Metabolomics to Assess Response to Immune Checkpoint Inhibitors in Patients with Non-Small-Cell Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
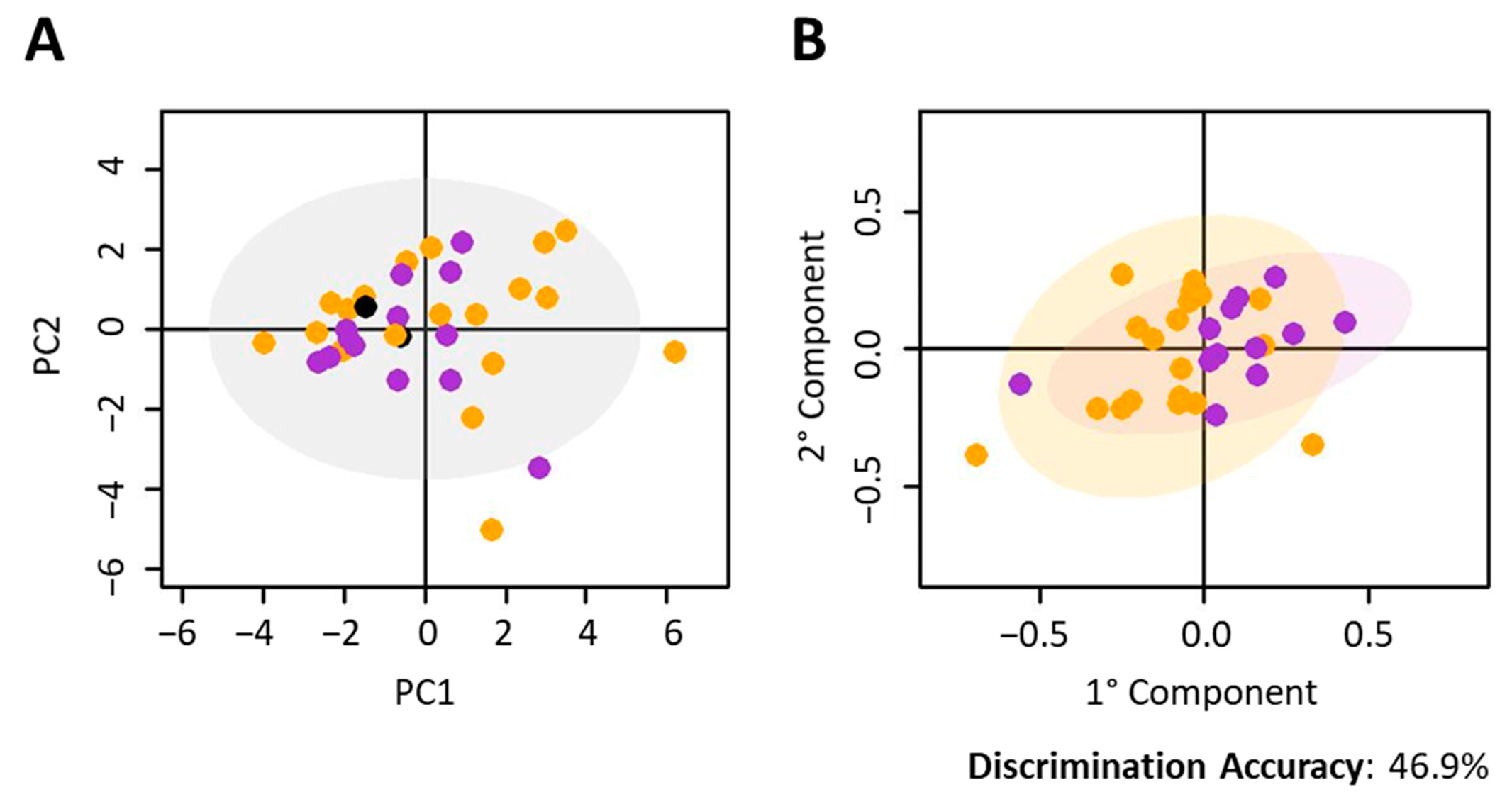
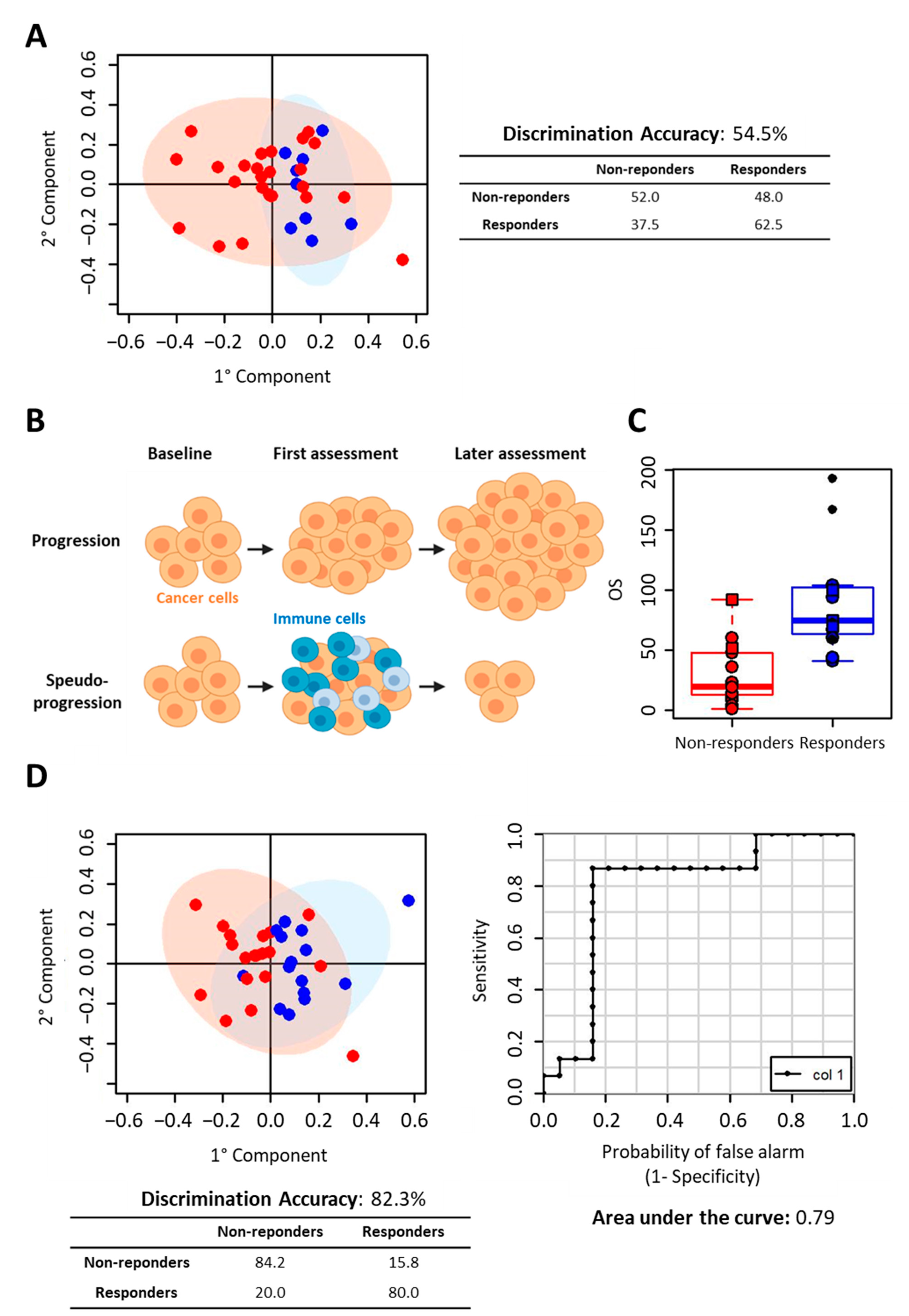
2.1. NSCLC Nivolumab-Treated Cohort
