, H. Different types of microsatellite instability in ovarian carcinoma. Int. J. Cancer 2004, 112, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Banno, K.; Yanokura, M.; Kobayashi, Y.; Kishimi, A.; Ogawa, S.; Kisu, I.; Nomura, H.; Hirasawa, A.; Susumu, N.; et al. Analysis of candidate target genes for mononucleotide repeat mutation in microsatellite instability-high (MSI-H) endometrial cancer. Int. J. Oncol. 2009, 35, 977–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvi, A.J.; Rader, J.S.; Broggini, M.; Latif, F.; Maher, E.R. Microsatellite instability and mutational analysis of transforming growth factor β receptor type II gene (TGFBR2) in sporadic ovarian cancer. Mol. Pathol. 2001, 54, 240–243. [Google Scholar] [CrossRef] [Green Version]
- Ozer, E.; Yuksel, E.; Kizildag, S.; Sercan, O.; Ozen, E.; Canda, T.; Sakizli, M. Microsatellite instability in early-onset breast cancer. Pathol. Res. Pract. 2002, 198, 525–530. [Google Scholar] [CrossRef]
- Dhillon, V.S.; Aslam, M.; Husain, S.A. The contribution of genetic and epigenetic changes in granulosa cell tumors of ovarian origin. Clin. Cancer Res. 2004, 10, 5537–5545. [Google Scholar] [CrossRef] [Green Version]
- Sood, A.K.; Holmes, R.; Hendrix, M.J.C.; Buller, R.E. Application of the National Cancer Institute International Criteria for Determination of Microsatellite Instability in Ovarian Cancer. Cancer Res. 2001, 61, 4371–4374. [Google Scholar]
- Wooster, R.; Cleton-Jansen, A.-M.; Collins, N.; Mangion, J.; Cornelis, R.S.; Cooper, C.S.; Gusterson, B.A.; Ponder, B.A.J.; von Deimling, A.; Wiestler, O.D.; et al. Instability of short tandem repeats (microsatellites) in human cancers. Nat. Genet. 1994, 6, 152–156. [Google Scholar] [CrossRef]
- Watson, M.M.C.; Berg, M.; Søreide, K. Prevalence and implications of elevated microsatellite alterations at selected tetranucleotides in cancer. Br. J. Cancer 2014, 111, 823–827. [Google Scholar] [CrossRef] [Green Version]
- Plisiecka-Hałasa, J.; Dansonka-Mieszkowska, A.; Kraszewska, E.; Dańska-Bidzińska, A.; Kupryjańczyk, J. Loss of heterozygosity, microsatellite instability and TP53 gene status in ovarian carcinomas. Anticancer Res. 2008, 28, 989–996. [Google Scholar]
- Ou, C.-Y.; Chang, J.-G.; Tseng, H.-H.; Wei, H.-J.; Su, T.-H.; Hsu, T.-Y.; Chang, C.-P.; Lee, H.-H. Analysis of microsatellite instability in cervical cancer. Int. J. Gynecol. Cancer 1999, 9, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Geisler, J.P.; Goodheart, M.J.; Sood, A.K.; Holmes, R.J.; Hatterman-Zogg, M.A.; Buller, R.E. Mismatch repair gene expression defects contribute to microsatellite instability in ovarian carcinoma. Cancer 2003, 98, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- You, J.-F.; Buhard, O.; Ligtenberg, M.J.L.; Kets, C.M.; Niessen, R.C.; Hofstra, R.M.W.; Wagner, A.; Dinjens, W.N.M.; Colas, C.; Lascols, O.; et al. Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br. J. Cancer 2010, 103, 1840–1845. [Google Scholar] [CrossRef] [PubMed]
- Suraweera, N.; Duval, A.; Reperant, M.; Vaury, C.; Furlan, D.; Leroy, K.; Seruca, R.; Iacopetta, B.; Hamelin, R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology 2002, 123, 1804–1811. [Google Scholar] [CrossRef] [PubMed]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; de la Chapelle, A.; Rüschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Libera, L.; Sahnane, N.; Carnevali, I.W.; Cimetti, L.; Cerutti, R.; Chiaravalli, A.M.; Riva, C.; Tibiletti, M.G.; Sessa, F.; Furlan, D. Microsatellite analysis of sporadic and hereditary gynaecological cancer in routine diagnostics. J. Clin. Pathol. 2017, 70, 792–797. [Google Scholar] [CrossRef]
