The Non-Coding Landscape of Cutaneous Malignant Melanoma: A Possible Route to Efficient Targeted Therapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. MicroRNAs Modulate Melanoma Invasion and Metastasis
3. LncRNAs Modulate Melanoma Invasion and Metastasis
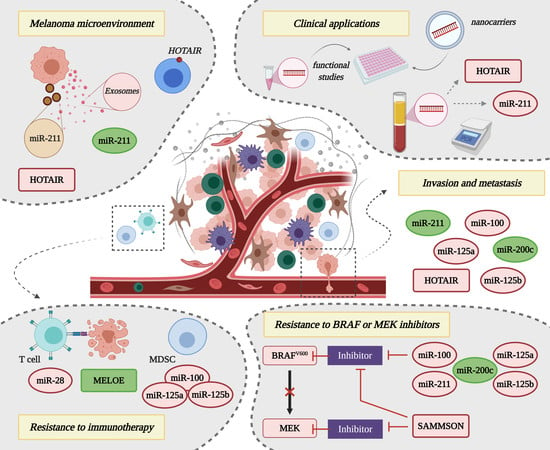
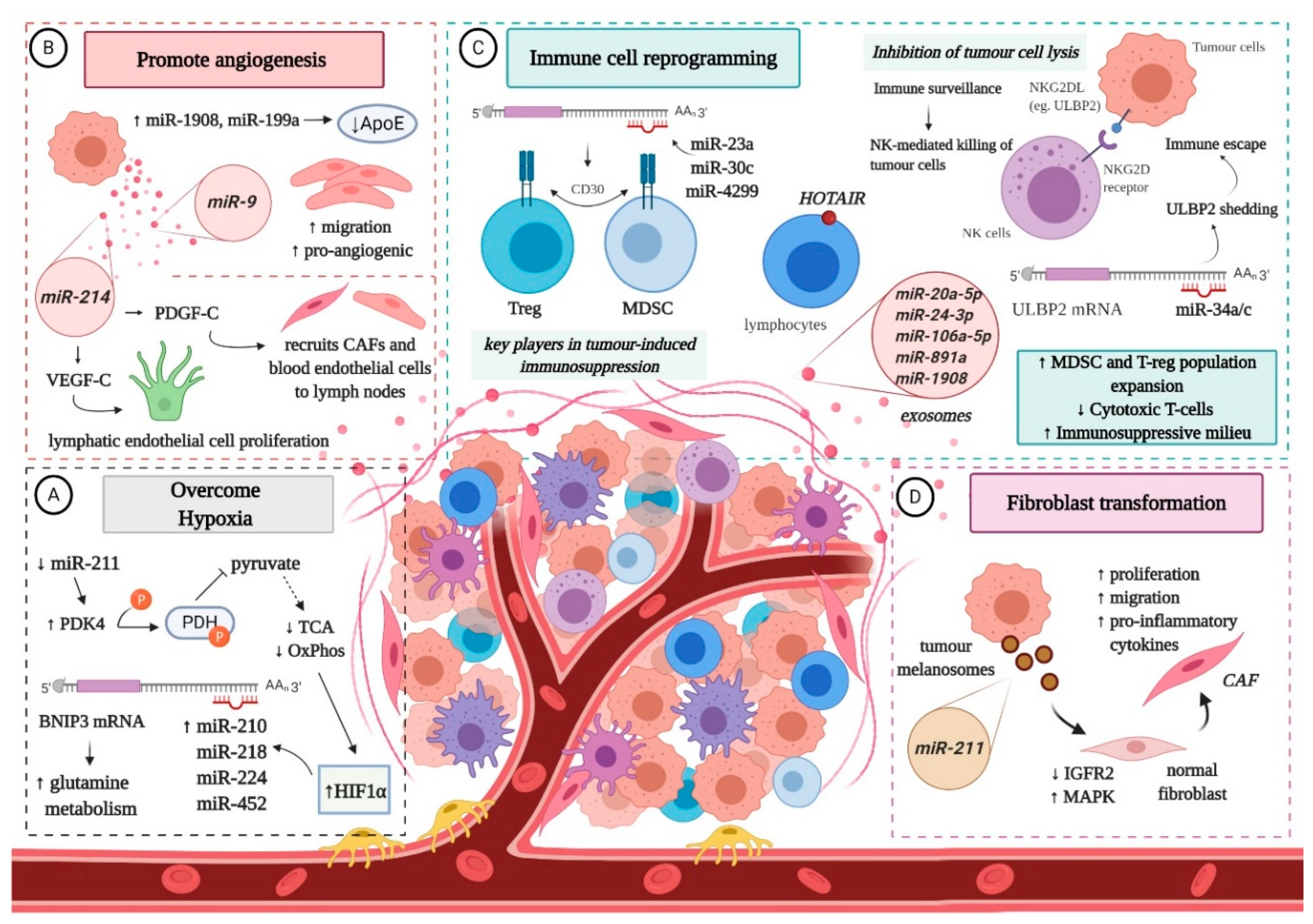
4. Non-Coding RNAs and the Interplay with the Melanoma Microenvironment
5. Non-Coding RNAs in Drug Resistance
5.1. Resistance to BRAF or MAPK Inhibitors
5.2. Resistance to Immunotherapy
6. Clinical Applications of Non-Coding RNA Molecules in Melanoma Management
6.1. Non-Coding RNAs as Circulating Biomarkers for Cutaneous Melanoma
6.2. Non-Coding RNAs as Targets for Promising Therapeutic Strategies
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Paluncic, J.; Kovacevic, Z.; Jansson, P.J.; Kalinowski, D.; Merlot, A.; Huang, M.L.-H.; Lok, H.; Sahni, S.; Lane, D.J.R.; Richardson, D.R. Roads to melanoma: Key pathways and emerging players in melanoma progression and oncogenic signaling. Biochim. Biophys. Acta 2016, 1863, 770–784. [Google Scholar] [CrossRef] [PubMed]
- Lazar, A.D.; Dinescu, S.; Costache, M. Deciphering the Molecular Landscape of Cutaneous Squamous Cell Carcinoma for Better Diagnosis and Treatment. J. Clin. Med. 2020, 9, 2228. [Google Scholar] [CrossRef] [PubMed]
- Sommer, L. Generation of melanocytes from neural crest cells. Pigment Cell Melanoma Res. 2011, 24, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.R. Ultraviolet radiation and skin cancer: Molecular mechanisms. J. Cutan Pathol. 2005, 32, 191–205. [Google Scholar] [CrossRef]
- Lopez, A.T.; Liu, L.; Geskin, L. Molecular Mechanisms and Biomarkers of Skin Photocarcinogenesis. In Human Skin Cancers: Pathways, Mechanisms, Targets and Treatments; Blumenberg, M., Ed.; IntechOpen: London, UK, 2017; pp. 175–200. [Google Scholar]
- Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Clark, W.H., Jr.; Elder, D.E.; Guerry, D.T.; Epstein, M.N.; Greene, M.H.; Van Horn, M. A study of tumor progression: The precursor lesions of superficial spreading and nodular melanoma. Hum. Pathol. 1984, 15, 1147–1165. [Google Scholar] [CrossRef]
- Damsky, W.E.; Theodosakis, N.; Bosenberg, M. Melanoma metastasis: New concepts and evolving paradigms. Oncogene 2014, 33, 2413–2422. [Google Scholar] [CrossRef] [Green Version]
- Hussussian, C.J.; Struewing, J.P.; Goldstein, A.M.; Higgins, P.A.; Ally, D.S.; Sheahan, M.D.; Clark, W.H., Jr.; Tucker, M.A.; Dracopoli, N.C. Germline p16 mutations in familial melanoma. Nat. Genet. 1994, 8, 15–21. [Google Scholar] [CrossRef]
- Sviderskaya, E.V.; Gray-Schopfer, V.C.; Hill, S.P.; Smit, N.P.; Evans-Whipp, T.J.; Bond, J.; Hill, L.; Bataille, V.; Peters, G.; Kipling, D.; et al. p16/cyclin-dependent kinase inhibitor 2A deficiency in human melanocyte senescence, apoptosis, and immortalization: Possible implications for melanoma progression. J. Natl. Cancer Inst. 2003, 95, 723–732. [Google Scholar] [CrossRef]
- Bennett, D.C. Human melanocyte senescence and melanoma susceptibility genes. Oncogene 2003, 22, 3063–3069. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.J.; Mihm, M.C., Jr. Melanoma. N. Engl. J. Med. 2006, 355, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Shepelin, D.; Korzinkin, M.; Vanyushina, A.; Aliper, A.; Borisov, N.; Vasilov, R.; Zhukov, N.; Sokov, D.; Prassolov, V.; Gaifullin, N.; et al. Molecular pathway activation features linked with transition from normal skin to primary and metastatic melanomas in human. Oncotarget 2016, 7, 656–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turajlic, S.; Swanton, C. Metastasis as an evolutionary process. Science 2016, 352, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging biological principles of metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, C.A. Selection and adaptation during metastatic cancer progression. Nature 2013, 501, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Stoecklein, N.H.; Klein, C.A. Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int. J. Cancer 2010, 126, 589–598. [Google Scholar] [CrossRef]
- Eyles, J.; Puaux, A.L.; Wang, X.; Tan, T.G.; Zheng, L.; Ong, L.C.; Jin, Y.; Kato, M.; Prevost-Blondel, A.; Chow, P.; et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J. Clin. Investig. 2010, 120, 2030–2039. [Google Scholar] [CrossRef]
- Klein, C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef]
- Brastianos, P.K.; Carter, S.L.; Santagata, S.; Cahill, D.P.; Taylor-Weiner, A.; Jones, R.T.; Van Allen, E.M.; Lawrence, M.S.; Horowitz, P.M.; Cibulskis, K.; et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015, 5, 1164–1177. [Google Scholar] [CrossRef] [Green Version]