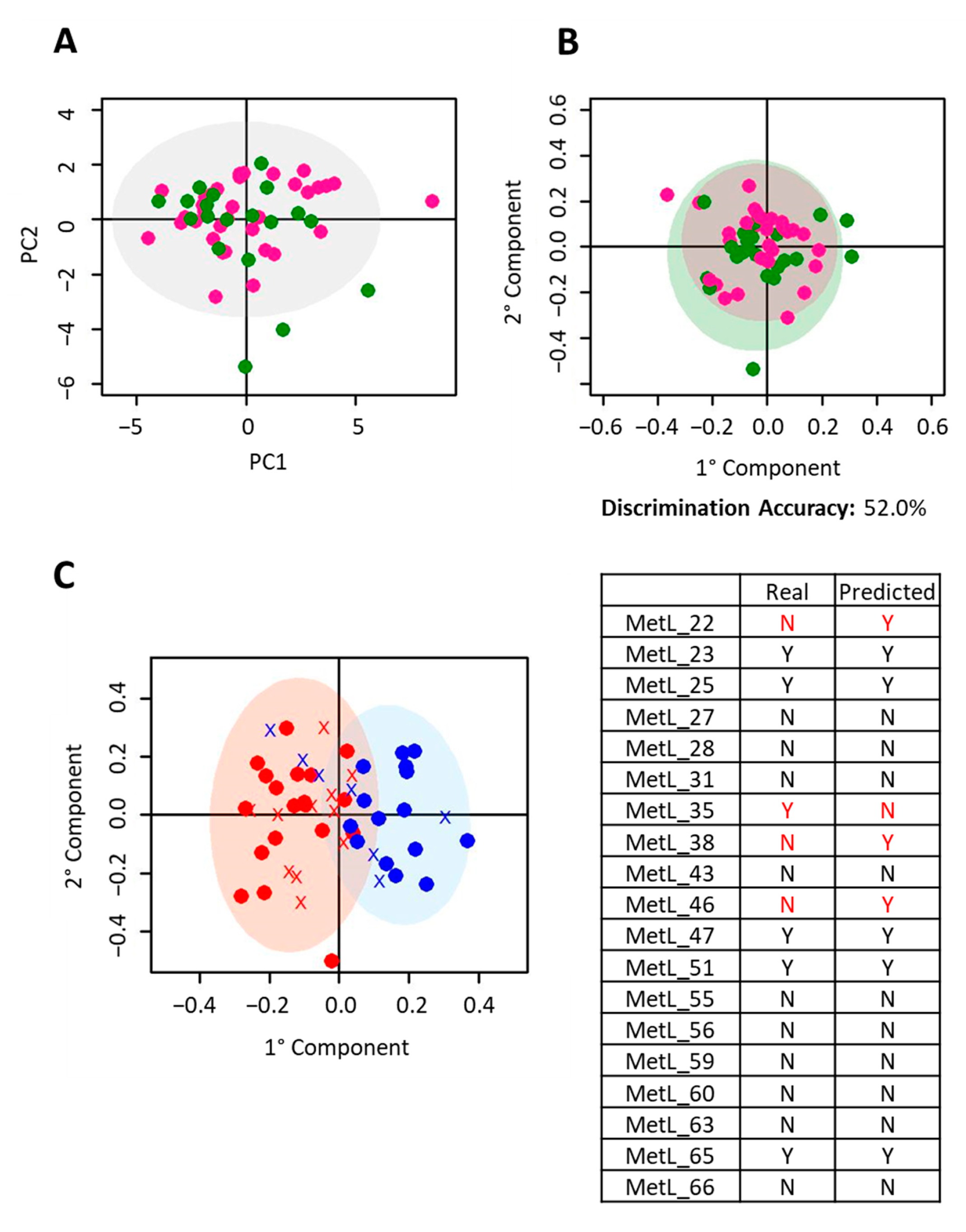
2.2. NSCLC Pembrolizumab-Treated Cohort
2.3. Metabolite Analysis
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment and Serum Sample Collection
4.2. NMR Analysis
- (i)
- A standard NOESY 1Dpresat (noesygppr1d.comp; Bruker BioSpin) pulse sequence (using 32 scans, 98,304 data points, a spectral width of 18,028 Hz, an acquisition time of 2.7 s, a relaxation delay of 4 s and a mixing time of 0.1 s.) to obtain a spectrum in which signals of both metabolites and high molecular weight molecules (lipids and lipoproteins) are visible.