- Larson, A.A.; Kern, S.; Sommers, R.L.; Yokota, J.; Cavenee, W.K.; Hampton, G.M. Analysis of replication error (RER+) phenotypes in cervical carcinoma. Cancer Res. 1996, 56, 1426–1431. [Google Scholar]
- Zhu, C.S.; Pinsky, P.F.; Cramer, D.W.; Ransohoff, D.F.; Hartge, P.; Pfeiffer, R.M.; Urban, N.; Mor, G.; Bast, R.C.; Moore, L.E.; et al. A Framework for Evaluating Biomarkers for Early Detection: Validation of Biomarker Panels for Ovarian Cancer. Cancer Prev. Res. (Phila.) 2011, 4, 375–383. [Google Scholar] [CrossRef] [Green Version]
- Metcalf, A.M.; Spurdle, A.B. Endometrial tumour BRAF mutations and MLH1 promoter methylation as predictors of germline mismatch repair gene mutation status: A literature review. Fam. Cancer 2014, 13, 1–12. [Google Scholar] [CrossRef]
- Johannsdottir, J.T.; Jonasson, J.G.; Bergthorsson, J.T.; Amundadottir, L.T.; Magnusson, J.; Egilsson, V.; Ingvarsson, S. The effect of mismatch repair deficiency on tumourigenesis; microsatellite instability affecting genes containing short repeated sequences. Int. J. Oncol. 2000, 16, 133–142. [Google Scholar] [CrossRef]
- Codegoni, A.M.; Bertoni, F.; Colella, G.; Caspani, G.; Grassi, L.; D’Incalci, M.; Broggini, M. Microsatellite instability and frameshift mutations in genes involved in cell cycle progression or apoptosis in ovarian cancer. Oncol. Res. 1999, 11, 297–301. [Google Scholar] [PubMed]
- Albacker, L.A.; Wu, J.; Smith, P.; Warmuth, M.; Stephens, P.J.; Zhu, P.; Yu, L.; Chmielecki, J. Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.K.; Bashashati, A.; Anglesio, M.S.; Cochrane, D.R.; Grewal, D.S.; Ha, G.; McPherson, A.; Horlings, H.M.; Senz, J.; Prentice, L.M.; et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat. Genet. 2017, 49, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhang, Y.; Liu, R.Z.; Fenstermacher, D.A.; Wright, K.L.; Teer, J.K.; Wu, J. JAK1 truncating mutations in gynecologic cancer define new role of cancer-associated protein tyrosine kinase aberrations. Sci. Rep. 2013, 3, 3042. [Google Scholar] [CrossRef] [Green Version]
- Vassileva, V.; Millar, A.; Briollais, L.; Chapman, W.; Bapat, B. Genes Involved in DNA Repair Are Mutational Targets in Endometrial Cancers with Microsatellite Instability. Cancer Res. 2002, 62, 4095–4099. [Google Scholar]
- Vassileva, V.; Millar, A.; Briollais, L.; Chapman, W.; Bapat, B. Apoptotic and growth regulatory genes as mutational targets in mismatch repair deficient endometrioid adenocarcinomas of young patients. Oncol. Rep. 2004, 11, 931–937. [Google Scholar] [CrossRef]
- Maruvka, Y.E.; Mouw, K.W.; Karlic, R.; Parasuraman, P.; Kamburov, A.; Polak, P.; Haradhvala, N.J.; Hess, J.M.; Rheinbay, E.; Brody, Y.; et al. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat. Biotechnol. 2017, 35, 951–959. [Google Scholar] [CrossRef]
- Bertoni, F.; Codegoni, A.M.; Furlan, D.; Tibiletti, M.G.; Capella, C.; Broggini, M. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes Chromosomes Cancer 1999, 26, 176–180. [Google Scholar] [CrossRef]
- Lin, E.I.; Tseng, L.-H.; Gocke, C.D.; Reil, S.; Le, D.T.; Azad, N.S.; Eshleman, J.R. Mutational profiling of colorectal cancers with microsatellite instability. Oncotarget 2015, 6, 42334–42344. [Google Scholar] [CrossRef] [Green Version]