- Werner-Klein, M.; Scheitler, S.; Hoffmann, M.; Hodak, I.; Dietz, K.; Lehnert, P.; Naimer, V.; Polzer, B.; Treitschke, S.; Werno, C.; et al. Genetic alterations driving metastatic colony formation are acquired outside of the primary tumour in melanoma. Nat. Commun. 2018, 9, 595. [Google Scholar] [CrossRef] [Green Version]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014, 371, 1877–1888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizos, H.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Fung, C.; Hyman, J.; Haydu, L.E.; Mijatov, B.; Becker, T.M.; Boyd, S.C.; et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin. Cancer Res. 2014, 20, 1965–1977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatzka, M.V. Targeted Tumor Therapy Remixed-An Update on the Use of Small-Molecule Drugs in Combination Therapies. Cancers 2018, 10, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, J.; Hodi, F.S.; Wolchok, J.D. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 1270–1271. [Google Scholar] [CrossRef] [Green Version]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. KEYNOTE-006 investigators. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [Green Version]
- Gajos-Michniewicz, A.; Czyz, M. Role of miRNAs in Melanoma Metastasis. Cancers 2019, 11, 326. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Zheng, H.; Tse, G.; Chan, M.T.V.; Wu, W.K.K. Long non-coding RNAs in melanoma. Cell Prolif. 2018, 51, e12457. [Google Scholar] [CrossRef] [Green Version]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- Luo, C.; Weber, C.E.M.; Osen, W.; Bosserhoff, A.-K.; Eichmüller, S.B. The role of microRNAs in melanoma. Eur. J. Cell Biol. 2014, 93, 11–22. [Google Scholar] [CrossRef]
- Hirota, K.; Miyoshi, T.; Kugou, K.; Hoffman, C.S.; Shibata, T.; Ohta, K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 2008, 456, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N. Noncoding RNA and Polycomb recruitment. RNA 2013, 19, 429–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Segura, M.F.; Greenwald, H.S.; Hanniford, D.; Osman, I.; Hernando, E. MicroRNA and cutaneous melanoma: From discovery to prognosis and therapy. Carcinogenesis 2012, 33, 1823–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannavola, F.; Tucci, M.; Felici, C.; Stucci, S.; Silvestris, F. miRNAs in melanoma: A defined role in tumor progression and metastasis. Exp. Rev. Clin. Immunol. 2016, 12, 79–89. [Google Scholar] [CrossRef]
- Varrone, F.; Caputo, E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 878. [Google Scholar] [CrossRef] [Green Version]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–222. [Google Scholar] [CrossRef]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef] [Green Version]
- Bell, R.E.; Levy, C. The three M’s: Melanoma, microphthalmia-associated transcription factor and microRNA. Pigment. Cell Melanoma Res. 2011, 24, 1088–1106. [Google Scholar] [CrossRef]
- Bemis, L.T.; Chen, R.; Amato, C.M.; Classen, E.H.; Robinson, S.E.; Coffey, D.G.; Erickson, P.F.; Shellman, G.; Robinson, W.A.F. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008, 68, 1362–1368. [Google Scholar] [CrossRef] [Green Version]
- Segura, M.F.; Hanniford, D.; Menendez, S.; Reavie, L.; Zou, X.; Alvarez-Diaz, S.; Zakrzewski, J.; Blochin, E.; Rose, A.; Bogunovic, D.; et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. USA 2009, 106, 1814–1819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyle, G.M.; Woods, S.L.; Bonazzi, V.F.; Stark, M.S.; Hacker, E.; Aoude, L.G.; Dutton-Regester, K.; Cook, A.L.; Sturm, R.A.; Hayward, N.K. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011, 24, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wei, Z.; Guo, Y. MicroRNA-107 is a novel tumor suppressor targeting POU3F2 in melanoma. Biol. Res. 2020, 53, 11. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Tetteh, P.W.; Merz, P.R.; Dickes, E.; Abukiwan, A.; Hotz-Wagenblatt, A.; Holland-Cunz, S.; Sinnberg, T.; Schittek, B.; Schadendorf, D.; et al. MiR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J. Investig. Derm. 2013, 133, 768–775. [Google Scholar] [CrossRef] [Green Version]
- Mueller, D.W.; Rehli, M.; Bosserhoff, A.K. MiRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J. Investig. Derm. 2009, 129, 1740–1751. [Google Scholar] [CrossRef] [Green Version]
- Bell, R.E.; Khaled, M.; Netanely, D.; Schubert, S.; Golan, T.; Buxbaum, A.; Janas, M.M.; Postolsky, B.; Goldberg, M.S.; Shamir, R.; et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J. Investig. Derm. 2014, 134, 441–451. [Google Scholar] [CrossRef] [Green Version]
