The Non-Coding RNA Landscape of Plasma Cell Dyscrasias
Abstract
:1. Introduction
2. ncRNAs: Molecular Features
2.1. sncRNAs
2.2. LncRNAs
2.3. CircRNAs
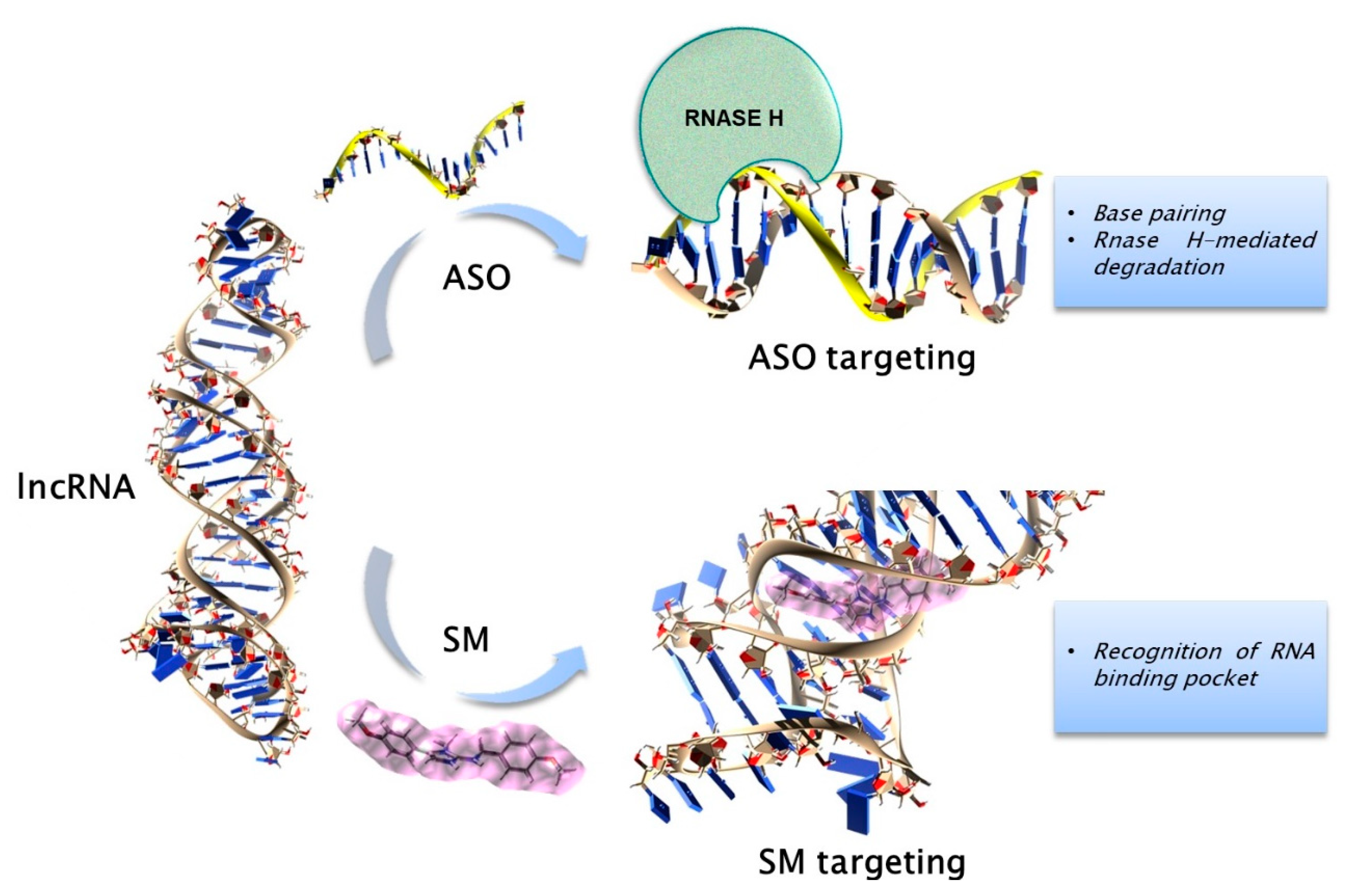
3. Therapeutic Targeting of ncRNAs in PC Dyscrasias
3.1. General Strategies for Targeting the ncRNAs
3.2. Preclinical Findings on ncRNAs in PC Dyscrasis
3.2.1. sncRNAs
3.2.2. LncRNAs
3.2.3. CircRNAs
4. Circulating ncRNAs in PC Dyscrasias
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Beason, T.S.; Chang, S.H.; Sanfilippo, K.M.; Luo, S.; Colditz, G.A.; Vij, R.; Tomasson, M.H.; Dipersio, J.F.; Stockerl-Goldstein, K.; Ganti, A.; et al. Influence of body mass index on survival in veterans with multiple myeloma. Oncologist 2013, 18, 1074–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, E.V.; Edwards, C.M. Adipokines, adiposity, and bone marrow adipocytes: Dangerous accomplices in multiple myeloma. J. Cell Physiol. 2018, 233, 9159–9166. [Google Scholar] [CrossRef] [PubMed]
- Kyle, R.A.; Larson, D.R.; Therneau, T.M.; Dispenzieri, A.; Kumar, S.; Cerhan, J.R.; Rajkumar, S.V. Long-Term Follow-up of Monoclonal Gammopathy of Undetermined Significance. N Engl. J. Med. 2018, 378, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Multiple myeloma: 2018 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2018, 93, 981–1114. [CrossRef] [Green Version]
- Wadhera, R.K.; Rajkumar, S.V. Prevalence of monoclonal gammopathy of undetermined significance: A systematic review. Mayo Clin. Proc. 2010, 85, 933–942. [Google Scholar] [CrossRef] [Green Version]
- Gámez, B.; Edwards, C.M. Contributions of the Bone Microenvironment to Monoclonal Gammopathy of Undetermined Significance Pathogenesis. Curr. Osteoporos. Rep. 2018, 16, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Sant, M.; Allemani, C.; Tereanu, C.; De Angelis, R.; Capocaccia, R.; Visser, O.; Marcos-Gragera, R.; Maynadié, M.; Simonetti, A.; Lutz, J.M.; et al. Incidence of hematologic malignancies in Europe by morphologic subtype: Results of the HAEMACARE project. Blood 2010, 116, 3724–3734. [Google Scholar] [CrossRef]
- Gundesen, M.T.; Lund, T.; Moeller, H.E.H.; Abildgaard, N. Plasma Cell Leukemia: Definition, Presentation, and Treatment. Curr. Oncol. Rep. 2019, 21, 8. [Google Scholar] [CrossRef] [Green Version]
- Cifola, I.; Lionetti, M.; Pinatel, E.; Todoerti, K.; Mangano, E.; Pietrelli, A.; Fabris, S.; Mosca, L.; Simeon, V.; Petrucci, M.T.; et al. Whole-exome sequencing of primary plasma cell leukemia discloses heterogeneous mutational patterns. Oncotarget 2015, 6, 17543–17558. [Google Scholar] [CrossRef]
- Chiecchio, L.; Dagrada, G.P.; White, H.E.; Towsend, M.R.; Protheroe, R.K.; Cheung, K.L.; Stockley, D.M.; Orchard, K.H.; Cross, N.C.; Harrison, C.J.; et al. Frequent upregulation of MYC in plasma cell leukemia. Genes Chromosomes Cancer 2009, 48, 624–636. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [Green Version]
- Rajkumar, S.V.; Dispenzieri, A.; Kyle, R.A. Monoclonal gammopathy of undetermined significance, Waldenström macroglobulinemia, AL amyloidosis, and related plasma cell disorders: Diagnosis and treatment. Mayo Clin. Proc. 2006, 81, 693–703. [Google Scholar] [CrossRef] [Green Version]
- Owen, R.G.; Treon, S.P.; Al-Katib, A.; Fonseca, R.; Greipp, P.R.; McMaster, M.L.; Morra, E.; Pangalis, G.A.; San Miguel, J.F.; Branagan, A.R.; et al. Clinicopathological definition of Waldenstrom’s macroglobulinemia: Consensus panel recommendations from the Second International Workshop on Waldenstrom’s Macroglobulinemia. Semin Oncol. 2003, 30, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R.; Hayman, S. Waldenström macroglobulinaemia. Br J. Haematol. 2007, 138, 700–720. [Google Scholar] [CrossRef] [PubMed]
- Vaxman, I.; Dispenzieri, A.; Muchtar, E.; Gertz, M. New developments in diagnosis, risk assessment and management in systemic amyloidosis. Blood Rev. 2019, 100636. [Google Scholar] [CrossRef] [PubMed]
- Jaccard, A. POEMS Syndrome: Therapeutic Options. Hematol. Oncol. Clin. North Am. 2018, 32, 141–151. [Google Scholar] [CrossRef]
- Amodio, N.; D’Aquila, P.; Passarino, G.; Tassone, P.; Bellizzi, D. Epigenetic modifications in multiple myeloma: Recent advances on the role of DNA and histone methylation. Expert Opin. Ther. Targets 2017, 21, 91–101. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Drusco, A.; Croce, C.M. MicroRNAs and Cancer: A Long Story for Short RNAs. Adv. Cancer Res. 2017, 135, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Van Roosbroeck, K.; Calin, G.A. Cancer Hallmarks and MicroRNAs: The Therapeutic Connection. Adv. Cancer Res. 2017, 135, 119–149. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Su, M.Y.; Maggi, L.B., Jr.; Lu, L.; Mullins, C.; Crosby, S.; Huang, G.; Chng, W.J.; Vij, R.; Tomasson, M.H. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J. Clin. Invest. 2012, 122, 2793–2806. [Google Scholar] [CrossRef]