- Novetsky, A.P.; Zighelboim, I.; Thompson, D.M.; Powell, M.A.; Mutch, D.G.; Goodfellow, P.J. Frequent Mutations in the RPL22 Gene and its Clinical and Functional Implications. Gynecol. Oncol. 2013, 128. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stelloo, E.; Versluis, M.A.; Nijman, H.W.; de Bruyn, M.; Plat, A.; Osse, E.M.; van Dijk, R.H.; Nout, R.A.; Creutzberg, C.L.; de Bock, G.H.; et al. Microsatellite instability derived JAK1 frameshift mutations are associated with tumor immune evasion in endometrioid endometrial cancer. Oncotarget 2016, 7, 39885–39893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davalos, V.; Dopeso, H.; Velho, S.; Ferreira, A.M.; Cirnes, L.; Díaz-Chico, N.; Bilbao, C.; Ramírez, R.; Rodríguez, G.; Falcón, O.; et al. High EPHB2 mutation rate in gastric but not endometrial tumors with microsatellite instability. Oncogene 2007, 26, 308–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.; Yamamoto, H.; Navarro, M.; Maestro, M.; Reventós, J.; Perucho, M. Frameshift Mutations at Mononucleotide Repeats in caspase-5 and Other Target Genes in Endometrial and Gastrointestinal Cancer of the Microsatellite Mutator Phenotype. Cancer Res. 1999, 59, 2995–3002. [Google Scholar] [PubMed]
- Jo, Y.S.; Kim, S.S.; Kim, M.S.; Yoo, N.J.; Lee, S.H. Candidate tumor suppressor gene MCPH1 is mutated in colorectal and gastric cancers. Int. J. Colorectal. Dis. 2017, 32, 161–162. [Google Scholar] [CrossRef] [PubMed]
- Giannini, G.; Rinaldi, C.; Ristori, E.; Ambrosini, M.I.; Cerignoli, F.; Viel, A.; Bidoli, E.; Berni, S.; D’Amati, G.; Scambia, G.; et al. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene 2004, 23, 2640–2647. [Google Scholar] [CrossRef] [Green Version]
- Ikenoue, T.; Togo, G.; Nagai, K.; Ijichi, H.; Kato, J.; Yamaji, Y.; Okamoto, M.; Kato, N.; Kawabe, T.; Tanaka, A.; et al. Frameshift Mutations at Mononucleotide Repeats in RAD50 Recombinational DNA Repair Gene in Colorectal Cancers with Microsatellite Instability. Jpn. J. Cancer Res. 2001, 92, 587–591. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Hirasawa, A.; Aoki, D.; Inoue, J.; Imoto, I.; Susumu, N.; Sugano, K.; Nozawa, S.; Inazawa, J. Unfavorable Prognostic Factors Associated with High Frequency of Microsatellite Instability and Comparative Genomic Hybridization Analysis in Endometrial Cancer. Clin. Cancer Res. 2003, 9, 5675–5682. [Google Scholar]
- Umar, A.; Boyer, J.C.; Thomas, D.C.; Nguyen, D.C.; Risinger, J.I.; Boyd, J.; Ionov, Y.; Perucho, M.; Kunkel, T.A. Defective mismatch repair in extracts of colorectal and endometrial cancer cell lines exhibiting microsatellite instability. J. Biol. Chem. 1994, 269, 14367–14370. [Google Scholar]
- Zhou, X.-P.; Kuismanen, S.; Nystrom-Lahti, M.; Peltomaki, P.; Eng, C. Distinct PTEN mutational spectra in hereditary non-polyposis colon cancer syndrome-related endometrial carcinomas compared to sporadic microsatellite unstable tumors. Hum. Mol. Genet. 2002, 11, 445–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, R.A.; Fleming, G.F.; Lastra, R.R.; Lee, N.K.; Moroney, J.W.; Son, C.H.; Tatebe, K.; Veneris, J.L. Current recommendations and recent progress in endometrial cancer. CA A Cancer J. Clin. 2019, 69, 258–279. [Google Scholar] [CrossRef] [PubMed]
- Egoavil, C.; Alenda, C.; Castillejo, A.; Paya, A.; Peiro, G.; Sánchez-Heras, A.-B.; Castillejo, M.-I.; Rojas, E.; Barberá, V.-M.; Cigüenza, S.; et al. Prevalence of Lynch Syndrome among Patients with Newly Diagnosed Endometrial Cancers. PLoS ONE 2013, 8, e79737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, A.C.; Blanchard, Z.; Maurer, K.A.; Gertz, J. Estrogen Signaling in Endometrial Cancer: A Key Oncogenic Pathway with Several Open Questions. Horm Cancer 2019, 10, 51–63. [Google Scholar] [CrossRef] [Green Version]
- Amankwah, E.K.; Friedenreich, C.M.; Magliocco, A.M.; Brant, R.; Speidel, T.; Rahman, W.; Cook, L.S. Hormonal and Reproductive Risk Factors for Sporadic Microsatellite Stable and Unstable Endometrial Tumors. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1325–1331. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, T.; Shiozawa, T.; Kashima, H.; Feng, Y.-Z.; Suzuki, A.; Kurai, M.; Nikaido, T.; Konishi, I. Estrogen Up-Regulates Mismatch Repair Activity in Normal and Malignant Endometrial Glandular Cells. Endocrinology 2006, 147, 4863–4870. [Google Scholar] [CrossRef]