- Mazar, J.; DeYoung, K.; Khaitan, D.; Meister, E.; Almodovar, A.; Goydos, J.; Ray, A.; Perera, R.J. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS ONE 2010, 5, e13779. [Google Scholar] [CrossRef] [Green Version]
- Arozarena, I.; Sanchez-Laorden, B.; Packer, L.; Hidalgo-Carcedo, C.; Hayward, R.; Viros, A.; Sahai, E.; Marais, R. Oncogenic BRAF induces melanoma cell invasion by downregulating the cGMP-specific phosphodiesterase PDE5A. Cancer Cell 2011, 19, 45–57. [Google Scholar] [CrossRef]
- Pan, Z.; Tian, Y.; Niu, G.; Cao, C. Role of microRNAs in remodeling the tumor microenvironment (Review). Int. J. Oncol. 2020, 56, 407–416. [Google Scholar] [CrossRef] [Green Version]
- Müller, D.W.; Bosserhoff, A.K. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 2008, 27, 6698–6706. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.Y.; Chang, C.C.; Lin, C.T.; Lai, C.H.; Peng, S.Y.; Ko, Y.J.; Tang, P.C. Let-7b-mediated suppression of basigin expression and metastasis in mouse melanoma cells. Exp. Cell Res. 2011, 317, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Martin del Campo, S.E.; Latchana, N.; Levine, K.M.; Grignol, V.P.; Fairchild, E.T.; Jaime-Ramirez, A.C.; Dao, T.V.; Karpa, V.I.; Carson, M.; Ganju, A.; et al. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: In vivo effects of MiR-21 inhibitor. PLoS ONE 2015, 10, e0115919. [Google Scholar] [CrossRef] [PubMed]
- Gaziel-Sovran, A.; Segura, M.F.; Di Micco, R.; Collins, M.K.; Hanniford, D.; Vega-Saenz de Miera, E.; Rakus, J.F.; Dankert, J.F.; Shang, S.; Kerbel, R.S.; et al. MiR30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 2011, 20, 104–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoll, S.; Fürst, K.; Kowtharapu, B.; Schmitz, U.; Marquardt, S.; Wolkenhauer, O.; Martin, H.; Pützer, B.M. E2F1 induces miR 224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep. 2014, 15, 1315–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, X.; Zheng, L.; Shen, J.; Zhang, D.; Xiong, M.; Zhang, Y.; He, X.; Tanyi, J.L.; Yang, F.; Montone, K.T.; et al. Suppression of MicroRNA 200 Family Expression by Oncogenic KRAS Activation Promotes Cell Survival and Epithelial-Mesenchymal Transition in KRAS-Driven Cancer. Mol. Cell. Biol. 2016, 36, 2742–2754. [Google Scholar] [CrossRef] [Green Version]
- Penna, E.; Orso, F.; Cimino, D.; Tenaglia, E.; Lembo, A.; Quaglino, E.; Poliseno, L.; Haimovic, A.; Osella-Abate, S.; De Pittà, C.; et al. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011, 30, 1990–2007. [Google Scholar] [CrossRef] [Green Version]
- Rambow, F.; Bechadergue, A.; Luciani, F.; Gros, G.; Domingues, M.; Bonaventure, J.; Meurice, G.; Marine, J.C.; Larue, L. Regulation of Melanoma Progression through the TCF4/miR-125b/NEDD9 Cascade. J. Investig. Derm. 2016, 136, 1229–1237. [Google Scholar] [CrossRef] [Green Version]
- Domingues, M.J.; Rambow, F.; Job, B.; Papon, L.; Liu, W.; Larue, L.; Bonaventure, J. Beta-catenin inhibitor ICAT modulates the invasive motility of melanoma cells. Cancer Res. 2014, 74, 1983–1995. [Google Scholar] [CrossRef] [Green Version]
- Pencheva, N.; Tran, H.; Buss, C.; Huh, D.; Drobnjak, M.; Busam, K.; Tavazoie, S.F. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8 dependent melanoma metastasis and angiogenesis. Cell 2012, 151, 1068–1082. [Google Scholar] [CrossRef] [Green Version]
- Migliore, C.; Petrelli, A.; Ghiso, E.; Corso, S.; Capparuccia, L.; Eramo, A.; Comoglio, P.M.; Giordano, S. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008, 68, 10128–10136. [Google Scholar] [CrossRef] [Green Version]
- Tucci, M.; Mannavola, F.; Passarelli, A.; Stucci, L.S.; Cives, M.; Silvestris, F. Exosomes in melanoma: A role in tumor progression, metastasis and impaired immune system activity. Oncotarget 2018, 9, 20826–20837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowda, R.; Robertson, B.M.; Iyer, S.; Barry, J.; Dinavahi, S.S.; Robertson, G.P. The role of exosomes in metastasis and progression of melanoma. Cancer Treat. Rev. 2020, 85, 101975. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hood, J.L. Melanoma exosomes enable tumor tolerance in lymph nodes. Med. Hypotheses 2016, 90, 11–13. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [Green Version]
- Dinescu, S.; Ignat, S.; Lazar, A.D.; Constantin, C.; Neagu, M.; Costache, M. Epitranscriptomic Signatures in lncRNAs and Their Possible Roles in Cancer. Genes 2019, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Dalay, N. Role of the lncRNAs in malignant melanoma and their involvement in metastasis. Transl Cancer Res. 2016, 5, S758–S764. [Google Scholar] [CrossRef]