- Taulli, R.; Pandolfi, P.P. “Snorkeling” for missing players in cancer. J. Clin. Invest. 2012, 122, 2765–2768. [Google Scholar] [CrossRef] [Green Version]
- Hashim, A.; Rizzo, F.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Santamaria, G.; Cordella, A.; Cantarella, C.; et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014, 5, 9901–9910. [Google Scholar] [CrossRef] [Green Version]
- Martinez, V.D.; Vucic, E.A.; Thu, K.L.; Hubaux, R.; Enfield, K.S.; Pikor, L.A.; Becker-Santos, D.D.; Brown, C.J.; Lam, S.; Lam, W.L. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci. Rep. 2015, 5, 10423. [Google Scholar] [CrossRef]
- Horwich, M.D.; Li, C.; Matranga, C.; Vagin, V.; Farley, G.; Wang, P.; Zamore, P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007, 17, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Carmell, M.A.; Girard, A.; van de Kant, H.J.; Bourc’his, D.; Bestor, T.H.; de Rooij, D.G.; Hannon, G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 2007, 12, 503–514. [Google Scholar] [CrossRef] [Green Version]
- Amodio, N.; Di Martino, M.T.; Neri, A.; Tagliaferri, P.; Tassone, P. Non-coding RNA: A novel opportunity for the personalized treatment of multiple myeloma. Expert Opin. Biol. Ther. 2013, 13 Suppl 1, S125–S137. [Google Scholar] [CrossRef]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.C.; Franco, B.; Robine, N.; Lai, E.C.; Pelisson, A.; Simonelig, M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellitto, A.; Geles, K.; D’Agostino, Y.; Conte, M.; Alexandrova, E.; Rocco, D.; Nassa, G.; Giurato, G.; Tarallo, R.; Weisz, A.; et al. Molecular and Functional Characterization of the Somatic PIWIL1/piRNA Pathway in Colorectal Cancer Cells. Cells 2019, 8, 1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Gao, Q.; Chen, K.; Xue, X.; Li, M.; Chen, Q.; Zhu, G.; Gao, Y. Hiwi facilitates chemoresistance as a cancer stem cell marker in cervical cancer. Oncol. Rep. 2014, 32, 1853–1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [Green Version]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu Rev. Biochem 2012, 81, 145–166. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [Green Version]
- Mohammad, F.; Mondal, T.; Kanduri, C. Epigenetics of imprinted long non-coding RNAs. Epigenetics 2009, 4, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Pontier, D.B.; Gribnau, J. Xist regulation and function explored. Hum. Genet. 2011, 130, 223–236. [Google Scholar] [CrossRef] [Green Version]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munschauer, M.; Nguyen, C.T.; Sirokman, K.; Hartigan, C.R.; Hogstrom, L.; Engreitz, J.M.; Ulirsch, J.C.; Fulco, C.P.; Subramanian, V.; Chen, J.; et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 2018, 561, 132–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, K. Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 2008, 129, 91–98. [Google Scholar] [CrossRef] [Green Version]
- Barrett, S.P.; Wang, P.L.; Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015, 4, e07540. [Google Scholar] [CrossRef]
- Schindewolf, C.; Braun, S.; Domdey, H. In vitro generation of a circular exon from a linear pre-mRNA transcript. Nucleic Acids Res. 1996, 24, 1260–1266. [Google Scholar] [CrossRef] [Green Version]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Nazarali, A.J.; Ji, S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016, 6, 1167–1176. [Google Scholar] [PubMed]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupaimoole, R.; Calin, G.A.; Lopez-Berestein, G.; Sood, A.K. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016, 6, 235–246. [Google Scholar] [CrossRef] [Green Version]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Chen, L.L.; Wang, Y.; Wong, C.C.; et al. Extensive translation of circular RNAs driven by N. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol. Life Sci. 2018, 75, 1071–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, J.; Li, J.; Gui, R.; Nie, X.; Huang, R. CircRNA_014511 affects the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p. Bosn. J. Basic Med. Sci. 2019, 19, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2018. [Google Scholar] [CrossRef] [Green Version]
- Trang, P.; Wiggins, J.F.; Daige, C.L.; Cho, C.; Omotola, M.; Brown, D.; Weidhaas, J.B.; Bader, A.G.; Slack, F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011, 19, 1116–1122. [Google Scholar] [CrossRef]
- Chery, J. RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc. J. 2016, 4, 35–50. [Google Scholar] [CrossRef] [Green Version]
- Petersen, M.; Wengel, J. LNA: A versatile tool for therapeutics and genomics. Trends Biotechnol. 2003, 21, 74–81. [Google Scholar] [CrossRef]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [Green Version]