- Lu, J.-Y.; Jin, P.; Gao, W.; Wang, D.-Z.; Sheng, J.-Q. Estrogen enhances mismatch repair by induction of MLH1 expression via estrogen receptor-β. Oncotarget 2017, 8, 38767–38779. [Google Scholar] [CrossRef]
- Dashti, S.G.; Chau, R.; Ouakrim, D.A.; Buchanan, D.D.; Clendenning, M.; Young, J.P.; Winship, I.M.; Arnold, J.; Ahnen, D.J.; Haile, R.W.; et al. Female Hormonal Factors and the Risk of Endometrial Cancer in Lynch Syndrome. JAMA 2015, 314, 61–71. [Google Scholar] [CrossRef]
- SGO Clinical Practice Statement: Screening for Lynch Syndrome in Endometrial Cancer. Available online: https://www.sgo.org/clinical-practice/guidelines/screening-for-lynch-syndrome-in-endometrial-cancer/ (accessed on 30 March 2020).
- Shia, J. Immunohistochemistry versus Microsatellite Instability Testing For Screening Colorectal Cancer Patients at Risk For Hereditary Nonpolyposis Colorectal Cancer Syndrome: Part I. The Utility of Immunohistochemistry. J. Mol. Diagn. 2008, 10, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, H.; Zaanan, A.; Sinicrope, F.A. MSI testing and its role in the management of colorectal cancer. Curr. Treat Options Oncol. 2015, 16, 30. [Google Scholar] [CrossRef]
- Shikama, A.; Minaguchi, T.; Matsumoto, K.; Akiyama-Abe, A.; Nakamura, Y.; Michikami, H.; Nakao, S.; Sakurai, M.; Ochi, H.; Onuki, M.; et al. Clinicopathologic implications of DNA mismatch repair status in endometrial carcinomas. Gynecol. Oncol. 2016, 140, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Reijnen, C.; Küsters-Vandevelde, H.V.N.; Prinsen, C.F.; Massuger, L.F.A.G.; Snijders, M.P.M.L.; Kommoss, S.; Brucker, S.Y.; Kwon, J.S.; McAlpine, J.N.; Pijnenborg, J.M.A. Mismatch repair deficiency as a predictive marker for response to adjuvant radiotherapy in endometrial cancer. Gynecol. Oncol. 2019, 154, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Møller, P.; Seppälä, T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Evans, D.G.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.; et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: First report from the prospective Lynch syndrome database. Gut 2017, 66, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Bonadona, V.; Bonaïti, B.; Olschwang, S.; Grandjouan, S.; Huiart, L.; Longy, M.; Guimbaud, R.; Buecher, B.; Bignon, Y.-J.; Caron, O.; et al. Cancer Risks Associated With Germline Mutations in MLH1, MSH2, and MSH6 Genes in Lynch Syndrome. JAMA 2011, 305, 2304–2310. [Google Scholar] [CrossRef] [PubMed]
- Ten Broeke, S.W.; van der Klift, H.M.; Tops, C.M.J.; Aretz, S.; Bernstein, I.; Buchanan, D.D.; de la Chapelle, A.; Capella, G.; Clendenning, M.; Engel, C.; et al. Cancer Risks for PMS2-Associated Lynch Syndrome. J. Clin. Oncol. 2018, 36, 2961–2968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Committee on Practice Bulletins-Gynecology; Society of Gynecologic Oncology. Practice Bulletin No. 147: Lynch Syndrome. Obstet. Gynecol. 2014, 124, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Renkonen-Sinisalo, L.; Bützow, R.; Leminen, A.; Lehtovirta, P.; Mecklin, J.-P.; Järvinen, H.J. Surveillance for endometrial cancer in hereditary nonpolyposis colorectal cancer syndrome. Int. J. Cancer 2007, 120, 821–824. [Google Scholar] [CrossRef]
- Schmeler, K.M.; Lynch, H.T.; Chen, L.; Munsell, M.F.; Soliman, P.T.; Clark, M.B.; Daniels, M.S.; White, K.G.; Boyd-Rogers, S.G.; Conrad, P.G.; et al. Prophylactic Surgery to Reduce the Risk of Gynecologic Cancers in the Lynch Syndrome. Available online: https://www.nejm.org/doi/10.1056/NEJMoa052627?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dwww.ncbi.nlm.nih.gov (accessed on 26 March 2020).