- Khaitan, D.; Dinger, M.E.; Mazar, J.; Crawford, J.; Smith, M.A.; Mattick, J.S.; Perera, R.J. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011, 71, 3852–3862. [Google Scholar] [CrossRef] [Green Version]
- Mazar, J.; Zhao, W.; Khalil, A.M.; Lee, B.; Shelley, J.; Govindarajan, S.S.; Yamamoto, F.; Ratnam, M.; Aftab, M.N.; Collins, S.; et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget 2014, 5, 8959–8969. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Liu, X.H.; Lu, K.H.; Nie, F.Q.; Xia, R.; Kong, R.; Yang, J.S.; Xi, T.P.; Liu, Y.W.; Zou, Y.F.; et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014, 5, e1298. [Google Scholar] [CrossRef] [PubMed]
- Siena, Á.D.D.; Plaça, J.R.; Araújo, L.F.; Ichihara de Barros, I.; Peronni, K.; Molfetta, G.; Oliveira de Biagi, C.A., Jr.; Espreafico, E.M.; Sousa, J.F.; Silva, W.A., Jr. Whole transcriptome analysis reveals correlation of long noncoding RNA ZEB1-AS1 with invasive profile in melanoma. Sci. Rep. 2019, 9, 11350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, K.; Joyce, C.E.; Buquicchio, F.; Brown, A.; Ritz, J.; Distel, R.J.; Yoon, C.H.; Novina, C.D. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep. 2016, 15, 2025–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hombach, S.; Kretz, M. The non-coding skin: Exploring the roles of long non-coding RNAs in epidermal homeostasis and disease. Bioessays 2013, 35, 1093–1100. [Google Scholar] [CrossRef]
- Flockhart, R.J.; Webster, D.E.; Qu, K.; Mascarenhas, N.; Kovalski, J.; Kretz, M.; Khavari, P.A. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012, 22, 1006–1014. [Google Scholar] [CrossRef] [Green Version]
- Akhbari, P.; Whitehouse, A.; Boyne, J.R. Long non-coding RNAs drive metastatic progression in melanoma. Int J. Oncol. 2014, 45, 2181–2186. [Google Scholar] [CrossRef] [Green Version]
- Pasmant, E.; Laurendeau, I.; Heron, D.; Vidaud, M.; Vidaud, D.; Bieche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Rachakonda, P.S.; Heidenreich, B.; Nagore, E.; Sucker, A.; Hemminki, K.; Schadendorf, D.; Kumar, R. Mapping of deletion breakpoints at the CDKN2A locus in melanoma: Detection of MTAP-ANRIL fusion transcripts. Oncotarget 2016, 7, 16490–16504. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Wang, H.; Pan, H.; Shi, Y.; Li, T.; Ge, S.; Jia, R.; Zhang, H.; Fan, X. ANRIL lncRNA triggers efficient therapeutic efficacy by reprogramming the aberrant INK4-hub in melanoma. Cancer Lett. 2016, 381, 41–48. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Tang, L.; Zhang, W.; Su, B.; Yu, B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed. Res. Int. 2013, 2013, 251098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Zhang, X.; Hao, Y.; Fang, Z.; He, Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014, 24, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Sun, Q.; Zhao, L.; Wu, J.; Chen, X.; Wang, Y.; Zang, W.; Zhao, G. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med. Oncol. 2016, 33, 88. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.M.; Lin, X.; Zhang, L.; Hong, W.; Zhang, Y. Long noncoding RNA X-inactive specific transcript promotes malignant melanoma progression and oxaliplatin resistance. Melanoma Res. 2019, 29, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, H.; Xiao, Y.; Tang, X.; Li, Y.; Han, Q.; Fu, J.; Yang, Y.; Zhu, Y. LncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo. OncoTargets 2016, 9, 4075–4087. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Zhu, L.; Li, Y.; Zheng, Z.; Lin, X.; Yang, C. LncRNA MEG3 promotes melanoma growth, metastasis and formation through modulating miR-21/E-cadherin axis. Cancer Cell Int. 2020, 20, 12. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Xiao, Y.; Tang, X.; Li, Y.; Han, Q.; Fu, J.; Yang, Y.; Zhu, Y. Lentiviral-mediated overexpression of long noncoding RNA GAS5 reduces invasion by mediating MMP2 expression and activity in human melanoma cells. Int J. Oncol. 2016, 48, 1509–1518. [Google Scholar] [CrossRef]
- Bian, D.; Shi, W.; Shao, Y.; Li, P.; Song, G. Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma. Am. J. Transl. Res. 2017, 9, 1509–1520. [Google Scholar]
- Lorusso, C.; De Summa, S.; Pinto, R.; Danza, K.; Tommasi, S. miRNAs as Key Players in the Management of Cutaneous Melanoma. Cells 2020, 9, 415. [Google Scholar] [CrossRef] [Green Version]
- Dror, S.; Sander, L.; Schwartz, H.; Sheinboim, D.; Barzilai, A.; Dishon, Y.; Apcher, S.; Golan, T.; Greenberger, S.; Barshack, I.; et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat. Cell Biol. 2016, 18, 1006–1017. [Google Scholar] [CrossRef]