- Geary, R.S. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin. Drug Metab. Toxicol. 2009, 5, 381–391. [Google Scholar] [CrossRef]
- Pavco, P.A.; Bouhana, K.S.; Gallegos, A.M.; Agrawal, A.; Blanchard, K.S.; Grimm, S.L.; Jensen, K.L.; Andrews, L.E.; Wincott, F.E.; Pitot, P.A.; et al. Antitumor and antimetastatic activity of ribozymes targeting the messenger RNA of vascular endothelial growth factor receptors. Clin. Cancer Res. 2000, 6, 2094–2103. [Google Scholar]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brosnan, C.A.; Voinnet, O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009, 21, 416–425. [Google Scholar] [CrossRef]
- Zhao, F.; Zhao, Q.; Blount, K.F.; Han, Q.; Tor, Y.; Hermann, T. Molecular recognition of RNA by neomycin and a restricted neomycin derivative. Angew. Chem. Int. Ed. Engl. 2005, 44, 5329–5334. [Google Scholar] [CrossRef]
- Gumireddy, K.; Young, D.D.; Xiong, X.; Hogenesch, J.B.; Huang, Q.; Deiters, A. Small-molecule inhibitors of microrna miR-21 function. Angew. Chem. Int. Ed. Engl. 2008, 47, 7482–7484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Zhang, B.; Chen, G.; Wu, W.; Zhou, L.; Shi, Y.; Zeng, Q.; Li, Y.; Sun, Y.; Deng, X.; et al. Targeting miR-21 with Sophocarpine Inhibits Tumor Progression and Reverses Epithelial-Mesenchymal Transition in Head and Neck Cancer. Mol. Ther. 2017, 25, 2129–2139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Disney, M.D. Precise Small Molecule Degradation of a Noncoding RNA Identifies Cellular Binding Sites and Modulates an Oncogenic Phenotype. ACS Chem. Biol. 2018, 13, 3065–3071. [Google Scholar] [CrossRef]
- Disney, M.D.; Winkelsas, A.M.; Velagapudi, S.P.; Southern, M.; Fallahi, M.; Childs-Disney, J.L. Inforna 2.0: A Platform for the Sequence-Based Design of Small Molecules Targeting Structured RNAs. ACS Chem. Biol. 2016, 11, 1720–1728. [Google Scholar] [CrossRef] [Green Version]
- Costales, M.G.; Hoch, D.G.; Abegg, D.; Childs-Disney, J.L.; Velagapudi, S.P.; Adibekian, A.; Disney, M.D. A Designed Small Molecule Inhibitor of a Non-Coding RNA Sensitizes HER2 Negative Cancers to Herceptin. J. Am. Chem. Soc. 2019, 141, 2960–2974. [Google Scholar] [CrossRef]
- Kligun, E.; Mandel-Gutfreund, Y. Conformational readout of RNA by small ligands. RNA Biol. 2013, 10, 982–989. [Google Scholar] [CrossRef] [Green Version]
- Kondo, J.; Westhof, E. Base pairs and pseudo pairs observed in RNA-ligand complexes. J. Mol. Recognit. 2010, 23, 241–252. [Google Scholar] [CrossRef]
- Warner, K.D.; Hajdin, C.E.; Weeks, K.M. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 2018, 17, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Deigan, K.E.; Ferré-D’Amaré, A.R. Riboswitches: Discovery of drugs that target bacterial gene-regulatory RNAs. Acc. Chem. Res. 2011, 44, 1329–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blount, K.F.; Breaker, R.R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006, 24, 1558–1564. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.Y.; Cui, M.; Heldsinger, A.; Lemrow, S.M.; Loo, J.A.; Sannes-Lowery, K.A.; Sharmeen, L.; Czarnik, A.W. Inhibitors of protein-RNA complexation that target the RNA: Specific recognition of human immunodeficiency virus type 1 TAR RNA by small organic molecules. Biochemistry 1998, 37, 14204–14212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, J.; Jayaraman, B.; Frankel, A. The HIV-1 Rev. response element: An RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex. RNA Biol. 2012, 9, 6–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connelly, C.M.; Moon, M.H.; Schneekloth, J.S. The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem. Biol. 2016, 23, 1077–1090. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Collie, G.W.; Parkinson, G.N.; Neidle, S.; Rosu, F.; De Pauw, E.; Gabelica, V. Electrospray mass spectrometry of telomeric RNA (TERRA) reveals the formation of stable multimeric G-quadruplex structures. J. Am. Chem. Soc. 2010, 132, 9328–9334. [Google Scholar] [CrossRef] [Green Version]
- Di Antonio, M.; Biffi, G.; Mariani, A.; Raiber, E.A.; Rodriguez, R.; Balasubramanian, S. Selective RNA versus DNA G-quadruplex targeting by in situ click chemistry. Angew. Chem. Int. Ed. Engl. 2012, 51, 11073–11078. [Google Scholar] [CrossRef] [Green Version]
- Shirude, P.S.; Gillies, E.R.; Ladame, S.; Godde, F.; Shin-Ya, K.; Huc, I.; Balasubramanian, S. Macrocyclic and helical oligoamides as a new class of G-quadruplex ligands. J. Am. Chem. Soc. 2007, 129, 11890–11891. [Google Scholar] [CrossRef] [Green Version]
