Simple Summary
NKG2C and its ligand HLA-E represent key molecules for NK cell-mediated immune responsiveness. However, the impact of genetic variants in NKG2C and HLA-E on clinical outcomes of aggressive B-cell non-Hodgkin lymphoma patients (B-NHL) has not been clarified. In this study, we analyzed the distribution of NKG2C deletion status and HLA-E variants in 441 patients and 192 healthy individuals. Homozygous deletion of NKG2C (NKG2C−/−) was more often found in high-risk patients compared to patients with a lower risk and consequently was associated with reduced 2-year progression-free survival. The HLA-E*01:01 allele frequency was increased in B-NHL patients and was strongly related with complete remission. Our results show that absence of NKG2C and HLA-E allelic variations is predictive for B-NHL outcome; while carriers of HLA-E*01:01 are characterized by high, complete remission rates, NKG2C−/− was rare, but associated with poorer outcome. Prospective validation of our results identifies patients that may benefit from risk-adapted therapy.
Abstract
Aggressive B-cell lymphomas account for the majority of non-Hodgkin lymphomas (B-NHL). NK cells govern the responses to anti-CD20 monoclonal antibodies and have emerged as attractive targets for immunotherapy in subtypes of B-NHL. NKG2C and its cognate ligand HLA-E represent key molecules for fine-tuning of NK cell-mediated immune responses. Here, we investigated the impact of genetic variants of NKG2C and HLA-E on clinical outcomes of 441 B-NHL patients. Homozygous deletion of NKG2C (NKG2C−/−) was three-fold increased in patients compared to 192 healthy controls. Among studied patients, NKG2C−/− was more abundant in International Prognostic Index (IPI) high-risk patients compared to patients with a lower IPI (p = 0.013). Strikingly, NKG2C−/− was associated with a significantly reduced 2-year PFS (progression-free survival) (p = 0.0062) and represented an independent risk factor for 2-year PFS in multivariate analysis (p = 0.005). For HLA-E, the cognate ligand of NKG2C, the HLA-E*01:01 allele frequency was increased in B-NHL patients compared to controls (p = 0.033) and was associated with complete remission in univariate (p = 0.034) and multivariate (p = 0.018) analysis. Our data suggest that NKG2C and HLA-E genotyping is a promising tool for both defining risk groups of aggressive B-NHL and predicting response to immune therapeutic approaches.
1. Introduction
Aggressive B-cell non-Hodgkin lymphoma (B-NHL) represents malignant tumors of the immune system [1]. First-line treatment consists of the CHOP regimen (cyclophosphamide, doxorubicin, vincristine, and prednisone). The combined treatment with the anti-CD20 antibody rituximab (R-CHOP) has improved the therapy outcome of CD20 positive B-NHL patients with long-term remission rates of 60% to 70%. Consequently, up to 40% of patients experience treatment failure or disease relapse [2]. Therefore, surrogate markers identifying subgroups of patients with high-risk of treatment failure or early disease progression are of paramount importance for precision medicine approaches. The established risk model for B-NHL is based on clinical parameters summarized in the International Prognostic Index (IPI) [3]. However, molecular markers of the primary tumor or of immune effectors are currently not implemented in routine clinical care of B-NHL patients.
In recent years, increasing evidence suggests that NK cells as effectors of the innate immune system are critically involved in surveillance of B-NHL [4]. Particularly, NK cells have been discussed to support the therapeutic effect of CD20-targeting treatment approaches in lymphoma patients by strong Fc receptor-mediated NK cell activation, boosting the antibody-dependent cellular cytotoxicity (ADCC) [4,5]. Immune reactivity of NK cells is largely governed by the inhibitory lectin like receptors NK group 2 member A (NKG2A), and the counterbalancing activating NKG2C receptor, both of which are expressed as heterodimers with the invariant CD94 on the cell surface [6,7,8,9,10]. Expression of NKG2C is a feature of so-called adaptive NK cells operative in inflammation and infections [11,12,13]. CD94-NKG2 receptors recognize the non-classical HLA-E molecule. While HLA-E is widely expressed at low levels under physiological conditions [14,15], it can be up-regulated during pathological processes, including cancer [16,17,18]. HLA-E expression is a critical component for modulating immune responses as it can interact with the activating NKG2C as well as the inhibitory NKG2A receptor.
Genetic variants in NKG2C and HLA-E have been identified and functionally characterized. It has been described that heterozygous or homozygous deletion of a 16-kb section within the NKG2C gene correlates with decrease or absence of NKG2C surface expression, respectively [19,20]. Thus, the copy number of variations affects the adaptive response of NKG2C positive NK cells [12,21,22]. For HLA-E, two main genetic variants, HLA-E*01:01 and HLA-E*01:03, are equally distributed in populations of different origins [23,24]. However, the HLA-E*01:03 encoded protein shows a higher peptide-binding affinity and higher surface expression compared to the HLA-E*01:01 encoded protein [24,25,26]. Currently, the role of genetic variations in NKG2C and HLA-E in balancing anti-tumor immune response or disease outcome in hematological malignancies has not been investigated.
As the NKG2C/HLA-E axis is a central element for fine-tuning the immune responsiveness of NK cells, we here addressed the question if genetic variations in the encoding genes were relevant for clinical outcome and therapy response of B-NHL. We determined allele and genotype frequencies of the NKG2C deletion and HLA-E variants in CD20 positive B-NHL patients (n = 441) and healthy controls (n = 192) and related the results of the patients with clinical characteristics as well as disease outcome.
2. Results
2.1. Homozygous Absence of NKG2C Is More Frequent in International Prognostic Index (IPI) High-Risk Than in IPI Non-High-Risk Patients
While NKG2A and NKG2C are both located on chromosome 12, only the NKG2C locus is deleted in 4–14% of individuals depending on the population analyzed. Here, NKG2C genotype distribution was determined in B-NHL patients (n = 441) and controls (n = 189), respectively. Neither NKG2C deletion nor genotype frequencies of copy number variations were different between B-NHL patients and healthy controls (Table 1). Although homozygous NKG2C deletion (NKG2C−/−) frequency was two-fold higher in B-NHL patients (4.5%) than in healthy controls (1.6%), this finding, however, did not reach statistical significance.

Table 1.
Distribution of NKG2C allelic and genotype frequencies of B-cell non-Hodgkin lymphoma (B-NHL) patients and healthy controls. The NKG2C homozygous deletion allele frequencies and genotype frequencies did not differ among B-NHL patients and healthy controls.