- (ii)
- A standard CPMG (cpmgpr1d.comp; Bruker BioSpin) pulse sequence (using 32 scans, 73,728 data points, a spectral width of 12,019 Hz and a relaxation delay of 4 s.), designed for the selective observation of small molecule components in solutions containing macromolecules.
- (iii)
- A standard diffusion-edited (ledbgppr2s1d.comp; Bruker BioSpin) pulse sequence (using 32 scans, 98,304 data points, a spectral width of 18,028 Hz and a relaxation delay of 4 s.), for the selective observation of macromolecule components in solutions containing small molecules.
4.3. Spectral Processing
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brahmer, J.R.; Govindan, R.; Anders, R.A.; Antonia, S.J.; Sagorsky, S.; Davies, M.J.; Dubinett, S.M.; Ferris, A.; Gandhi, L.; Garon, E.B.; et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of non-small cell lung cancer (NSCLC). J. Immunother. Cancer 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.A.; Callahan, M.K.; Wolchok, J.D. Immune Checkpoint Blockade in Cancer Therapy. J. Clin. Oncol. 2015, 33, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Kosumi, K.; Nakai, Y.; Koike, K. Surrogate study endpoints in the era of cancer immunotherapy. Ann. Transl. Med. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J.; Wu, Y.-L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Non-squamous Non-small Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Rittmeyer, A.; Barlesi, F.; Waterkamp, D.; Park, K.; Ciardiello, F.; von Pawel, J.; Gadgeel, S.M.; Hida, T.; Kowalski, D.M.; Dols, M.C.; et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 2017, 389, 255–265. [Google Scholar] [CrossRef]
- Gettinger, S.N.; Horn, L.; Gandhi, L.; Spigel, D.R.; Antonia, S.J.; Rizvi, N.A.; Powderly, J.D.; Heist, R.S.; Carvajal, R.D.; Jackman, D.M.; et al. Overall Survival and Long-Term Safety of Nivolumab (Anti–Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients with previously Treated Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2015, 33, 2004–2012. [Google Scholar] [CrossRef]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Yokoi, T.; Chiappori, A.; Lee, K.H.; de Wit, M.; et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.R. Immune checkpoint blockade: The hope for immunotherapy as a treatment of lung cancer? Semin. Oncol. 2014, 41, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.A.; Weeder, B.R.; David, J.K.; Nellore, A.; Thompson, R.F. Burden of tumor mutations, neoepitopes, and other variants are weak predictors of cancer immunotherapy response and overall survival. Genome Med. 2020, 12, 33. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, J.; Wu, X. Pseudoprogression and hyperprogression after checkpoint blockade. Int. Immunopharmacol. 2018, 58, 125–135. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef]
- Nicholson, G.; Rantalainen, M.; Maher, A.D.; Li, J.V.; Malmodin, D.; Ahmadi, K.R.; Faber, J.H.; Hallgrímsdóttir, I.B.; Barrett, A.; Toft, H.; et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol. Syst. Biol. 2011, 7, 525. [Google Scholar] [CrossRef]