- Hanna, S.C.; Krishnan, B.; Bailey, S.T.; Moschos, S.J.; Kuan, P.F.; Shimamura, T.; Osborne, L.D.; Siegel, M.B.; Duncan, L.M.; O’Brien, E.T., 3rd; et al. HIF1α and HIF2α independently activate SRC to promote melanoma metastases. J. Clin. Investig. 2013, 123, 2078–9203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wozniak, M.; Peczek, L.; Czernek, L.; Düchler, M. Analysis of the miRNA profiles of melanoma exosomes derived under normoxic and hypoxic culture conditions. Anticancer Res. 2017, 37, 6779–6789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, H.W.; Baxter, L.L.; Loftus, S.K.; Cronin, J.C.; Trivedi, N.S.; Borate, B.; Pavan, W.J. Distinct microRNA expression signatures are associated with melanoma subtypes and are regulated by HIF1A. Pigment Cell Melanoma Res. 2014, 27, 777–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, H.; Van Eygen, S.; Krysko, D.V.; Vandenabeele, P.; Nys, K.; Rillaerts, K.; Garg, A.D.; Verfaillie, T.; Agostinis, P. BNIP3 supports melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton. Cell Death Dis. 2014, 5, e1127. [Google Scholar] [CrossRef] [Green Version]
- Vara-Perez, M.; Maes, H.; Van Dingenen, S.; Agostinis, P. BNIP3 contributes to the glutamine-driven aggressive behavior of melanoma cells. Biol. Chem. 2019, 400, 187–193. [Google Scholar] [CrossRef]
- Mazar, J.; Qi, F.; Lee, B.; Marchica, J.; Govindarajan, S.; Shelley, J.; Li, J.-L.; Ray, A.; Perera, R.J. MicroRNA 211 Functions as a Metabolic Switch in Human Melanoma Cells. Mol. Cell. Biol. 2016, 36, 1090–1108. [Google Scholar] [CrossRef] [Green Version]
- Chung, A.S.; Lee, J.; Ferrara, N. Targeting the tumour vasculature: Insights from physiological angiogenesis. Nat. Rev. Cancer 2010, 10, 505–514. [Google Scholar] [CrossRef]
- Bergers, G.; Benjamin, L.E. Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer 2003, 3, 401–410. [Google Scholar] [CrossRef]
- Zhuang, G.; Wu, X.; Jiang, Z.; Kasman, I.; Yao, J.; Guan, Y.; Oeh, J.; Modrusan, Z.; Bais, C.; Sampath, D.; et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012, 31, 3513–3523. [Google Scholar] [CrossRef] [PubMed]
- Crawford, Y.; Kasman, I.; Yu, L.; Zhong, C.; Wu, X.; Modrusan, Z.; Kaminker, J.; Ferrara, N. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell 2009, 15, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderberg, C.; Li, H.; Fredriksson, L.; Andrae, J.; Betsholtz, C.; Li, X.; Eriksson, U.; Pietras, K. Paracrine signaling by platelet-derived growth factor-CC promotes tumor growth by recruitment of cancer-associated fibroblasts. Cancer Res. 2009, 69, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallacchi, V.; Camisaschi, C.; Dugo, M.; Vergani, E.; Deho, P.; Gualeni, A.; Huber, V.; Gloghini, A.; Maurichi, A.; Santinami, M.; et al. MicroRNA Expression in Sentinel Nodes from Progressing Melanoma Patients Identifies Networks Associated with Dysfunctional Immune Response. Genes 2016, 7, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noman, M.Z.; Buart, S.; Romero, P.; Ketari, S.; Janji, B.; Mari, B.; Mami-Chouaib, F.; Chouaib, S. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res. 2012, 72, 4629–4641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Wang, L.; Fan, J.; Ye, C.; Dominguez, D.; Zhang, Y.; Curiel, T.J.; Fang, D.; Kuzel, T.M.; Zhang, B. Host miR155 promotes tumor growth through a myeloid-derived suppressor cell-dependent mechanism. Cancer Res. 2015, 75, 519–531. [Google Scholar] [CrossRef] [Green Version]
- Valenti, R.; Huber, V.; Iero, M.; Filipazzi, P.; Parmiani, G.; Rivoltini, L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007, 67, 2912–2915. [Google Scholar] [CrossRef] [Green Version]
- Ding, H.; Yang, X.; Wei, Y. Fusion Proteins of NKG2D/NKG2DL in Cancer Immunotherapy. Int. J. Mol. Sci. 2018, 19, 177. [Google Scholar] [CrossRef] [Green Version]
- Heinemann, A.; Zhao, F.; Pechlivanis, S.; Eberle, J.; Steinle, A.; Diederichs, S.; Schadendorf, D.; Paschen, A. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012, 72, 460–471. [Google Scholar] [CrossRef] [Green Version]
- Paschen, A.; Sucker, A.; Hill, B.; Moll, I.; Zapatka, M.; Nguyen, X.D.; Sim, G.C.; Gutmann, I.; Hassel, J.; Becker, J.C.; et al. Differential clinical significance of individual NKG2D ligands in melanoma: Soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin. Cancer Res. 2009, 15, 5208–5215. [Google Scholar] [CrossRef] [Green Version]