- Rivera, C.M.; Grossardt, B.R.; Rhodes, D.J.; Brown, R.D.; Roger, V.L.; Melton, L.J.; Rocca, W.A. Increased cardiovascular mortality following early bilateral oophorectomy. Menopause 2009, 16, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Hibler, E.; Kauderer, J.; Greene, M.H.; Rodriguez, G.C.; Alberts, D.S. Bone Loss Following Oophorectomy Among High-Risk Women: An NRG Oncology/Gynecologic Oncology Group study. Menopause 2016, 23, 1228–1232. [Google Scholar] [CrossRef] [Green Version]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. Available online: https://www.nejm.org/doi/10.1056/NEJMoa1200690?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dwww.ncbi.nlm.nih.gov (accessed on 20 April 2020).
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.M.; Hwu, W.-J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Chang, M.; Chang, H.M.; Chang, F. Microsatellite Instability: A Predictive Biomarker for Cancer Immunotherapy. Appl. Immunohistochem. Mol. Morphol. 2018, 26, e15. [Google Scholar] [CrossRef] [PubMed]
- Mandal, R.; Samstein, R.M.; Lee, K.-W.; Havel, J.J.; Wang, H.; Krishna, C.; Sabio, E.Y.; Makarov, V.; Kuo, F.; Blecua, P.; et al. Genetic diversity of tumors with mismatch repair deficiency influences anti–PD-1 immunotherapy response. Science 2019, 364, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wick, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, L.; Fan, H.; Wang, H.; Luber, B.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef]
- Yamashita, H.; Nakayama, K.; Ishikawa, M.; Nakamura, K.; Ishibashi, T.; Sanuki, K.; Ono, R.; Sasamori, H.; Minamoto, T.; Iida, K.; et al. Microsatellite instability is a biomarker for immune checkpoint inhibitors in endometrial cancer. Oncotarget 2017, 9, 5652–5664. [Google Scholar] [CrossRef] [Green Version]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. Available online: https://www.nejm.org/doi/10.1056/NEJMoa1500596?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dwww.ncbi.nlm.nih.gov (accessed on 26 March 2020).
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.-P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair–Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. JCO 2019, 38, 1–10. [Google Scholar] [CrossRef]
| Gene Mutation | Endometrial Cancer | Ovarian Cancer |
|---|---|---|
| MLH1 | 34–54% | 11% |
| MSH2 * | 21–51% | 15% |
| MSH6 | 16–49% | 0–1% |
| PMS2 | 13–24% | 0–1% |
| Cancer Type | Number Enrolled (n) | Complete Response (n) (%) | Partial Response (n) (%) | Objective Response Rate, Months (95% CI) | Median Progression Free Survival, Months (95% CI) |
|---|---|---|---|---|---|
| Endometrial | 49 | 8 (16.3%) | 20 (40.8%) | 57.1 (42.2–71.2) | 25.7 (4.9–DNR) |
| Ovarian | 15 | 3 (20%) | 2 (13.3%) | 33.3 (11.8–61.6) | 2.3 (1.9–6.2) |
| Cervical | 6 | NR | NR | NR | NR |
| Vaginal | 1 | NR | NR | NR | NR |
| Vulvar | 1 | NR | NR | NR | NR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deshpande, M.; Romanski, P.A.; Rosenwaks, Z.; Gerhardt, J. Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability. Cancers 2020, 12, 3319. https://doi.org/10.3390/cancers12113319
Deshpande M, Romanski PA, Rosenwaks Z, Gerhardt J. Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability. Cancers. 2020; 12(11):3319. https://doi.org/10.3390/cancers12113319
Chicago/Turabian StyleDeshpande, Madhura, Phillip A. Romanski, Zev Rosenwaks, and Jeannine Gerhardt. 2020. "Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability" Cancers 12, no. 11: 3319. https://doi.org/10.3390/cancers12113319
APA StyleDeshpande, M., Romanski, P. A., Rosenwaks, Z., & Gerhardt, J. (2020). Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability. Cancers, 12(11), 3319. https://doi.org/10.3390/cancers12113319