- Ghini, V.; Tenori, L.; Capozzi, F.; Luchinat, C.; Bub, A.; Malpuech-Brugere, C.; Orfila, C.; Ricciardiello, L.; Bordoni, A. DHA-Induced Perturbation of Human Serum Metabolome. Role of the Food Matrix and Co-Administration of Oat β-glucan and Anthocyanins. Nutrients 2019, 12, 86. [Google Scholar] [CrossRef]
- Ghini, V.; Saccenti, E.; Tenori, L.; Assfalg, M.; Luchinat, C. Allostasis and Resilience of the Human Individual Metabolic Phenotype. J. Proteome Res. 2015, 14, 2951–2962. [Google Scholar] [CrossRef]
- Antonaros, F.; Ghini, V.; Pulina, F.; Ramacieri, G.; Cicchini, E.; Mannini, E.; Martelli, A.; Feliciello, A.; Lanfranchi, S.; Onnivello, S.; et al. Plasma metabolome and cognitive skills in Down syndrome. Sci. Rep. 2020, 10, 10491. [Google Scholar] [CrossRef]
- Ghini, V.; Unger, F.T.; Tenori, L.; Turano, P.; Juhl, H.; David, K.A. Metabolomics profiling of pre-and post-anesthesia plasma samples of colorectal patients obtained via Ficoll separation. Metabolomics 2015, 11, 1769–1778. [Google Scholar] [CrossRef]
- Meoni, G.; Lorini, S.; Monti, M.; Madia, F.; Corti, G.; Luchinat, C.; Zignego, A.L.; Tenori, L.; Gragnani, L. The metabolic fingerprints of HCV and HBV infections studied by Nuclear Magnetic Resonance Spectroscopy. Sci. Rep. 2019, 9, 4128. [Google Scholar] [CrossRef] [PubMed]
- Dani, C.; Bresci, C.; Berti, E.; Ottanelli, S.; Mello, G.; Mecacci, F.; Breschi, R.; Hu, X.; Tenori, L.; Luchinat, C. Metabolomic profile of term infants of gestational diabetic mothers. J. Matern. Fetal Neonatal Med. 2014, 27, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Hart, C.D.; Vignoli, A.; Tenori, L.; Uy, G.L.; Van To, T.; Adebamowo, C.; Hossain, S.M.; Biganzoli, L.; Risi, E.; Love, R.R.; et al. Serum Metabolomic Profiles Identify ER-Positive Early Breast Cancer Patients at Increased Risk of Disease Recurrence in a Multicenter Population. Clin. Cancer Res. 2017, 23, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Cacciatore, S.; Jensen, B.V.; Schou, J.V.; Johansen, J.S.; Kruhøffer, M.; Luchinat, C.; Nielsen, D.L.; Turano, P. Metabolomic NMR Fingerprinting to Identify and Predict Survival of Patients with Metastatic Colorectal Cancer. Cancer Res. 2012, 72, 356–364. [Google Scholar] [CrossRef]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, G.S. Clinical Applications of Metabolomics in Oncology: A Review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Takis, P.G.; Ghini, V.; Tenori, L.; Turano, P.; Luchinat, C. Uniqueness of the NMR approach to metabolomics. TrAC Trends Anal. Chem. 2019, 120, 115300. [Google Scholar] [CrossRef]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. Engl. 2019, 58, 968–994. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Everett, J.R. NMR-based pharmacometabonomics: A new paradigm for personalised or precision medicine. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 102-103, 1–14. [Google Scholar] [CrossRef]