- Pucci, F.; Garris, C.; Lai, C.P.; Newton, A.; Pfirschke, C.; Engblom, C.; Alvarez, D.; Sprachman, M.; Evavold, C.; Magnuson, A.; et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science 2016, 352, 242–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Chen, Y.; Li, A.; Tan, C.; Liu, X. Exosomes Play Roles in Sequential Processes of Tumor Metastasis. Int J. Cancer 2019, 144, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Cantile, M.; Scognamiglio, G.; Marra, L.; Aquino, G.; Botti, C.; Falcone, M.; Malzone, M.G.; Liguori, G.; Di Bonito, M.; Franco, R.; et al. HOTAIR role in melanoma progression and its identification in the blood of patients with advanced disease. J. Cell Physiol. 2017, 232, 3422–3432. [Google Scholar] [CrossRef]
- Obaid, M.; Udden, S.M.N.; Deb, P.; Shihabeddin, N.; Zaki, M.H.; Mandal, S.S. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Sci. Rep. 2018, 8, 15670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botti, G.; Scognamiglio, G.; Aquino, G.; Liguori, G.; Cantile, M. LncRNA HOTAIR in Tumor Microenvironment: What Role? Int. J. Mol. Sci. 2019, 20, 2279. [Google Scholar] [CrossRef] [Green Version]
- Vergani, E.; Di Guardo, L.; Dugo, M.; Rigoletto, S.; Tragni, G.; Ruggeri, R.; Perrone, F.; Tamborini, E.; Gloghini, A.; Arienti, F.; et al. Overcoming melanoma resistance to vemurafenib by targeting CCL2-induced miR-34a, miR-100 and miR-125b. Oncotarget 2016, 7, 4428–4441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koetz-Ploch, L.; Hanniford, D.; Dolgalev, I.; Sokolova, E.; Zhong, J.; Diaz-Martinez, M.; Bernstein, E.; Darvishian, F.; Flaherty, K.T.; Chapman, P.B.; et al. MicroRNA-125a promotes resistance to BRAF inhibitors through suppression of the intrinsic apoptotic pathway. Pigment Cell Melanoma Res. 2017, 30, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Martínez, M.; Benito-Jardón, L.; Alonso, L.; Koetz-Ploch, L.; Hernando, E.; Teixidó, J. MiR-204-5p and miR-211-5p Contribute to BRAF Inhibitor Resistance in Melanoma. Cancer Res. 2018, 78, 1017–1030. [Google Scholar] [CrossRef] [Green Version]
- Vitiello, M.; D’Aurizio, R.; Poliseno, L. Biological role of miR-204 and miR-211 in melanoma. Oncoscience 2018, 5, 248–251. [Google Scholar] [CrossRef]
- Stark, M.S.; Bonazzi, V.F.; Boyle, G.M.; Palmer, J.M.; Symmons, J.; Lanagan, C.M.; Schmidt, C.W.; Herington, A.C.; Ballotti, R.; Pollock, P.M.; et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget 2015, 6, 17753–17763. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Li, J.; Sun, Y.; Zhang, Y.; Dong, L.; Shen, C.; Yang, L.; Yang, M.; Li, Y.; Shen, G.; et al. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget 2016, 7, 53558–53570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, P.J.; Merlino, G. Integrated Genomics Identifies miR-32/MCL-1 Pathway as a Critical Driver of Melanomagenesis: Implications for miR-Replacement and Combination Therapy. PLoS ONE 2016, 11, e0165102. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tetzlaff, M.T.; Wang, T.; Yang, R.; Xie, L.; Zhang, G.; Krepler, C.; Xiao, M.; Beqiri, M.; Xu, W.; et al. miR-200c/Bmi1 axis and epithelial-mesenchymal transition contribute to acquired resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res. 2015, 28, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Fattore, L.; Mancini, R.; Acunzo, M.; Romano, G.; Lagana, A.; Pisanu, M.E.; Malpicci, D.; Madonna, G.; Mallardo, D.; Capone, M.; et al. miR-579-3p controls melanoma progression and resistance to target therapy. Proc. Natl. Acad. Sci. USA 2016, 113, E5005–E5013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanlorenzo, M.; Vujic, I.; Esteve-Puig, R.; Lai, K.; Vujic, M.; Lin, K.; Posch, C.; Dimon, M.; Moy, A.; Zekhtser, M.; et al. The lincRNA MIRAT binds to IQGAP1 and modulates the MAPK pathway in NRAS mutant melanoma. Sci. Rep. 2018, 8, 10902. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 2017, 548, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Vishnubalaji, R.; Hibah, S.; Elango, R.; Alajez, N.M. Noncoding RNAs as potential mediators of resistance to cancer immunotherapy. Semin. Cancer Biol. 2020, 65, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhu, Y.; Xie, Y.; Ma, X. The Role of Long Non-coding RNAs in Immunotherapy Resistance. Front. Oncol. 2019, 9, 1292. [Google Scholar] [CrossRef] [Green Version]
- Huber, V.; Vallacchi, V.; Fleming, V.; Hu, X.; Cova, A.; Dugo, M.; Shahaj, E.; Sulsenti, R.; Vergani, E.; Filipazzi, P.; et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Investig. 2018, 128, 5505–5516. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Johnston, N.; Zheng, X.; Wang, H.; Zhang, X.; Gao, D.; Min, W. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget 2016, 7, 53735–53750. [Google Scholar] [CrossRef] [Green Version]