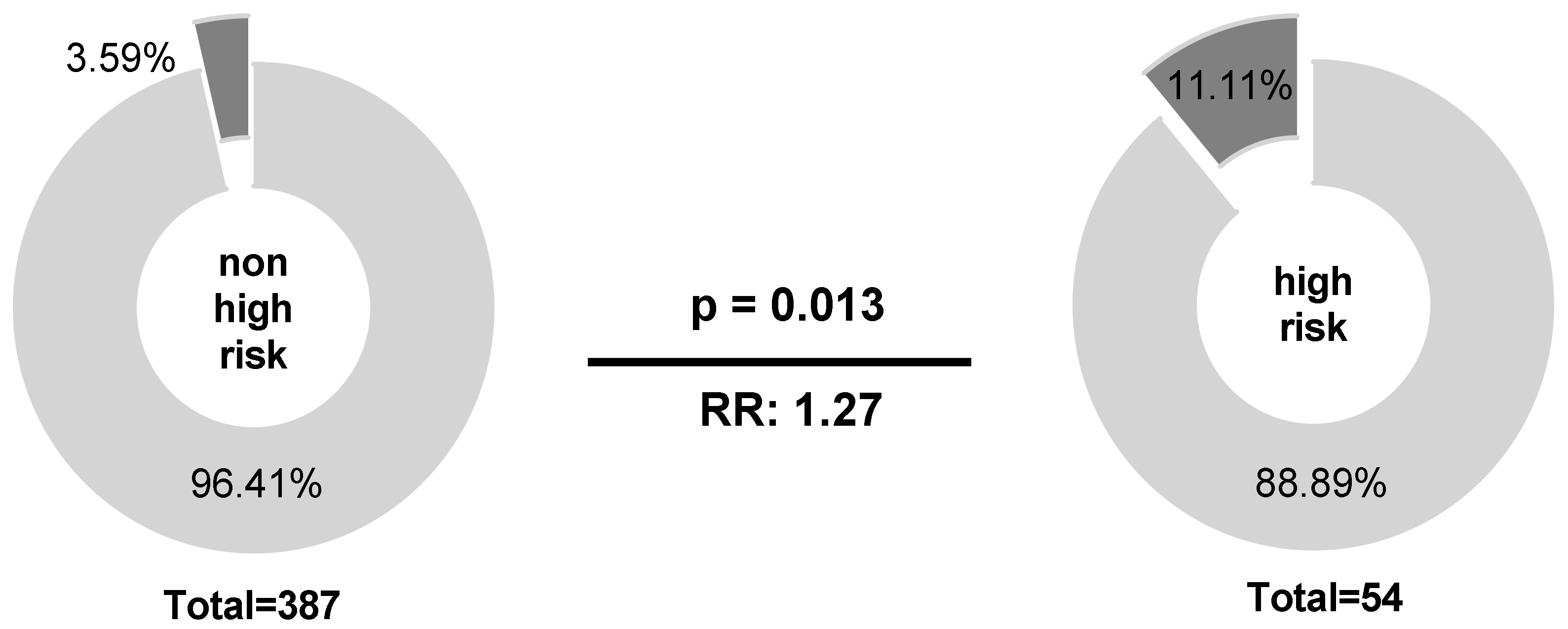
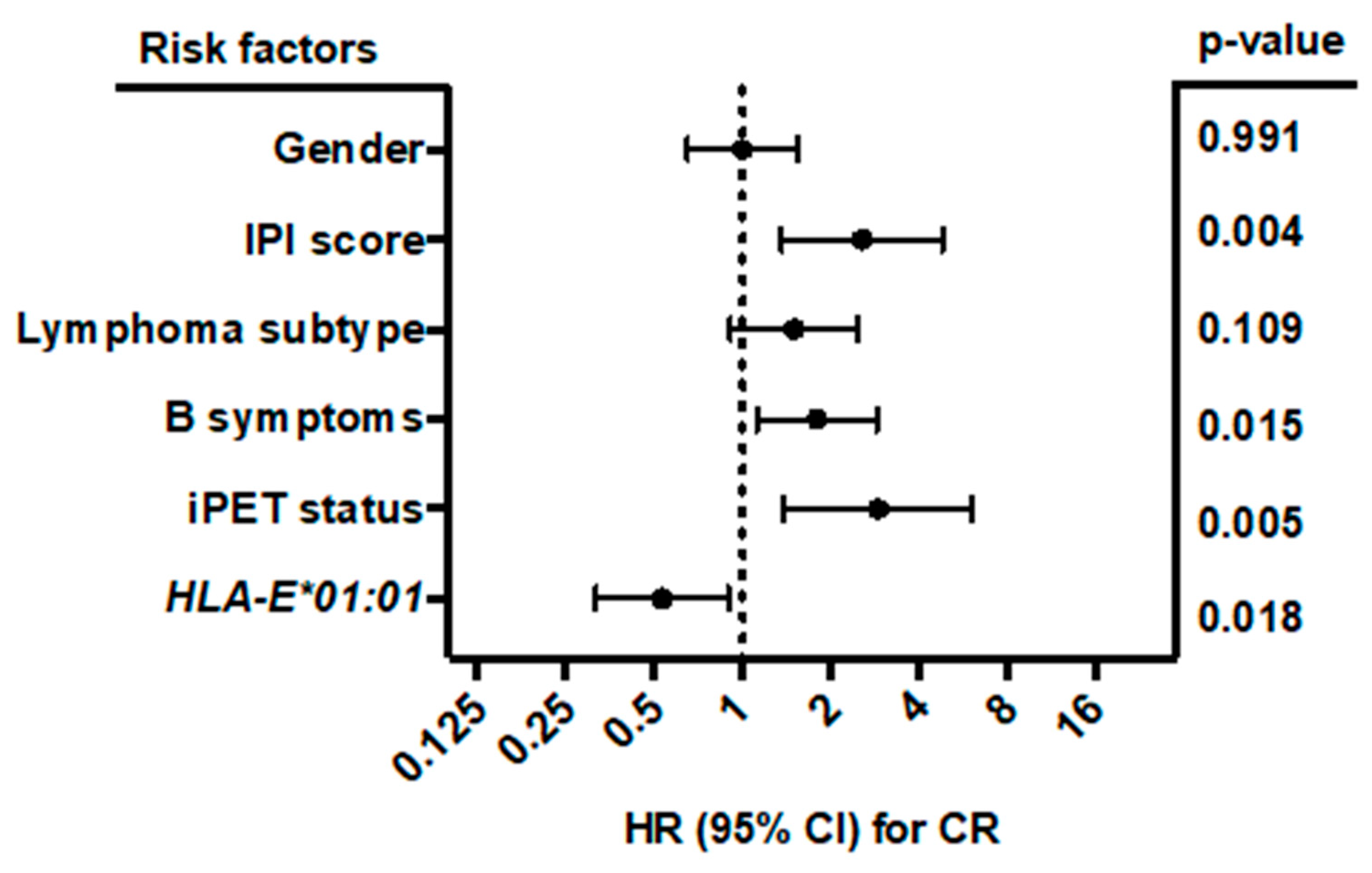
Among B-NHL patients, IPI score was the only clinical parameter associated with NKG2C−/− status (p = 0.013; n = 441). Here, NKG2C−/− status was 3-fold increased in the IPI high-risk group compared to patients with a lower IPI score (p = 0.013; RR: 1.27; 95% CI: 1.032 to 1.844; Figure 1).
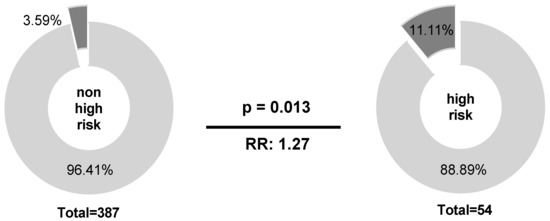
Figure 1.
Correlation of IPI score and the NKG2C−/− status. Higher frequencies of the NKG2C−/− could be observed for patients in the IPI high-risk group (IPI 4–5) compared to those in the non-high-risk group (IPI 0–3; low to high-intermediate risk). NKG2C−/− (dark grey), NKG2C+/− or NKG2C+/+ (light grey). Clinical data were not available for all patients.