- Rocca, R.; Talarico, C.; Moraca, F.; Costa, G.; Romeo, I.; Ortuso, F.; Alcaro, S.; Artese, A. Molecular recognition of a carboxy pyridostatin toward G-quadruplex structures: Why does it prefer RNA? Chem. Biol. Drug Des. 2017, 90, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Rocca, R.; Moraca, F.; Costa, G.; Nadai, M.; Scalabrin, M.; Talarico, C.; Distinto, S.; Maccioni, E.; Ortuso, F.; Artese, A.; et al. Identification of G-quadruplex DNA/RNA binders: Structure-based virtual screening and biophysical characterization. Biochim. Biophys Acta Gen. Subj. 2017, 1861, 1329–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Yang, M.; Tian, J.; Wang, X.; Li, Z. MALAT-1: A long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 2011, 39, 169–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.A.; Bulkley, D.; Wang, J.; Valenstein, M.L.; Yario, T.A.; Steitz, T.A.; Steitz, J.A. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014, 21, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; JnBaptiste, C.K.; Lu, L.Y.; Kuhn, C.D.; Joshua-Tor, L.; Sharp, P.A. A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012, 26, 2392–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donlic, A.; Morgan, B.S.; Xu, J.L.; Liu, A.; Roble, C.; Hargrove, A.E. Corrigendum: Discovery of Small Molecule Ligands for MALAT1 by Tuning an RNA-Binding Scaffold. Angew. Chem. Int. Ed. Engl. 2019, 58, 5482. [Google Scholar] [CrossRef] [Green Version]
- Abulwerdi, F.A.; Xu, W.; Ageeli, A.A.; Yonkunas, M.J.; Arun, G.; Nam, H.; Schneekloth, J.S.; Dayie, T.K.; Spector, D.; Baird, N.; et al. Selective Small-Molecule Targeting of a Triple Helix Encoded by the Long Noncoding RNA, MALAT1. ACS Chem. Biol. 2019, 14, 223–235. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, Y.F.; Zhang, J.; Wang, Q.X.; Han, L.; Mei, M.; Kang, C.S. Targeted design and identification of AC1NOD4Q to block activity of HOTAIR by abrogating the scaffold interaction with EZH2. Clin. Epigenetics 2019, 11, 29. [Google Scholar] [CrossRef]
- Li, Y.; Ren, Y.; Wang, Y.; Tan, Y.; Wang, Q.; Cai, J.; Zhou, J.; Yang, C.; Zhao, K.; Yi, K.; et al. A Compound AC1Q3QWB Selectively Disrupts HOTAIR-Mediated Recruitment of PRC2 and Enhances Cancer Therapy of DZNep. Theranostics 2019, 9, 4608–4623. [Google Scholar] [CrossRef]
- Lionetti, M.; Biasiolo, M.; Agnelli, L.; Todoerti, K.; Mosca, L.; Fabris, S.; Sales, G.; Deliliers, G.L.; Bicciato, S.; Lombardi, L.; et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood 2009, 114, e20-26. [Google Scholar] [CrossRef] [Green Version]
- Lionetti, M.; Musto, P.; Di Martino, M.T.; Fabris, S.; Agnelli, L.; Todoerti, K.; Tuana, G.; Mosca, L.; Gallo Cantafio, M.E.; Grieco, V.; et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clin. Cancer Res. 2013, 19, 3130–3142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roccaro, A.M.; Sacco, A.; Chen, C.; Runnels, J.; Leleu, X.; Azab, F.; Azab, A.K.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. microRNA expression in the biology, prognosis, and therapy of Waldenstrom macroglobulinemia. Blood 2009, 113, 4391–4402. [Google Scholar] [CrossRef]
- Weng, L.; Spencer, B.H.; SoohHoo, P.T.; Connors, L.H.; O’Hara, C.J.; Seldin, D.C. Dysregulation of miRNAs in AL amyloidosis. Amyloid 2011, 18, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichiorri, F.; Suh, S.S.; Ladetto, M.; Kuehl, M.; Palumbo, T.; Drandi, D.; Taccioli, C.; Zanesi, N.; Alder, H.; Hagan, J.P.; et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 12885–12890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roccaro, A.M.; Sacco, A.; Thompson, B.; Leleu, X.; Azab, A.K.; Azab, F.; Runnels, J.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009, 113, 6669–6680. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.T.; Gulla, A.; Cantafio, M.E.; Lionetti, M.; Leone, E.; Amodio, N.; Guzzi, P.H.; Foresta, U.; Conforti, F.; Cannataro, M.; et al. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget 2013, 4, 242–255. [Google Scholar] [CrossRef] [Green Version]
- Gulla, A.; Di Martino, M.T.; Gallo Cantafio, M.E.; Morelli, E.; Amodio, N.; Botta, C.; Pitari, M.R.; Lio, S.G.; Britti, D.; Stamato, M.A.; et al. A 13 mer LNA-i-miR-221 Inhibitor Restores Drug Sensitivity in Melphalan-Refractory Multiple Myeloma Cells. Clin. Cancer Res. 2016, 22, 1222–1233. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Su, Y.; Xu, A.; Fan, F.; Mu, S.; Chen, L.; Chu, Z.; Zhang, B.; Huang, H.; Zhang, J.; et al. miR-221/222-Mediated Inhibition of Autophagy Promotes Dexamethasone Resistance in Multiple Myeloma. Mol. Ther. 2019, 27, 559–570. [Google Scholar] [CrossRef] [Green Version]
- Di Martino, M.T.; Gulla, A.; Gallo Cantafio, M.E.; Altomare, E.; Amodio, N.; Leone, E.; Morelli, E.; Lio, S.G.; Caracciolo, D.; Rossi, M.; et al. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS ONE 2014, 9, e89659. [Google Scholar] [CrossRef]
- Di Martino, M.T.; Leone, E.; Amodio, N.; Foresta, U.; Lionetti, M.; Pitari, M.R.; Cantafio, M.E.; Gulla, A.; Conforti, F.; Morelli, E.; et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: In vitro and in vivo evidence. Clin. Cancer Res. 2012, 18, 6260–6270. [Google Scholar] [CrossRef] [Green Version]