- Vignoli, A.; Tenori, L.; Giusti, B.; Valente, S.; Carrabba, N.; Balzi, D.; Barchielli, A.; Marchionni, N.; Gensini, G.F.; Marcucci, R.; et al. Differential Network Analysis Reveals Metabolic Determinants Associated with Mortality in Acute Myocardial Infarction Patients and Suggests Potential Mechanisms Underlying Different Clinical Scores Used to Predict Death. J. Proteome Res. 2020, 19, 949–961. [Google Scholar] [CrossRef]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Ciuleanu, T.-E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef]
- De Vries, R.; Muller, M.; van der Noort, V.; Theelen, W.S.M.E.; Schouten, R.D.; Hummelink, K.; Muller, S.H.; Wolf-Lansdorf, M.; Dagelet, J.W.F.; Monkhorst, K.; et al. Prediction of response to anti-PD-1 therapy in patients with non-small-cell lung cancer by electronic nose analysis of exhaled breath. Ann. Oncol. 2019, 30, 1660–1666. [Google Scholar] [CrossRef]
- Duruisseaux, M.; Martínez-Cardús, A.; Calleja-Cervantes, M.E.; Moran, S.; de Moura, C.M.; Davalos, V.; Piñeyro, D.; Sanchez-Cespedes, M.; Girard, N.; Brevet, M.; et al. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: A multicentre, retrospective analysis. Lancet Respir. Med. 2018, 6, 771–781. [Google Scholar] [CrossRef]
- Hatae, R.; Chamoto, K.; Kim, Y.H.; Sonomura, K.; Taneishi, K.; Kawaguchi, S.; Yoshida, H.; Ozasa, H.; Sakamori, Y.; Akrami, M.; et al. Combination of host immune metabolic biomarkers for the PD-1 blockade cancer immunotherapy. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Botticelli, A.; Vernocchi, P.; Marini, F.; Quagliariello, A.; Cerbelli, B.; Reddel, S.; Del Chierico, F.; Di Pietro, F.; Giusti, R.; Tomassini, A.; et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J. Transl. Med. 2020, 18, 49. [Google Scholar] [CrossRef]
- Li, H.; Bullock, K.; Gurjao, C.; Braun, D.; Shukla, S.A.; Bossé, D.; Lalani, A.-K.A.; Gopal, S.; Jin, C.; Horak, C.; et al. Metabolomic adaptations and correlates of survival to immune checkpoint blockade. Nat. Commun. 2019, 10, 1–6. [Google Scholar] [CrossRef]
- Lim, A.R.; Rathmell, W.K.; Rathmell, J.C. The tumor microenvironment as a metabolic barrier to effector T cells and immunotherapy. Elife 2020, 9, e55185. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Harris, A.L. Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef]
- Wilson, W.R.; Hay, M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef]
- Bernacchioni, C.; Ghini, V.; Cencetti, F.; Japtok, L.; Donati, C.; Bruni, P.; Turano, P. NMR metabolomics highlights sphingosine kinase-1 as a new molecular switch in the orchestration of aberrant metabolic phenotype in cancer cells. Mol. Oncol. 2017, 11, 517–533. [Google Scholar] [CrossRef]
- D’Alessandro, G.; Quaglio, D.; Monaco, L.; Lauro, C.; Ghirga, F.; Ingallina, C.; De Martino, M.; Fucile, S.; Porzia, A.; Di Castro, M.A.; et al. 1H-NMR metabolomics reveals the Glabrescione B exacerbation of glycolytic metabolism beside the cell growth inhibitory effect in glioma. Cell Commun. Signal. 2019, 17, 108. [Google Scholar] [CrossRef]