2.2. Homozygous Absence of NKG2C Is Associated With Reduced Two-Year PFS
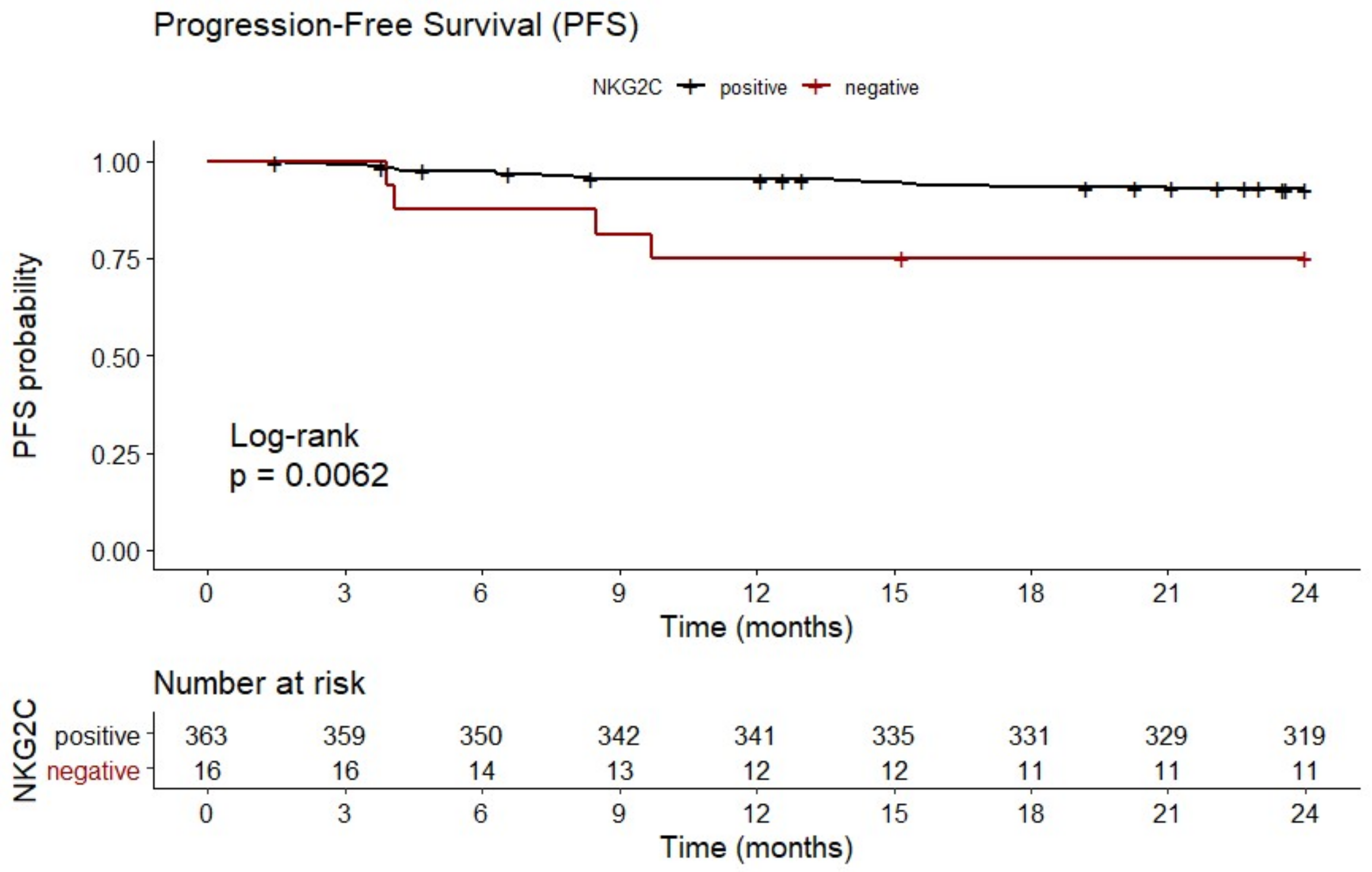
In line with enhanced IPI risk scores, patients with NKG2C−/− experienced a reduced progression-free survival (PFS) (19.1 ± 7.9 months) compared to NKG2C positive patients with NKG2C+/− or NKG2C+/+ (22.5 ± 4.9 months; p = 0.0062; HR: 3.88; 95% CI: 0.60 to 25.09; n = 379; Figure 2).
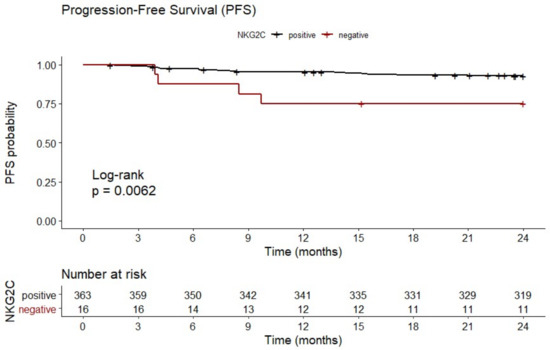
Figure 2.
Reduced 2-year progression-free survival (PFS) in patients with NKG2C−/−. B-NHL patients with homozygous deletion of NKG2C showed a reduced 2-year PFS compared to NKG2C positive patients. NKG2C−/− (negative, red); NKG2C+/− or NKG2C+/+ (positive, black). Data were not available for the total number of patients due to loss of follow up.