- Zarone, M.R.; Misso, G.; Grimaldi, A.; Zappavigna, S.; Russo, M.; Amler, E.; Di Martino, M.T.; Amodio, N.; Tagliaferri, P.; Tassone, P.; et al. Evidence of novel miR-34a-based therapeutic approaches for multiple myeloma treatment. Sci. Rep. 2017, 7, 17949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morelli, E.; Leone, E.; Cantafio, M.E.; Di Martino, M.T.; Amodio, N.; Biamonte, L.; Gulla, A.; Foresta, U.; Pitari, M.R.; Botta, C.; et al. Selective targeting of IRF4 by synthetic microRNA-125b-5p mimics induces anti-multiple myeloma activity in vitro and in vivo. Leukemia 2015, 29, 2173–2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Roccaro, A.M.; Rombaoa, C.; Flores, L.; Obad, S.; Fernandes, S.M.; Sacco, A.; Liu, Y.; Ngo, H.; Quang, P.; et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood 2012, 120, 1678–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aktas Samur, A.; Minvielle, S.; Shammas, M.; Fulciniti, M.; Magrangeas, F.; Richardson, P.G.; Moreau, P.; Attal, M.; Anderson, K.C.; Parmigiani, G.; et al. Deciphering the chronology of copy number alterations in Multiple Myeloma. Blood Cancer J. 2019, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Misiewicz-Krzeminska, I.; Krzeminski, P.; Corchete, L.A.; Quwaider, D.; Rojas, E.A.; Herrero, A.B.; Gutierrez, N.C. Factors Regulating microRNA Expression and Function in Multiple Myeloma. Noncoding RNA 2019, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amodio, N.; Leotta, M.; Bellizzi, D.; Di Martino, M.T.; D’Aquila, P.; Lionetti, M.; Fabiani, F.; Leone, E.; Gulla, A.M.; Passarino, G.; et al. DNA-demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget 2012, 3, 1246–1258. [Google Scholar] [CrossRef] [Green Version]
- Tam, W.; Dahlberg, J.E. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer 2006, 45, 211–212. [Google Scholar] [CrossRef]
- Krzeminski, P.; Sarasquete, M.E.; Misiewicz-Krzeminska, I.; Corral, R.; Corchete, L.A.; Martin, A.A.; Garcia-Sanz, R.; San Miguel, J.F.; Gutierrez, N.C. Insights into epigenetic regulation of microRNA-155 expression in multiple myeloma. Biochim. Biophys Acta 2015, 1849, 353–366. [Google Scholar] [CrossRef]
- Amodio, N.; Gallo Cantafio, M.E.; Botta, C.; Agosti, V.; Federico, C.; Caracciolo, D.; Ronchetti, D.; Rossi, M.; Driessen, C.; Neri, A.; et al. Replacement of miR-155 Elicits Tumor Suppressive Activity and Antagonizes Bortezomib Resistance in Multiple Myeloma. Cancers (Basel) 2019, 11, 236. [Google Scholar] [CrossRef] [Green Version]
- Amodio, N.; Rossi, M.; Raimondi, L.; Pitari, M.R.; Botta, C.; Tagliaferri, P.; Tassone, P. miR-29s: A family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget 2015, 6, 12837–12861. [Google Scholar] [CrossRef] [Green Version]
- Stamato, M.A.; Juli, G.; Romeo, E.; Ronchetti, D.; Arbitrio, M.; Caracciolo, D.; Neri, A.; Tagliaferri, P.; Tassone, P.; Amodio, N. Inhibition of EZH2 triggers the tumor suppressive miR-29b network in multiple myeloma. Oncotarget 2017, 8, 106527–106537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amodio, N.; Stamato, M.A.; Gulla, A.M.; Morelli, E.; Romeo, E.; Raimondi, L.; Pitari, M.R.; Ferrandino, I.; Misso, G.; Caraglia, M.; et al. Therapeutic Targeting of miR-29b/HDAC4 Epigenetic Loop in Multiple Myeloma. Mol. Cancer Ther. 2016, 15, 1364–1375. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Di Martino, M.T.; Foresta, U.; Leone, E.; Lionetti, M.; Leotta, M.; Gulla, A.M.; Pitari, M.R.; Conforti, F.; Rossi, M.; et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012, 3, e436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roccaro, A.M.; Sacco, A.; Jia, X.; Azab, A.K.; Maiso, P.; Ngo, H.T.; Azab, F.; Runnels, J.; Quang, P.; Ghobrial, I.M. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood 2010, 116, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Calura, E.; Bisognin, A.; Manzoni, M.; Todoerti, K.; Taiana, E.; Sales, G.; Morgan, G.J.; Tonon, G.; Amodio, N.; Tassone, P.; et al. Disentangling the microRNA regulatory milieu in multiple myeloma: Integrative genomics analysis outlines mixed miRNA-TF circuits and pathway-derived networks modulated in t(4;14) patients. Oncotarget 2016, 7, 2367–2378. [Google Scholar] [CrossRef] [PubMed]
- Pichiorri, F.; Suh, S.S.; Rocci, A.; De Luca, L.; Taccioli, C.; Santhanam, R.; Zhou, W.; Benson, D.M., Jr.; Hofmainster, C.; Alder, H.; et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010, 18, 367–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Fulciniti, M.; Amodio, N.; Bandi, R.L.; Cagnetta, A.; Samur, M.K.; Acharya, C.; Prabhala, R.; D’Aquila, P.; Bellizzi, D.; Passarino, G.; et al. miR-23b/SP1/c-myc forms a feed-forward loop supporting multiple myeloma cell growth. Blood Cancer J. 2016, 6, e380. [Google Scholar] [CrossRef]