- Prasad, V.; Kaestner, V. Nivolumab and pembrolizumab: Monoclonal antibodies against programmed cell death-1 (PD-1) that are interchangeable. Semin. Oncol. 2017, 44, 132–135. [Google Scholar] [CrossRef]
- Seymour, L.; Bogaerts, J.; Perrone, A.; Ford, R.; Schwartz, L.H.; Mandrekar, S.; Lin, N.U.; Litière, S.; Dancey, J.; Chen, A.; et al. iRECIST: Guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017, 18, e143–e152. [Google Scholar] [CrossRef]
- Sridhara, R.; Zhou, J.; Theoret, M.R.; Mishra-Kalyani, P.S. Time to treatment failure (TTF) as a potential clinical endpoint in real-world evidence (RWE) studies of melanoma. J. Clin. Oncol. 2018, 36, 9578. [Google Scholar] [CrossRef]
- Pazdur, R. Endpoints for assessing drug activity in clinical trials. Oncologist 2008, 13, 19–21. [Google Scholar] [CrossRef]
- Ghini, V.; Quaglio, D.; Luchinat, C.; Turano, P. NMR for sample quality assessment in metabolomics. New Biotechnol. 2019, 52, 25–34. [Google Scholar] [CrossRef]
- Bernini, P.; Bertini, I.; Luchinat, C.; Nincheri, P.; Staderini, S.; Turano, P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J. Biomol. NMR 2011, 49, 231–243. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; van Velzen, E.J.J.; Hoefsloot, H.C.J.; Smilde, A.K. Discriminant Q2 (DQ2) for improved discrimination in PLSDA models. Metabolomics 2008, 4, 293–296. [Google Scholar] [CrossRef]

| Subject Number | N° Cycles | TTF (Weeks) | OS (Weeks) | 1 RA | 2 RA | 3 RA | 4 RA | 5 RA | 6 RA | 7 RA | 8 RA | 9 RA | 10 RA | 11 RA | 12 RA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MetL01 | 4 | 8 | 10 | na (HP) | |||||||||||
| MetL05 | 24 | 59 | 94 | iSD/PR | iSD | iUPD | iCPD | ||||||||
| MetL06 | 4 | 8 | 13 | iUPD | |||||||||||
| MetL07 | 84 | 186+ | 193+ | iPR | iPR | iPR | iPR | iPR | iPR | iPR | iPR | iPR | iPR | iPR | iPR |
| MetL09 | 19 | 38 | 67 | iSD | iUPD | iUPD | iCPD | ||||||||
| MetL10 | 10 | 18 | 20 | iSD | iSD | ||||||||||
| MetL11 | 9 | 23 | 60 | iUPD | iCPD | ||||||||||
| MetL12 | 23 | 45 | 60 | iUPD | iPR | iSD | iPD | ||||||||
| MetL13 | 29 | 140+ | 167+ | iUPD | iUPD | iPR | iPR | iUPD | iUPD | iPR | iSD | iSD | iSD | ||
| MetL15 | 2 | 2 | 5 | na (HP) | |||||||||||
| MetL17 | 19 | 41 | 103 | iUPD | iUPD | iCPD | |||||||||
| MetL19 | 27 | 66 | 104 | iUPD | iUPD | iSD | iSD | iCPD | |||||||
| MetL20 | 4 | 6 | 8 | na (HP) | |||||||||||
| MetL24 | 6 | 10 | 48 | iPD | |||||||||||
| MetL26 | 6 | 10 | 36 | iPD | |||||||||||
| MetL29 | 3 | 5 | 60 | na (HP) | |||||||||||
| MetL30 | 2 | 4 | 14 | HP | |||||||||||
| MetL32 | 8 | 17 | 19 | iUPD | |||||||||||
| MetL33 | 13 | 35 | 41 | iSD | iSD | iUPD | iCPD | ||||||||
| MetL34 | 2 | 2 | 4 | na (HP) | |||||||||||
| MetL36 | 8 | 16 | 21 | iUPD | iCPD | ||||||||||
| MetL37 | 17 | 38 | 44 | iUPD | iUPD | iCPD | |||||||||
| MetL39 | 4 | 7 | 14 | iUPD | |||||||||||
| MetL40 | 47 | 99+ | 102+ | iPR | iPR | iPR | iPR | iPR | iPR | iPR | |||||
| MetL41 | 6 | 12 | 23 | iUPD | iCPD | ||||||||||
| MetL42 | 18 | 39 | 100+ | iUPD | iUPD | iUPD | iCPD | ||||||||