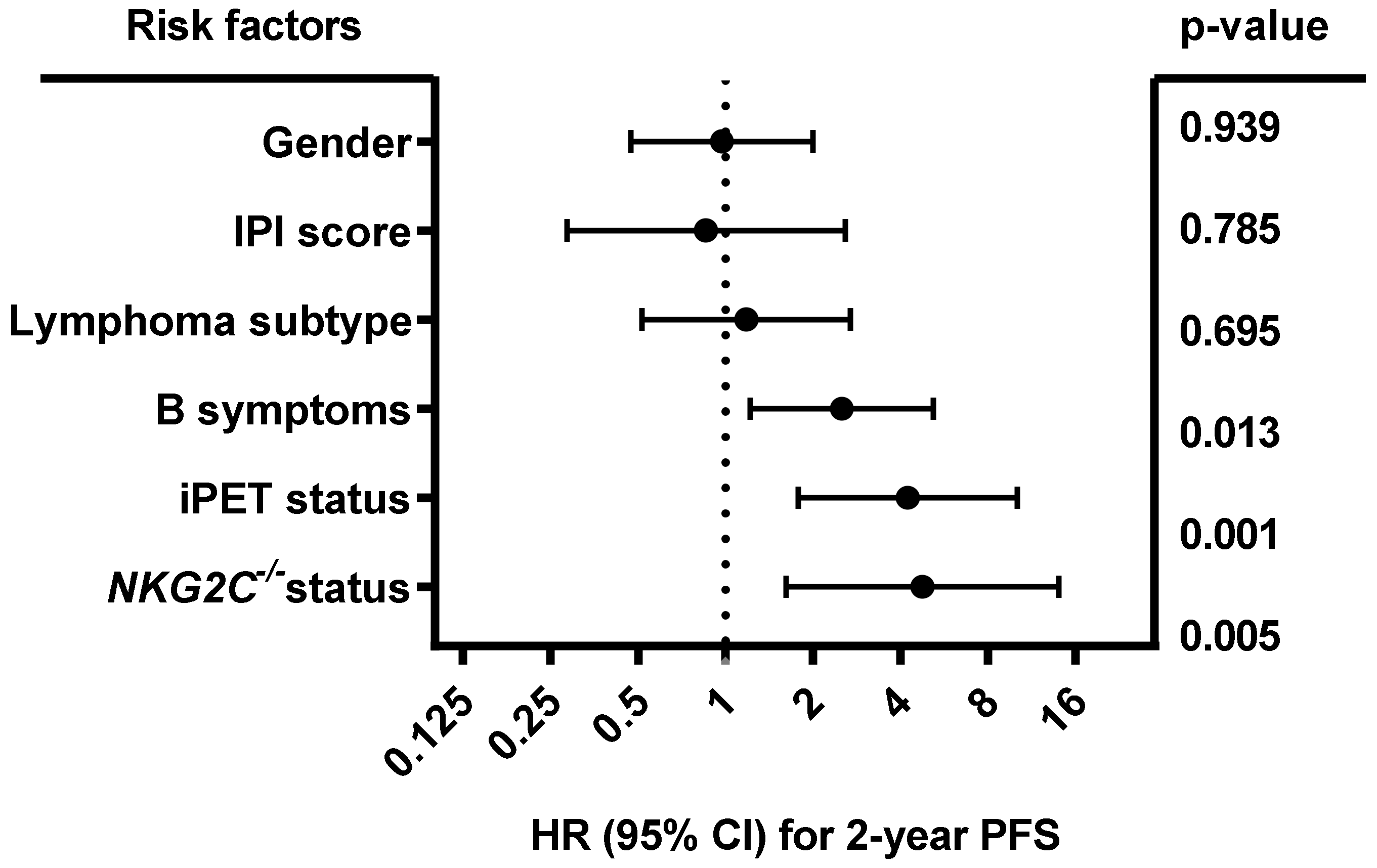
As shown in Figure 3, multivariate analysis including gender (male vs. female), IPI (0–3 vs. 4–5), lymphoma subtype (diffuse large B-cell lymphoma (DLBCL) vs. other), B symptoms (A vs. B), iPET status (favorable iPET vs. unfavorable iPET) and NKG2C−/− status (NKG2C−/− vs. NKG2C+/− or NKG2C+/+) showed that besides B symptoms (p = 0.013; HR:2.51; 95% CI: 1.218 to 5.173) and iPET (p = 0.001; HR: 4.242; 95% CI: 1.781 to 10.102), the NKG2C−/− status was an independent predictive factor for 2-year PFS in B-NHL patients (p = 0.005; HR: 4.758; 95% CI: 1.617 to 13.999; n = 379). No association of NKG2C absence with the overall survival was found in univariate analysis (data not shown).
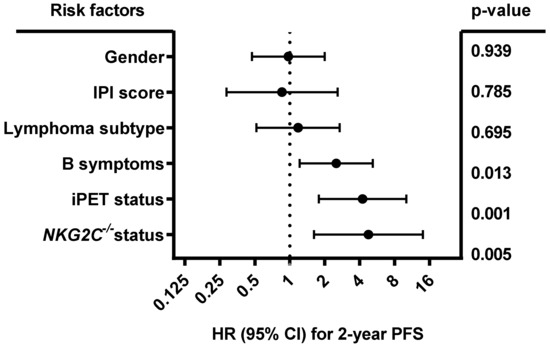
Figure 3.
Forest plot of risk factors for 2-year PFS. The forest plot visualizes the multivariate analysis of the following parameters: Gender (male vs. female), IPI (0–3 vs. 4–5), lymphoma subtype (DLBCL vs. other), B symptoms (A vs. B), iPET status (favorable vs. unfavorable iPET), and NKG2C−/− status (NKG2C−/− vs. NKG2C+/− or NKG2C+/+). The analysis showed that besides B symptoms and iPET status, the NKG2C status is an independent prognostic marker of 2-year PFS in B-NHL patients. Due to loss of follow up, data were available for 379 out of 441 patients. CI (confidence interval), HR (hazard ratio), PFS (progression-free survival at end of treatment).
2.3. HLA-E*01:01 Allele Frequencies Are Increased in B-NHL Patients
To analyze the impact of HLA-E as cognate ligand of NKG2C, HLA-E genotyping was performed in patients (n = 441) and controls (n = 192), respectively (Table 2). HLA-E*01:01 allele frequencies were increased in B-NHL patients compared to healthy controls (57.3% vs. 50.8%; RR: 1.083, 95% CI: 1.006 to 1.168; OR: 1.298; p = 0.0332), whereas the HLA-E genotypes were not found to be different between patients and controls. Furthermore, HLA-E allele or HLA-E genotype frequencies were not associated with clinical or prognostic parameters (data not shown).

Table 2.
Distribution of HLA-E allelic and genotype frequencies of patients and controls. HLA-E*01:01 allele frequencies were increased in B-NHL patients compared to healthy controls. The genotype frequencies did not differ between patients and controls.
2.4. HLA-E*01:01 Is Associated with CR in B-NHL Patients
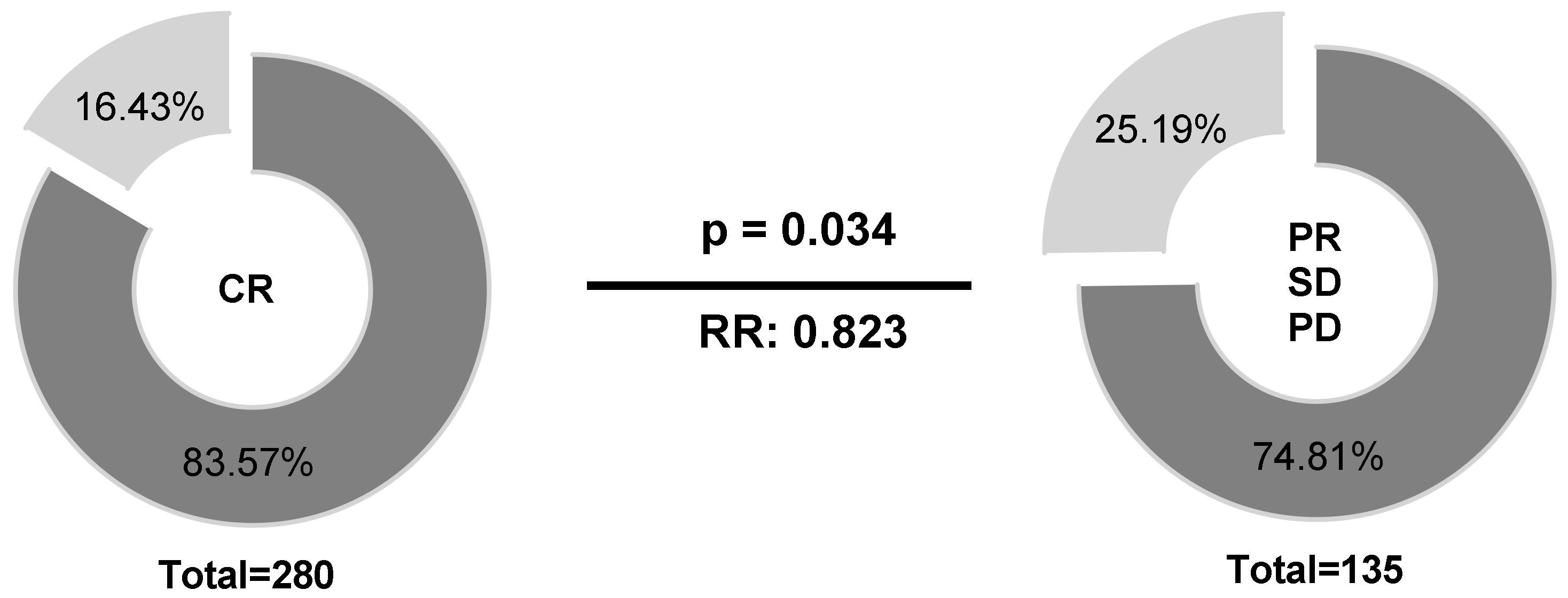
Regarding therapy response at the end of treatment, patients (n = 441) were stratified into two groups according to their HLA-E*01:01 allele carrier status (HLA-E*01:01/*01:01 or HLA-E*01:01/01:03 vs. HLA-E*01:03/01:03). Univariate analysis (Figure 4) revealed higher frequencies of HLA-E*01:01 allele carriers (83.57% vs. 74.81%) in patients with complete remission (CR) compared to patients with limited therapy response (p = 0.034; RR: 0.823; 95% CI: 0.66 to 0.987).