- Morelli, E.; Biamonte, L.; Federico, C.; Amodio, N.; Di Martino, M.T.; Gallo Cantafio, M.E.; Manzoni, M.; Scionti, F.; Samur, M.K.; Gulla, A.; et al. Therapeutic vulnerability of multiple myeloma to MIR17PTi, a first-in-class inhibitor of pri-miR-17-92. Blood 2018, 132, 1050–1063. [Google Scholar] [CrossRef]
- Pyzer, A.R.; Cole, L.; Rosenblatt, J.; Avigan, D.E. Myeloid-derived suppressor cells as effectors of immune suppression in cancer. Int. J. Cancer 2016, 139, 1915–1926. [Google Scholar] [CrossRef] [Green Version]
- Malek, E.; de Lima, M.; Letterio, J.J.; Kim, B.G.; Finke, J.H.; Driscoll, J.J.; Giralt, S.A. Myeloid-derived suppressor cells: The green light for myeloma immune escape. Blood Rev. 2016, 30, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Zhang, J.; Lwin, S.T.; Edwards, J.R.; Edwards, C.M.; Mundy, G.R.; Yang, X. Osteoclasts in multiple myeloma are derived from Gr-1+CD11b+myeloid-derived suppressor cells. PLoS ONE 2012, 7, e48871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, L.; Mu, S.; Sun, C.; Fan, F.; Yan, H.; Qin, Y.; Cui, G.; Wang, Y.; Guo, T.; Mei, H.; et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol. Cancer 2019, 18, 88. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, L.; De Luca, A.; Morelli, E.; Giavaresi, G.; Tagliaferri, P.; Tassone, P.; Amodio, N. MicroRNAs: Novel Crossroads between Myeloma Cells and the Bone Marrow Microenvironment. Biomed. Res. Int. 2016, 2016, 6504593. [Google Scholar] [CrossRef] [Green Version]
- Bellavia, D.; Salamanna, F.; Raimondi, L.; De Luca, A.; Carina, V.; Costa, V.; Alessandro, R.; Fini, M.; Giavaresi, G. Deregulated miRNAs in osteoporosis: Effects in bone metastasis. Cell Mol. Life Sci. 2019, 76, 3723–3744. [Google Scholar] [CrossRef]
- Rossi, M.; Pitari, M.R.; Amodio, N.; Di Martino, M.T.; Conforti, F.; Leone, E.; Botta, C.; Paolino, F.M.; Del Giudice, T.; Iuliano, E.; et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: Implications for the treatment of multiple myeloma-related bone disease. J. Cell Physiol. 2013, 228, 1506–1515. [Google Scholar] [CrossRef]
- Pitari, M.R.; Rossi, M.; Amodio, N.; Botta, C.; Morelli, E.; Federico, C.; Gulla, A.; Caracciolo, D.; Di Martino, M.T.; Arbitrio, M.; et al. Inhibition of miR-21 restores RANKL/OPG ratio in multiple myeloma-derived bone marrow stromal cells and impairs the resorbing activity of mature osteoclasts. Oncotarget 2015, 6, 27343–27358. [Google Scholar] [CrossRef] [Green Version]
- Leone, E.; Morelli, E.; Di Martino, M.T.; Amodio, N.; Foresta, U.; Gulla, A.; Rossi, M.; Neri, A.; Giordano, A.; Munshi, N.C.; et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin. Cancer Res. 2013, 19, 2096–2106. [Google Scholar] [CrossRef] [Green Version]
- Amodio, N.; Bellizzi, D.; Leotta, M.; Raimondi, L.; Biamonte, L.; D’Aquila, P.; Di Martino, M.T.; Calimeri, T.; Rossi, M.; Lionetti, M.; et al. miR-29b induces SOCS-1 expression by promoter demethylation and negatively regulates migration of multiple myeloma and endothelial cells. Cell Cycle 2013, 12, 3650–3662. [Google Scholar] [CrossRef] [Green Version]
- Raimondi, L.; Amodio, N.; Di Martino, M.T.; Altomare, E.; Leotta, M.; Caracciolo, D.; Gulla, A.; Neri, A.; Taverna, S.; D’Aquila, P.; et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: In vitro and in vivo anti-tumor activity. Oncotarget 2014, 5, 3039–3054. [Google Scholar] [CrossRef] [Green Version]
- Umezu, T.; Tadokoro, H.; Azuma, K.; Yoshizawa, S.; Ohyashiki, K.; Ohyashiki, J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 2014, 124, 3748–3757. [Google Scholar] [CrossRef]
- Botta, C.; Cuce, M.; Pitari, M.R.; Caracciolo, D.; Gulla, A.; Morelli, E.; Riillo, C.; Biamonte, L.; Gallo Cantafio, M.E.; Prabhala, R.; et al. MiR-29b antagonizes the pro-inflammatory tumor-promoting activity of multiple myeloma-educated dendritic cells. Leukemia 2018, 32, 1003–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronchetti, D.; Todoerti, K.; Tuana, G.; Agnelli, L.; Mosca, L.; Lionetti, M.; Fabris, S.; Colapietro, P.; Miozzo, M.; Ferrarini, M.; et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2012, 2, e96. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, N.; Wu, H.J.; Bennett, R.L.; Troche, C.; Licht, J.D.; Weber, J.D.; Maggi, L.B., Jr.; Tomasson, M.H. Sabotaging of the oxidative stress response by an oncogenic noncoding RNA. FASEB J. 2017, 31, 482–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Wu, Q.L.; Sun, C.Y.; Ai, L.S.; Deng, J.; Zhang, L.; Chen, L.; Chu, Z.B.; Tang, B.; Wang, K.; et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015, 29, 196–206. [Google Scholar] [CrossRef]
- Li, B.; Hong, J.; Hong, M.; Wang, Y.; Yu, T.; Zang, S.; Wu, Q. piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene 2019, 38, 5227–5238. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Ronchetti, D.; Agnelli, L.; Taiana, E.; Galletti, S.; Manzoni, M.; Todoerti, K.; Musto, P.; Strozzi, F.; Neri, A. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget 2016, 7, 14814–14830. [Google Scholar] [CrossRef] [Green Version]