- Audrito, V.; Serra, S.; Stingi, A.; Orso, F.; Gaudino, F.; Bologna, C.; Neri, F.; Garaffo, G.; Nassini, R.; Baroni, G.; et al. PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget 2017, 8, 15894–15911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, W.; Gao, Y.; Tang, Z.; Zhang, Y.; Yang, R. The pseudogene Olfr29-ps1 promotes the suppressive function and differentiation of monocytic MDSCs. Cancer Immunol. Res. 2019, 7, 813–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Wang, T.; Li, Y.; Zhang, Y.; Yang, R. Lnc-chop promotes immunosuppressive function of myeloid-derived suppressor cells in tumor and inflammatory environments. J. Immunol. 2018, 200, 2603–2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charpentier, M.; Croyal, M.; Carbonnelle, D.; Fortun, A.; Florenceau, L.; Rabu, C.; Krempf, M.; Labarriere, N.; Lang, F. IRES-dependent translation of the long non coding RNA meloe in melanoma cells produces the most immunogenic MELOE antigens. Oncotarget 2016, 7, 59704–59713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godet, Y.; Moreau-Aubry, A.; Guilloux, Y.; Vignard, V.; Khammari, A.; Dreno, B.; Jotereau, F.; Labarriere, N. MELOE-1 is a new antigen overexpressed in melanomas and involved in adoptive T cell transfer efficiency. J. Exp. Med. 2008, 205, 2673–2682. [Google Scholar] [CrossRef] [Green Version]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin. Cancer Biol. 2019, 58, 100–108. [Google Scholar] [CrossRef]
- Leidinger, P.; Keller, A.; Borries, A.; Reichrath, J.; Rass, K.; Jager, S.V.; Lenhof, H.-P.; Meeseet, E. High-throughput miRNA profiling of human melanoma blood samples. BMC Cancer 2010, 10, 262. [Google Scholar] [CrossRef] [Green Version]
- Mumford, S.L.; Towler, B.P.; Pashler, A.L.; Gilleard, O.; Martin, Y.; Newbury, S.F. Circulating MicroRNA Biomarkers in Melanoma: Tools and Challenges in Personalised Medicine. Biomolecules 2018, 8, 21. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, S.R.; Grossmann, K.F.; Cassidy, P.B.; Yang, C.H.; Fan, M.; Kopelovich, L.; Leachman, S.A.; Pfeffer, L.M. Detection of Exosomal miRNAs in the Plasma of Melanoma Patients. J. Clin. Med. 2015, 4, 2012–2027. [Google Scholar] [CrossRef] [PubMed]
- Mannavola, F.; D’Oronzo, S.; Cives, M.; Stucci, L.S.; Ranieri, G.; Silvestris, F.; Tucci, M. Extracellular Vesicles and Epigenetic Modifications Are Hallmarks of Melanoma Progression. Int. J. Mol. Sci. 2020, 21, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armand-Labit, V.; Meyer, N.; Casanova, A.; Bonnabau, H.; Platzer, V.; Tournier, E.; Sansas, B.; Verdun, S.; Thouvenot, B.; Hilselberger, B.; et al. Identification of a Circulating MicroRNA Profile as a Biomarker of Metastatic Cutaneous Melanoma. Acta Derm. Venereol. 2016, 96, 29–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Laar, R.; Lincoln, M.; Van Laar, B. Development and validation of a plasma-based melanoma biomarker suitable for clinical use. Br. J. Cancer 2018, 118, 857–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; He, Q.Y.; Luo, C.Q.; Qian, L.Y. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med. Sci Monit. 2014, 20, 2472–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, N.H.; Zhong, J.; Da Silva, I.P.; De Miera, E.V.S.; Brady, B.; Han, S.W.; Hanniford, D.; Wang, J.; Shapiro, R.L.; Hernando, E.; et al. Serum-based miRNAs in the prediction and detection of recurrence in melanoma patients. Cancer 2015, 121, 51–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, M.S.; Klein, K.; Weide, B.; Haydu, L.E.; Pflugfelder, A.; Tang, Y.H.; Palmer, J.M.; Whiteman, D.C.; Scolyer, R.A.; Mann, G.J.; et al. The Prognostic and Predictive Value of Melanoma-related MicroRNAs Using Tissue and Serum: A MicroRNA Expression Analysis. EBioMedicine 2015, 2, 671–680. [Google Scholar] [CrossRef]
- Liu, T.; Shen, S.K.; Xiong, J.G.; Xu, Y.; Zhang, H.Q.; Liu, H.J.; Lu, Z.G. Clinical significance of long noncoding RNA SPRY4-IT1 in melanoma patients. FEBS Open Bio 2016, 6, 147–154. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Hulstaert, E.; Brochez, L.; Volders, P.J.; Vandesompele, J.; Mestdagh, P. Long non-coding RNAs in cutaneous melanoma: Clinical perspectives. Oncotarget 2017, 8, 43470–43480. [Google Scholar] [CrossRef] [Green Version]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef] [PubMed]
- Fattore, L.; Campani, V.; Ruggiero, C.F.; Salvati, V.; Liguoro, D.; Scotti, L.; Botti, G.; Ascierto, P.A.; Mancini, R.; De Rosa, G.; et al. In Vitro Biophysical and Biological Characterization of Lipid Nanoparticles Co-Encapsulating Oncosuppressors miR-199b-5p and miR-204-5p as Potentiators of Target Therapy in Metastatic Melanoma. Int. J. Molec. Sci. 2020, 21, 1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aftab, M.N.; Dinger, M.E.; Perera, R.J. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch. Biochem. Biophys. 2014, 563, 60–70. [Google Scholar] [CrossRef] [Green Version]