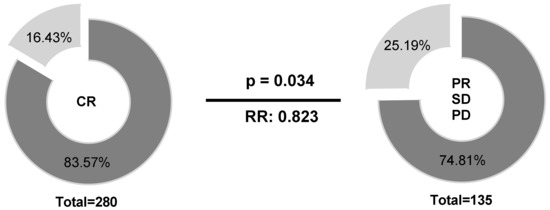
Figure 4.
Association of the HLA-E*01:01 allele carrier status according to therapy response. Higher frequencies of HLA-E*01:01 allele carriers could be observed in patients with CR compared to patients with limited therapy response. HLA-E*01:01/01:01 or HLA-E*01:01/01:03, dark grey; HLA-E*01:03/01:03; light grey; PD = progressive disease, SD = stable disease, PR = partial remission, CR = complete remission; Clinical data were available for 415 out of 441 patients.
Multivariate analysis (Figure 5) including gender (male vs. female), B symptoms (A vs. B), IPI score (0–3 vs. 4–5), lymphoma subtype (DLBCL vs. other), iPET status (favorable iPET vs. unfavorable iPET), and HLA-E*01:01 allele carrier status (HLA-E*01:01/*01:01 or HLA-E*01:01/01:03 vs. HLA-E*01:03/01:03) showed that the HLA-E*01:01 allele carrier status was an independent prognostic factor correlating with complete remission in B-NHL patients at the end of treatment (p = 0.018; HR: 0.534; 95% CI: 0.318 to 0.898; n = 414). Both homozygous and heterozygous HLA-E*01:01 allele carriers had the same clinical correlation. By contrast, B symptoms (p = 0.015; HR: 1.796; 95% CI: 1.12 to 2.88), higher IPI score (p = 0.004; HR: 2.561; 95% CI: 1.361 to 4.819) and unfavorable iPET status (p = 0.005; HR: 2.902; 1.386 to 6.076) were significantly associated with less favorable treatment outcome.
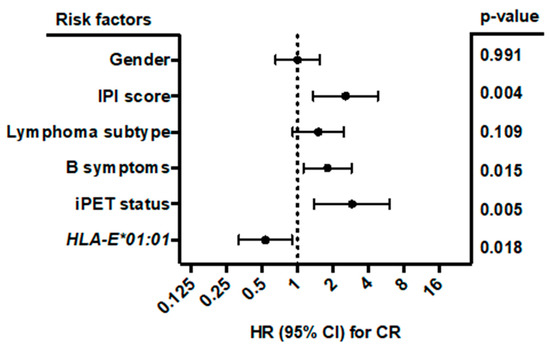
Figure 5.
Forest plot of risk factors for CR. The forest plot visualizes the multivariate analysis (binomial logistic regression) of the following parameters: gender (male vs. female), IPI score (0–3 vs. 4–5), lymphoma subtype (DLBCL vs. other), B symptoms (A vs. B), iPET status (favorable vs. unfavorable iPET), and HLA-E*01:01 allele carrier status (HLA-E*01:01/*01:01 or HLA-E*01:01/01:03, vs. HLA-E*01:03/01:03). All clinical data were available for 414 out of 441 patients. The analysis showed that besides B symptoms, IPI score, and iPET status, the HLA-E*01:01 allele carrier status was an independent prognostic risk factor for the prediction of CR. CI = confidence interval, HR = hazard ratio, CR = complete remission.
3. Discussion
The interplay of NKG2C/A receptors with their ligand HLA-E is thought to play a crucial role in the fine-tuning of immune responses [9,10]. Genetic variations of these molecules are known to affect the expression or function of encoded proteins [12,19,24] and may have an impact on NKG2C/A-HLA-E-mediated immune reactions.
In our study, we focused on the diagnostic and prognostic potential of NKG2C gene deletion and HLA-E allelic variations in patients with B-NHL. We here demonstrate for the first time that the homozygous NKG2C deletion status (i) is increased in high risk B-NHL patients; (ii) is associated with a reduced 2-year PFS in univariate analysis, and (iii) represents an independent prognostic factor for 2-year PFS in multivariate analysis. For HLA-E, the corresponding ligand of NKG2C, frequencies of the HLA-E*01:01 allele are increased in B-NHL patients, and the HLA-E*01:01 status is related with complete remission in both univariate and multivariate analysis.
The low frequency of homozygous NKG2C deletion (4.5%) in B-NHL patients and healthy controls (1.6%) in this study are comparable to other population studies including Japanese, Dutch [20], Mexican [27], German [12] and Spanish investigations [28]. So far, the impact of the NKG2C gene deletion has been shown to be associated with the susceptibility to psoriasis [11] and viral infections [12,29,30], but this has not yet been described in the context of hematological malignancies. In our study, the homozygous NKG2C deletion was found at a higher frequency in B-NHL patients compared to healthy controls without reaching statistical significance. By contrast, NKG2C deletion was detected more often in high-risk than low-risk patients (11.11% vs. 3.59%). Here, the absence of NKG2C may favor the risk of progression in B-NHL patients as HLA-E bearing tumor cells are not being recognized by NK cells, while the probability of an inhibition by the NKG2A receptor may rise. Noticeably, parameters being incorporated in the IPI score, including age, performance status (ECOG), Ann Arbor stage, extra nodal manifestation, and elevated LDH are independent from the homozygous NKG2C deletion status (data not shown).