- Ronchetti, D.; Agnelli, L.; Pietrelli, A.; Todoerti, K.; Manzoni, M.; Taiana, E.; Neri, A. A compendium of long non-coding RNAs transcriptional fingerprint in multiple myeloma. Sci. Rep. 2018, 8, 6557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samur, M.K.; Minvielle, S.; Gulla, A.; Fulciniti, M.; Cleynen, A.; Aktas Samur, A.; Szalat, R.; Shammas, M.; Magrangeas, F.; Tai, Y.T.; et al. Long intergenic non-coding RNAs have an independent impact on survival in multiple myeloma. Leukemia 2018, 32, 2626–2635. [Google Scholar] [CrossRef]
- Hu, Y.; Lin, J.; Fang, H.; Fang, J.; Li, C.; Chen, W.; Liu, S.; Ondrejka, S.; Gong, Z.; Reu, F.; et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia 2018, 32, 2250–2262. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handa, H.; Kuroda, Y.; Kimura, K.; Masuda, Y.; Hattori, H.; Alkebsi, L.; Matsumoto, M.; Kasamatsu, T.; Kobayashi, N.; Tahara, K.I.; et al. Long non-coding RNA MALAT1 is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. Br J. Haematol. 2017, 179, 449–460. [Google Scholar] [CrossRef] [Green Version]
- Taiana, E.; Favasuli, V.; Ronchetti, D.; Todoerti, K.; Pelizzoni, F.; Manzoni, M.; Barbieri, M.; Fabris, S.; Silvestris, I.; Gallo Cantafio, M.E.; et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia 2019. [Google Scholar] [CrossRef]
- Taiana, E.; Ronchetti, D.; Favasuli, V.; Todoerti, K.; Manzoni, M.; Amodio, N.; Tassone, P.; Agnelli, L.; Neri, A. Long non-coding RNA NEAT1 shows high expression unrelated to molecular features and clinical outcome in multiple myeloma. Haematologica 2019, 104, e72–e76. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, N.; Wang, C.; Zhao, H.; Gu, Y. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle 2018, 17, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Pan, J.; Zhang, N.; Wei, W.; Yu, S.; Ai, L. Knockdown of long non-coding RNA H19 inhibits multiple myeloma cell growth via NF-kappaB pathway. Sci. Rep. 2017, 7, 18079. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Zhang, Y.; Liu, W.; Huang, Y.; Shen, X.; Jing, R.; Pu, J.; Wang, X.; Ju, S.; Cong, H.; et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019, 10, 106. [Google Scholar] [CrossRef] [Green Version]
- Shang, Q.; Yang, Z.; Jia, R.; Ge, S. The novel roles of circRNAs in human cancer. Mol. Cancer 2019, 18, 6. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Hansen, T.B.; Venø, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef] [Green Version]
- Ji, T.; Chen, Q.; Tao, S.; Shi, Y.; Chen, Y.; Shen, L.; Wang, C.; Yu, L. The research progress of circular RNAs in hematological malignancies. Hematology 2019, 24, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhang, L.; Wu, J.; Khadka, B.; Fang, Z.; Gu, J.; Tang, B.; Xiao, R.; Pan, G.; Liu, J. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J. Exp Clin. Cancer Res. 2019, 38, 54. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, Y.; Wang, S.; Jiang, J.; Zhang, C.; Jiang, Y.; Wang, X.; Hong, L.; Huang, H. Circ-SMARCA5 suppresses progression of multiple myeloma by targeting miR-767-5p. BMC Cancer 2019, 19, 937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, W.; Wei, Q.; Toden, S.; Yoshida, K.; Nagasaka, T.; Fujiwara, T.; Cai, S.; Qin, H.; Ma, Y.; Goel, A. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin. Cancer Res. 2017, 23, 3918–3928. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Okholm, T.L.H.; Nielsen, M.M.; Hamilton, M.P.; Christensen, L.L.; Vang, S.; Hedegaard, J.; Hansen, T.B.; Kjems, J.; Dyrskjøt, L.; Pedersen, J.S. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ. Genom. Med. 2017, 2, 36. [Google Scholar] [CrossRef]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.M.; Ragusa, M.; et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef] [Green Version]
- Yao, Z.; Luo, J.; Hu, K.; Lin, J.; Huang, H.; Wang, Q.; Zhang, P.; Xiong, Z.; He, C.; Huang, Z.; et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol. Oncol. 2017, 11, 422–437. [Google Scholar] [CrossRef] [Green Version]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [Green Version]
- Dahl, M.; Daugaard, I.; Andersen, M.S.; Hansen, T.B.; Grønbæk, K.; Kjems, J.; Kristensen, L.S. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Investig. 2018, 98, 1657–1669. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Li, C.; Xiao, H.; Dong, H.; Jiang, S.; Fu, Y.; Gong, L. hsa_circ_0007841: A Novel Potential Biomarker and Drug Resistance for Multiple Myeloma. Front. Oncol. 2019, 9, 1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 79. [Google Scholar] [CrossRef] [PubMed]