| Induced Effect | Overexpression of ncRNAs | Target(s) | Drugs | References |
|---|---|---|---|---|
| Drug resistance | miR-34a, miR-100 and miR-125b | CCL-2 | Vemurafenib | [117] |
| miR-125a | BAK1, MLK3 | [118] | ||
| miR-204 and miR-211 | NUAK1/ARK5, IGFBP5, TGF-bRII, Slug, CHD5 | [119,120] | ||
| miR-514a | NF1 | [121] | ||
| MIRAT | IQGAP1/MAPK signalling | Trametinib (MEKi) and PLX4720 (BRAFi) | [126] | |
| SAMMSON | p32 mitochondrial protein | Vemurafenib and pimasertib (MEKi) | [81] | |
| EMICERI | MOB3B/LATS1/Hippo signalling axis | Vemurafenib | [127] | |
| Drug sensitivity | miR-7 | EGFR/IGF-1R/CRAF | Vemurafenib | [122] |
| miR-32 | MCL-1 | [123] | ||
| miR-200c | Bmi-1 | Vemurafenib or analog PLX4720 | [124] | |
| miR-579-3p | BRAF, MDM2 | Vemurafenib and trametinib | [125] |
| Name of ncRNAs | Function | Immune Response | References |
|---|---|---|---|
| miR-28 | Blocks immune checkpoint Silences PD-1 by binding to its 3′UTR region | Negative | [131] |
| miR-17-5p | Blocks immune checkpoint ligand Binds to PD-L1 and contributes to melanoma resistance | Negative | [132] |
| Let-7e, miR-99b, miR-100, miR-125a, miR-125b, miR-146a, miR-146b, miR-155 | Control of MDSC Favours myeloid cell differentiation and polarization towards an immunosuppressive phenotype | Negative | [130] |
| Olfr29-ps1 | Control of MDSC Promotes MDSCs’ differentiation and function via de m6A-modified Olfr29-ps1/miR-214-3p/MyD88 regulatory network | Negative | [133] |
| Lnc-chop | Control of MDSC Promotes the differentiation and function of MDSCs | Negative | [134] |
| MELOE | Antigen presentation Produces immunogenic antigens (MELOE-1 and -2) that are recognized by cytotoxic T cells | Positive | [135] |
| Potential Role | Non-Coding RNAs | Sample Type | References |
|---|---|---|---|
| Circulating biomarkers for prognosis and diagnosis | 16 miRNA panel (among them miR-30d and miR-17) | Blood | [140] |
| miR-19a miR-126 miR-149 | Plasma | [141,142,143] | |
| miR-185 miR-1246 | [144] | ||
| 18 miRNA panel (among them miR-199b-5p and let-7e) | [145] | ||
| miR-221 | Serum | [146] | |
| miR-15b miR-30d miR-150 miR-425 | [147] | ||
| 7 miRNA panel (among them miR-16, miR-211-5p, miR-4706 and miR-509) | [148] | ||
| SPRY4-IT | Plasma | [149] | |
| HOTAIR | Serum | [82,114] |
| Potential Role | Functional Studies | Research Model | Therapeutic Effect(s) | References |
|---|---|---|---|---|
| Targets for promising therapeutic strategies | Lentiviral overexpression of miR-200c | BRAFi-resistant cell lines | Restores sensitivity to BRAFi therapies | [124] |
| Lipid nanoparticles loaded with miR-204-5p and/or miR-199b-5p | In vitro drug resistant models | Impair melanoma cell proliferation and viability Positively influence the effect of MAPKi | [154] | |
| siRNA-mediated knockdown of SPRY4-IT1 | Malignant melanoma cell lines | Prevents tumour cell growth and limits invasion | [70,71] | |
| siRNA-mediated knockdown of HOTAIR | Inhibits cell motility and decreases invasion | [82] | ||
| siRNA-mediated knockdown of UCA1 | Inhibits cell proliferation and invasion Induces cell cycle arrest | [84] | ||
| siRNA-mediated knockdown of MALAT1 | Impairs melanoma cell migration | [83] | ||
| siRNA-mediated knockdown of ANRIL | Diminishes colony formation and metastatic ability | [80] | ||
| siRNA-mediated knockdown of SLNCR1 | Decreases invasiveness of melanoma cells | [74] | ||
| Lentiviral overexpression of GAS5 | In vitro and in vivo models | Inhibits melanoma growth and cell migration | [86] | |
| SAMMSON-specific antisense oligonucleotide | Patient- derived xenograft | Induces apoptosis Exerts a synergistic anti-tumour effect with dabrafenib | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazăr, A.D.; Dinescu, S.; Costache, M. The Non-Coding Landscape of Cutaneous Malignant Melanoma: A Possible Route to Efficient Targeted Therapy. Cancers 2020, 12, 3378. https://doi.org/10.3390/cancers12113378
Lazăr AD, Dinescu S, Costache M. The Non-Coding Landscape of Cutaneous Malignant Melanoma: A Possible Route to Efficient Targeted Therapy. Cancers. 2020; 12(11):3378. https://doi.org/10.3390/cancers12113378
Chicago/Turabian StyleLazăr, Andreea D., Sorina Dinescu, and Marieta Costache. 2020. "The Non-Coding Landscape of Cutaneous Malignant Melanoma: A Possible Route to Efficient Targeted Therapy" Cancers 12, no. 11: 3378. https://doi.org/10.3390/cancers12113378