The successful implementation of the IPI score as a model to identify high-risk subgroups of B-NHL patients with unfavorable PFS or OS (overall survival) in clinical trials enrolling patients from different health care centers is controversially discussed [3,31,32,33]. Based on our results, homozygous NKG2C deletion could serve as a molecular marker, which may be considered for risk stratification of aggressive B-NHL, although this is restricted to a minor fraction of patients. However, patients with a homozygous NKG2C deletion also suffer from a reduced 2-year PFS at end of treatment, supporting the notion that the NKG2C genotype could be clinically relevant. Multivariate analysis revealed that homozygous NKG2C deletion was an independent prognostic risk factor for the prediction of 2-year PFS, whereas IPI was not. A recent report also emphasized that distinct molecular alterations, structural variants, and recurrent mutations, were predictive of disease outcome independent of the clinical IPI score in DLBCL subtypes [34] and thereby were a meaningful complementary parameter for risk stratification.
Accumulating evidence suggests that NK cells contribute to successful elimination of malignant B-cells during rituximab therapy [35,36]. Of note, all patients in our cohort were treated with the antibody rituximab. So-called adaptive NK cells expressing high level of the activating receptor NKG2C [21,22] and CD57 NK cells display enhanced capacity to produce IFN-γ in response to antibody-coated target cells, suggesting that NKG2C positive NK cells are better equipped for cytokine production upon rituximab treatment. Although the functional consequences of NKG2C absence in B-NHL patients have to be investigated in further studies, it is tempting to speculate that absence of the NKG2C positive cells may impair NK cell-mediated support of rituximab treatment, especially since previous studies demonstrated that low copy numbers of NKG2C were associated with impaired NK cell activation or proliferation [12,13]. In the absence of NKG2C-mediated activation, NK cells may be more prone to inhibition by NKG2A/HLA-E interactions, leading to early progression. Thus, NKG2C deficient B-NHL patients may benefit from NKG2A blocking. For these patients, the monoclonal antibody monalizumab, which blocks NKG2A-mediated signaling, may serve as an additional immune therapeutic option [37].
For the cognate ligand of NKG2C, HLA-E, two main alleles exist that are equally present in all populations analyzed [23]. HLA-E*01:01 is linked to comparatively low HLA-E surface expression, whereas substantially higher HLA-E expression is found in the presence of at least one HLA-E*01:03 allele in healthy individuals [6,24,25] as well as in hematological disorders such as acute leukemia [9] or chronic lymphocytic leukemia [10]. Several studies reported a higher prevalence of the HLA-E*01:03 allele or the HLA-E*01:03/01:03 genotype in infectious diseases [38] and several cancer types including nasopharyngeal carcinoma, serous ovarian cancer, and acute leukemia [9,39,40]. Interestingly, the HLA-E*01:01 allele seems to be underrepresented in classical Hodgkin lymphomas, especially among Epstein Barr virus (EBV)-positive cases compared to healthy controls [41]. We observed a slightly higher prevalence of the HLA-E*01:01 genotype in aggressive B-NHL. Importantly, increased frequency of HLA-E*01:01 allele carriers was related with complete remission after the end of treatment in univariate and multivariate analysis. Here it is likely that the low surface expression of the HLA-E*01:01 allele might attenuate the inhibition of NK cells via NKG2A/HLA-E interactions, which in turn supports the antibody-dependent cellular cytotoxicity of NK cells during rituximab treatment favoring a complete remission. Nevertheless, this observation has to be confirmed in independent investigations. Notably, genome wide analyses show that classical HLA is associated with lymphoma and is suggested to be an independent factor for disease risk [42].
4. Materials and Methods
4.1. Patients and Controls
The multicenter study PETAL (Positron Emission Tomography-Guided Therapy of Aggressive Non-Hodgkin Lymphomas) randomized controlled trial (EudraCT 2006-001641-33, ClinicalTrials.gov NCT00554164) enrolled patients with newly diagnosed aggressive B-cell or T-cell lymphomas (age 18–80 years) and an Eastern Co-operative Oncology Group (ECOG) performance status ≤ 3 [43]. All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of BfArM, Germany (61-3910-4032976; date of approval: 11 June 2007) and ethics committee of all participating centers coordinated by the Ethics Committee of the Medical Faculty of the University of Duisburg-Essen (07-3366; date of approval: 25 July 2007).
Here, we focused on a subgroup of this study cohort including patients with CD20 positive aggressive B-cell lymphoma (B-NHL) and a positive baseline positron emission tomography (PET, n = 441) receiving two cycles of R-CHOP. After the pretreatment, patients were randomized and allocated according to the interim PET (iPET) scan result in groups, with favorable iPET (n = 396) being treated either with four cycles of R-CHOP or with the same the number of cycles but with two additional doses of rituximab, and with unfavorable iPET (n = 42) receiving either six additional cycles of R-CHOP or six blocks of an intensive protocol originally designed for Burkitt’s lymphoma [44]. PET scans were evaluated using the ΔSUVmax method [43,45]. The demographic profile of the B-NHL patients included in this sub-study is summarized in Table 3. Seventy-seven male and 115 female healthy blood donors (n = 192) with a mean age of 51.31 ± 9.95 years were used as controls.

Table 3.
Demographic profile of B-NHL patients. iPET (interim positron emission tomography); International Prognostic Index (IPI) stratifying patients in four risk groups (0–1 factor positive = low risk, 2 factors positive = low-intermediate risk, 3 factors positive = high-intermediate risk, 4–5 factors positive = high risk); ECOG performance status (0–1, 2–3); B symptoms (A = no B symptoms; B ≥ 1 B symptom including fever > 38 °C, drenching night sweats, weight loss > 10 kg); diffuse large B-cell lymphoma (DLBCL); other included: follicular lymphoma (FL) with FL grade 1–2, grade 3 with areas of grades 1 or 2, grade 3; primary mediastinal B-cell lymphoma (PMBL); mantle cell lymphoma, unclassified B-cell lymphoma and mixed histologies; complete remission (CR), partial remission (PR), progressive disease (PD), stable disease (SD); progression-free survival (PFS); overall survival (OS); * data were not available for the total number of patients.
4.2. NKG2C and HLA-E Genotyping
For patients and controls, genotyping of the NKG2C deletion or HLA-E was performed as previously described [20,46]. NKG2C and HLA-E genotype distributions in patients and controls did not significantly deviate from the Hardy-Weinberg equilibrium.
4.3. Statistical Analysis
Metric data are given as mean ± standard deviation. Chi-square tests were used to analyze categorical data. Binomial logistic regression was used to identify clinical parameters influencing the therapy response after ending of treatment. Progression-free survival (PFS) and overall survival (OS), respectively, were defined as the time from the date of iPET to disease progression or death from any cause. Probabilities of OS and PFS were analyzed using the Kaplan–Meier method in combination with the log-rank test implemented in the R package survminer (version 0.4.0; https://CRAN.R-project.or/package=survminer). Multivariate Cox regression according to proportional hazards assumption was used to identify prognostic factors for PFS or OS. p values < 0.05 were considered statistically significant. Remaining statistical analyses were performed using either SPSS 23.0 (SPSS Inc, Chicago, IL, USA), GraphPad Prism 8.3.1 (GraphPad Software, San Diego, CA, USA) or GENEPOP 4.7 (https://genepop.curtin.edu.au/).
5. Conclusions
In conclusion, our results provide substantial evidence that absence of NKG2C and HLA-E allelic variations are predictive of outcomes in B-NHL; while allele carriers of HLA-E*01:01 are characterized by high complete remission rates, NKG2C absence is rare but significantly associated with poorer outcomes. Prospective validation of our results is warranted to identify patient subgroups, which may provide benefits from risk-adapted therapy.