| MetL44 | 11 | 20 | 92+ | iUPD | iCPD | ||||||||||
| MetL45 | 8 | 15 | 19 | iUPD | iCPD | ||||||||||
| MetL48 | 19 | 37 | 75+ | iUPD | iPR | iPD | |||||||||
| MetL49 | 1 | 1 | 1 | na (HP) | |||||||||||
| MetL50 | 35 | 70+ | 73+ | iPR | iPR | iPR | iPR | ||||||||
| MetL52 | 29 | 58+ | 70+ | iUPD | iUPD | iUPD | iUPD | iUPD | iCPD | ||||||
| MetL53 | 19 | 54+ | 59+ | iPR | iPR | iSD | iPR | ||||||||
| MetL57 | 7 | 15 | 52+ | iUPD | iCPD |
| Subject Number | N° Cycles | TTF (Week) | OS (Week) | PD-L1 | 1 RA | 2 RA | 3 RA | 4 RA | 5 RA | 6 RA | 7 RA | 8 RA | 9 RA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MetL22 | 35 | 115+ | 118+ | 80 | iPR | iPR | iPR | iPR | iPR | iSD | iSD | iUPD | iSD |
| MetL23 | 36 | 115+ | 116+ | 60 | iPR | iPR | iCR | iCR | iCR | iSD | iSD | iSD | |
| MetL25 | 33 | 106+ | 109+ | 70 | iSD | iSD | iSD | iSD | iSD | iSD | iSD | iSD | iSD |
| MetL27 | 3 | 6 | 14 | 80 | |||||||||
| MetL28 | 8 | 24 | 39 | 70 | iPR | iUPD | iCPD | ||||||
| MetL31 | 3 | 8 | 10 | 70 | |||||||||
| MetL35 | 11 | 37 | 39 | 70 | iSD | iUPD | iUPD | iCPD | |||||
| MetL38 | 26 | 82+ | 87+ | 60 | iPR | iPR | iSD | iSD | iSD | ||||
| MetL43 | 1 | 5 | 5 | 60 | |||||||||
| MetL46 | 17 | 62+ | 66+ | 60 | iUPD | iPR | iPR | iPR | iPR | ||||
| MetL47 | 17 | 54+ | 56+ | 80 | iPR | iUPD | iSD | ||||||
| MetL51 | 14 | 45+ | 49+ | 70 | iPR | iPR | iSD | ||||||
| MetL55 | 1 | 1 | 1 | 80 | |||||||||
| MetL56 | 3 | 9 | 17 | 90 | iPD | ||||||||
| MetL59 | 3 | 6 | xx | 80 | iPD | ||||||||
| MetL60 | 5 | 15 | 19 | 60 | iSD | ||||||||
| MetL63 | 4 | 20 | 23 | 50 | iUPD | iCPD | |||||||
| MetL65 | 8 | 22 | 30+ | 90 | iPR | iUPD | |||||||
| MetL66 | 7 | 26 | 90 | iUPD | iCPD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghini, V.; Laera, L.; Fantechi, B.; del Monte, F.; Benelli, M.; McCartney, A.; Tenori, L.; Luchinat, C.; Pozzessere, D. Metabolomics to Assess Response to Immune Checkpoint Inhibitors in Patients with Non-Small-Cell Lung Cancer. Cancers 2020, 12, 3574. https://doi.org/10.3390/cancers12123574
Ghini V, Laera L, Fantechi B, del Monte F, Benelli M, McCartney A, Tenori L, Luchinat C, Pozzessere D. Metabolomics to Assess Response to Immune Checkpoint Inhibitors in Patients with Non-Small-Cell Lung Cancer. Cancers. 2020; 12(12):3574. https://doi.org/10.3390/cancers12123574
Chicago/Turabian StyleGhini, Veronica, Letizia Laera, Beatrice Fantechi, Francesca del Monte, Matteo Benelli, Amelia McCartney, Leonardo Tenori, Claudio Luchinat, and Daniele Pozzessere. 2020. "Metabolomics to Assess Response to Immune Checkpoint Inhibitors in Patients with Non-Small-Cell Lung Cancer" Cancers 12, no. 12: 3574. https://doi.org/10.3390/cancers12123574
APA StyleGhini, V., Laera, L., Fantechi, B., del Monte, F., Benelli, M., McCartney, A., Tenori, L., Luchinat, C., & Pozzessere, D. (2020). Metabolomics to Assess Response to Immune Checkpoint Inhibitors in Patients with Non-Small-Cell Lung Cancer. Cancers, 12(12), 3574. https://doi.org/10.3390/cancers12123574