Author Contributions
Conceptualization, B.W. and V.R.; Data curation, B.W. and V.R.; Formal analysis, B.W. and V.R.; Funding acquisition, V.R.; Investigation, B.W., H.R., and S.S.; Methodology, R.T.M.; Project administration, U.D., A.H., and H.N.; Supervision, V.R.; Writing—original draft, B.W.; Writing—review and editing, U.D., P.A.H., and V.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Deutsche Krebshilfe (grant nos. 107592, 109816, and 110515). We acknowledge support from the Open Access Publication Fund of the University of Duisburg-Essen.
Conflicts of Interest
U.D.: institutional research funding and honoraria from Roche and Amgen.
References
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; WHO: Geneva, Switzerland, 2008; Volume 2. [Google Scholar]
- Sehn, L.H.; Donaldson, J.; Chhanabhai, M.; Fitzgerald, C.; Gill, K.; Klasa, R.; MacPherson, N.; O’Reilly, S.; Spinelli, J.J.; Sutherland, J.; et al. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J. Clin. Oncol. 2005, 23, 5027–5033. [Google Scholar] [CrossRef] [PubMed]
- International Non-Hodgkin’s Lymphoma Prognostic Factors Project. A predictive model for aggressive non-Hodgkin’s lymphoma. N. Engl. J. Med. 1993, 329, 987–994. [CrossRef] [PubMed]
- Guillerey, C.; Smyth, M.J. NK Cells and Cancer Immunoediting. Curr. Top. Microbiol. Immunol. 2016, 395, 115–145. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Gardenswartz, A.; Termuhlen, A.M.; Cairo, M.S. Advances in cellular and humoral immunotherapy—Implications for the treatment of poor risk childhood, adolescent, and young adult B-cell non-Hodgkin lymphoma. Br. J. Haematol. 2019, 185, 1055–1070. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Soderstrom, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- Posch, P.E.; Borrego, F.; Brooks, A.G.; Coligan, J.E. HLA-E is the ligand for the natural killer cell CD94/NKG2 receptors. J. Biomed. Sci. 1998, 5, 321–331. [Google Scholar] [CrossRef]
- Guma, M.; Busch, L.K.; Salazar-Fontana, L.I.; Bellosillo, B.; Morte, C.; Garcia, P.; Lopez-Botet, M. The CD94/NKG2C killer lectin-like receptor constitutes an alternative activation pathway for a subset of CD8+ T cells. Eur. J. Immunol. 2005, 35, 2071–2080. [Google Scholar] [CrossRef]
- Xu, Y.P.; Wieten, L.; Wang, S.X.; Cai, Y.; Olieslagers, T.; Zhang, L.; He, L.M.; Tilanus, M.G.J.; Hong, W.X. Clinical significance of HLA-E genotype and surface/soluble expression levels between healthy individuals and patients with acute leukemia. Leuk. Lymphoma 2019, 60, 208–215. [Google Scholar] [CrossRef]
- Wagner, B.; da Silva, N.F.; Schramm, S.; Kraemer, T.; Celik, A.A.; Durig, J.; Horn, P.A.; Duhrsen, U.; Nuckel, H.; Rebmann, V. HLA-E allelic genotype correlates with HLA-E plasma levels and predicts early progression in chronic lymphocytic leukemia. Cancer 2017, 123, 814–823. [Google Scholar] [CrossRef]
- Zeng, X.; Chen, H.; Gupta, R.; Paz-Altschul, O.; Bowcock, A.M.; Liao, W. Deletion of the activating NKG2C receptor and a functional polymorphism in its ligand HLA-E in psoriasis susceptibility. Exp. Dermatol. 2013, 22, 679–681. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Low, H.Z.; Kniesch, K.; Jacobs, R.; Schmidt, R.E.; Witte, T. NKG2C deletion is a risk factor of HIV infection. AIDS Res. Hum. Retrovir. 2012, 28, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Muntasell, A.; Lopez-Montanes, M.; Vera, A.; Heredia, G.; Romo, N.; Penafiel, J.; Moraru, M.; Vila, J.; Vilches, C.; Lopez-Botet, M. NKG2C zygosity influences CD94/NKG2C receptor function and the NK-cell compartment redistribution in response to human cytomegalovirus. Eur. J. Immunol. 2013, 43, 3268–3278. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.H.; Orr, H.T. Differential expression of HLA-E, HLA-F, and HLA-G transcripts in human tissue. Hum. Immunol. 1990, 29, 131–142. [Google Scholar] [CrossRef]
- Coupel, S.; Moreau, A.; Hamidou, M.; Horejsi, V.; Soulillou, J.P.; Charreau, B. Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood 2007, 109, 2806–2814. [Google Scholar] [CrossRef]
- Sasaki, T.; Kanaseki, T.; Shionoya, Y.; Tokita, S.; Miyamoto, S.; Saka, E.; Kochin, V.; Takasawa, A.; Hirohashi, Y.; Tamura, Y.; et al. Microenvironmental stresses induce HLA-E/Qa-1 surface expression and thereby reduce CD8(+) T-cell recognition of stressed cells. Eur. J. Immunol. 2016, 46, 929–940. [Google Scholar] [CrossRef]
- Joosten, S.A.; Sullivan, L.C.; Ottenhoff, T.H. Characteristics of HLA-E Restricted T-Cell Responses and Their Role in Infectious Diseases. J. Immunol. Res. 2016, 2016, 2695396. [Google Scholar] [CrossRef]
- Marin, R.; Ruiz-Cabello, F.; Pedrinaci, S.; Mendez, R.; Jimenez, P.; Geraghty, D.E.; Garrido, F. Analysis of HLA-E expression in human tumors. Immunogenetics 2003, 54, 767–775. [Google Scholar] [CrossRef]
- Hikami, K.; Tsuchiya, N.; Yabe, T.; Tokunaga, K. Variations of human killer cell lectin-like receptors: Common occurrence of NKG2-C deletion in the general population. Genes Immun. 2003, 4, 160–167. [Google Scholar] [CrossRef]
- Miyashita, R.; Tsuchiya, N.; Hikami, K.; Kuroki, K.; Fukazawa, T.; Bijl, M.; Kallenberg, C.G.; Hashimoto, H.; Yabe, T.; Tokunaga, K. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int. Immunol. 2004, 16, 163–168. [Google Scholar] [CrossRef]
- Guma, M.; Angulo, A.; Vilches, C.; Gomez-Lozano, N.; Malats, N.; Lopez-Botet, M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004, 104, 3664–3671. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Botet, M.; Muntasell, A.; Vilches, C. The CD94/NKG2C+ NK-cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection. Semin. Immunol. 2014, 26, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Felicio, L.P.; Porto, I.O.; Mendes-Junior, C.T.; Veiga-Castelli, L.C.; Santos, K.E.; Vianello-Brondani, R.P.; Sabbagh, A.; Moreau, P.; Donadi, E.A.; Castelli, E.C. Worldwide HLA-E nucleotide and haplotype variability reveals a conserved gene for coding and 3’ untranslated regions. Tissue Antigens 2014, 83, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Strong, R.K.; Holmes, M.A.; Li, P.; Braun, L.; Lee, N.; Geraghty, D.E. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J. Biol. Chem. 2003, 278, 5082–5090. [Google Scholar] [CrossRef] [PubMed]
- Ulbrecht, M.; Couturier, A.; Martinozzi, S.; Pla, M.; Srivastava, R.; Peterson, P.A.; Weiss, E.H. Cell surface expression of HLA-E: Interaction with human beta2-microglobulin and allelic differences. Eur. J. Immunol. 1999, 29, 537–547. [Google Scholar] [CrossRef]
- Kaiser, B.K.; Barahmand-Pour, F.; Paulsene, W.; Medley, S.; Geraghty, D.E.; Strong, R.K. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J. Immunol. 2005, 174, 2878–2884. [Google Scholar] [CrossRef]
- Rangel-Ramirez, V.V.; Garcia-Sepulveda, C.A.; Escalante-Padron, F.; Perez-Gonzalez, L.F.; Rangel-Castilla, A.; Aranda-Romo, S.; Noyola, D.E. NKG2C gene deletion in the Mexican population and lack of association to respiratory viral infections. Int. J. Immunogenet. 2014, 41, 126–130. [Google Scholar] [CrossRef]
- Moraru, M.; Canizares, M.; Muntasell, A.; de Pablo, R.; Lopez-Botet, M.; Vilches, C. Assessment of copy-number variation in the NKG2C receptor gene in a single-tube and characterization of a reference cell panel, using standard polymerase chain reaction. Tissue Antigens 2012, 80, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Vietzen, H.; Pollak, K.; Honsig, C.; Jaksch, P.; Puchhammer-Stockl, E. NKG2C Deletion Is a Risk Factor for Human Cytomegalovirus Viremia and Disease After Lung Transplantation. J. Infect. Dis. 2018, 217, 802–806. [Google Scholar] [CrossRef]
- Moraru, M.; Cisneros, E.; Gomez-Lozano, N.; de Pablo, R.; Portero, F.; Canizares, M.; Vaquero, M.; Roustan, G.; Millan, I.; Lopez-Botet, M.; et al. Host genetic factors in susceptibility to herpes simplex type 1 virus infection: Contribution of polymorphic genes at the interface of innate and adaptive immunity. J. Immunol. 2012, 188, 4412–4420. [Google Scholar] [CrossRef]
- Hong, F.; Kahl, B.S.; Gray, R. Incremental value in outcome prediction with gene expression-based signatures in diffuse large B-cell lymphoma. Blood 2013, 121, 156–158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ziepert, M.; Hasenclever, D.; Kuhnt, E.; Glass, B.; Schmitz, N.; Pfreundschuh, M.; Loeffler, M. Standard International prognostic index remains a valid predictor of outcome for patients with aggressive CD20+ B-cell lymphoma in the rituximab era. J. Clin. Oncol. 2010, 28, 2373–2380. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Sehn, L.H.; Rademaker, A.W.; Gordon, L.I.; Lacasce, A.S.; Crosby-Thompson, A.; Vanderplas, A.; Zelenetz, A.D.; Abel, G.A.; Rodriguez, M.A.; et al. An enhanced International Prognostic Index (NCCN-IPI) for patients with diffuse large B-cell lymphoma treated in the rituximab era. Blood 2014, 123, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Enqvist, M.; Jacobs, B.; Junlen, H.R.; Schaffer, M.; Melen, C.M.; Friberg, D.; Wahlin, B.E.; Malmberg, K.J. Systemic and Intra-Nodal Activation of NK Cells After Rituximab Monotherapy for Follicular Lymphoma. Front. Immunol. 2019, 10, 2085. [Google Scholar] [CrossRef]
- Golay, J.; Manganini, M.; Facchinetti, V.; Gramigna, R.; Broady, R.; Borleri, G.; Rambaldi, A.; Introna, M. Rituximab-mediated antibody-dependent cellular cytotoxicity against neoplastic B cells is stimulated strongly by interleukin-2. Haematologica 2003, 88, 1002–1012. [Google Scholar]
- Andre, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Blery, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731.e13–1743.e13. [Google Scholar] [CrossRef]
- Guberina, H.; da Silva, N.F.; Michita, R.T.; Dolff, S.; Bienholz, A.; Heinemann, F.M.; Wilde, B.; Trilling, M.; Horn, P.A.; Kribben, A.; et al. Susceptibility of HLA-E*01:03 Allele Carriers to Develop Cytomegalovirus Replication After Living-Donor Kidney Transplantation. J. Infect. Dis. 2018, 217, 1918–1922. [Google Scholar] [CrossRef]
- Zheng, H.; Lu, R.; Xie, S.; Wen, X.; Wang, H.; Gao, X.; Guo, L. Human leukocyte antigen-E alleles and expression in patients with serous ovarian cancer. Cancer Sci. 2015, 106, 522–528. [Google Scholar] [CrossRef]
- Hirankarn, N.; Kimkong, I.; Mutirangura, A. HLA-E polymorphism in patients with nasopharyngeal carcinoma. Tissue Antigens 2004, 64, 588–592. [Google Scholar] [CrossRef]
- Martin, P.; Krsnik, I.; Navarro, B.; Provencio, M.; Garcia, J.F.; Bellas, C.; Vilches, C.; Gomez-Lozano, N. HLA Allele E*01:01 Is Associated with a Reduced Risk of EBV-Related Classical Hodgkin Lymphoma Independently of HLA-A*01/*02. PLoS ONE 2015, 10, e0135512. [Google Scholar] [CrossRef] [PubMed]
- McAulay, K.A.; Jarrett, R.F. Human leukocyte antigens and genetic susceptibility to lymphoma. Tissue Antigens 2015, 86, 98–113. [Google Scholar] [CrossRef] [PubMed]
- Duhrsen, U.; Muller, S.; Hertenstein, B.; Thomssen, H.; Kotzerke, J.; Mesters, R.; Berdel, W.E.; Franzius, C.; Kroschinsky, F.; Weckesser, M.; et al. Positron Emission Tomography-Guided Therapy of Aggressive Non-Hodgkin Lymphomas (PETAL): A Multicenter, Randomized Phase III Trial. J. Clin. Oncol. 2018, 36, 2024–2034. [Google Scholar] [CrossRef] [PubMed]
- Hoelzer, D.; Walewski, J.; Dohner, H.; Viardot, A.; Hiddemann, W.; Spiekermann, K.; Serve, H.; Duhrsen, U.; Huttmann, A.; Thiel, E.; et al. Improved outcome of adult Burkitt lymphoma/leukemia with rituximab and chemotherapy: Report of a large prospective multicenter trial. Blood 2014, 124, 3870–3879. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Itti, E.; Haioun, C.; Petegnief, Y.; Luciani, A.; Dupuis, J.; Paone, G.; Talbot, J.N.; Rahmouni, A.; Meignan, M. Early 18F-FDG PET for prediction of prognosis in patients with diffuse large B-cell lymphoma: SUV-based assessment versus visual analysis. J. Nucl. Med. 2007, 48, 1626–1632. [Google Scholar] [CrossRef]
- Grimsley, C.; Kawasaki, A.; Gassner, C.; Sageshima, N.; Nose, Y.; Hatake, K.; Geraghty, D.E.; Ishitani, A. Definitive high resolution typing of HLA-E allelic polymorphisms: Identifying potential errors in existing allele data. Tissue Antigens 2002, 60, 206–212. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).