- Manier, S.; Powers, J.T.; Sacco, A.; Glavey, S.V.; Huynh, D.; Reagan, M.R.; Salem, K.Z.; Moschetta, M.; Shi, J.; Mishima, Y.; et al. The LIN28B/let-7 axis is a novel therapeutic pathway in multiple myeloma. Leukemia 2017, 31, 853–860. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, B.; Li, W.; Wang, L.; Yan, Z.; Li, H.; Yao, Y.; Yao, R.; Xu, K.; Li, Z. MiR-15a/16 regulates the growth of myeloma cells, angiogenesis and antitumor immunity by inhibiting Bcl-2, VEGF-A and IL-17 expression in multiple myeloma. Leuk. Res. 2016, 49, 73–79. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, L.; Shi, M.; Kuang, Y.; Fang, L. Downregulation of miRNA-15a and miRNA-16 promote tumor proliferation in multiple myeloma by increasing CABIN1 expression. Oncol. Lett. 2018, 15, 1287–1296. [Google Scholar] [CrossRef] [Green Version]
- Caracciolo, D.; Di Martino, M.T.; Amodio, N.; Morelli, E.; Montesano, M.; Botta, C.; Scionti, F.; Talarico, D.; Altomare, E.; Gallo Cantafio, M.E.; et al. miR-22 suppresses DNA ligase III addiction in multiple myeloma. Leukemia 2019, 33, 487–498. [Google Scholar] [CrossRef] [Green Version]
- Leotta, M.; Biamonte, L.; Raimondi, L.; Ronchetti, D.; Di Martino, M.T.; Botta, C.; Leone, E.; Pitari, M.R.; Neri, A.; Giordano, A.; et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J. Cell Physiol. 2014, 229, 2106–2116. [Google Scholar] [CrossRef]
- Wong, K.Y.; Liang, R.; So, C.C.; Jin, D.Y.; Costello, J.F.; Chim, C.S. Epigenetic silencing of MIR203 in multiple myeloma. Br J. Haematol. 2011, 154, 569–578. [Google Scholar] [CrossRef]
- Misiewicz-Krzeminska, I.; Sarasquete, M.E.; Quwaider, D.; Krzeminski, P.; Ticona, F.V.; Paino, T.; Delgado, M.; Aires, A.; Ocio, E.M.; Garcia-Sanz, R.; et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica 2013, 98, 640–648. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Zhang, Y.; Huang, C.; Lin, S. miR-215-5p is an anticancer gene in multiple myeloma by targeting RUNX1 and deactivating the PI3K/AKT/mTOR pathway. J. Cell Biochem. 2020, 121, 1475–1490. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Federico, C.; Sacco, A.; Belotti, A.; Ribolla, R.; Cancelli, V.; Giacomini, A.; Ronca, R.; Chiarini, M.; Imberti, L.; Marini, M.; et al. Circulating microRNAs and Their Role in Multiple Myeloma. Noncoding RNA 2019, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.I.; Zabolotskaya, M.V.; King, A.J.; Stewart, H.J.; Horne, G.A.; Chevassut, T.J.; Newbury, S.F. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br J. Cancer 2012, 107, 1987–1996. [Google Scholar] [CrossRef]
- Kubiczkova, L.; Kryukov, F.; Slaby, O.; Dementyeva, E.; Jarkovsky, J.; Nekvindova, J.; Radova, L.; Greslikova, H.; Kuglik, P.; Vetesnikova, E.; et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica 2014, 99, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Hao, M.; Zang, M.; Wendlandt, E.; Xu, Y.; An, G.; Gong, D.; Li, F.; Qi, F.; Zhang, Y.; Yang, Y.; et al. Low serum miR-19a expression as a novel poor prognostic indicator in multiple myeloma. Int. J. Cancer 2015, 136, 1835–1844. [Google Scholar] [CrossRef] [Green Version]
- Yoshizawa, S.; Ohyashiki, J.H.; Ohyashiki, M.; Umezu, T.; Suzuki, K.; Inagaki, A.; Iida, S.; Ohyashiki, K. Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood Cancer J. 2012, 2, e53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevcikova, S.; Kubiczkova, L.; Sedlarikova, L.; Slaby, O.; Hajek, R. Serum miR-29a as a marker of multiple myeloma. Leuk. Lymphoma 2013, 54, 189–191. [Google Scholar] [CrossRef]
- Rocci, A.; Hofmeister, C.C.; Geyer, S.; Stiff, A.; Gambella, M.; Cascione, L.; Guan, J.; Benson, D.M.; Efebera, Y.A.; Talabere, T.; et al. Circulating miRNA markers show promise as new prognosticators for multiple myeloma. Leukemia 2014, 28, 1922–1926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, X.; Zhao, M.; Wu, S.; Yu, W.; Xu, J.; Li, J.; Chen, L. Circulating microRNA 483-5p as a novel biomarker for diagnosis survival prediction in multiple myeloma. Med. Oncol. 2014, 31, 219. [Google Scholar] [CrossRef] [PubMed]
- Besse, L.; Sedlarikova, L.; Kryukov, F.; Nekvindova, J.; Radova, L.; Slaby, O.; Kuglik, P.; Almasi, M.; Penka, M.; Krejci, M.; et al. Circulating Serum MicroRNA-130a as a Novel Putative Marker of Extramedullary Myeloma. PLoS ONE 2015, 10, e0137294. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Zang, M.; Zhao, L.; Deng, S.; Xu, Y.; Qi, F.; An, G.; Qin, Y.; Sui, W.; Li, F.; et al. Serum high expression of miR-214 and miR-135b as novel predictor for myeloma bone disease development and prognosis. Oncotarget 2016, 7, 19589–19600. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Zhao, C.; Li, Y.; Wang, L.; Nie, G.; Peng, J.; Wang, A.; Zhang, P.; Tian, W.; Li, Q.; et al. Osteoclast-derived microRNA-containing exosomes selectively inhibit osteoblast activity. Cell Discov. 2016, 2, 16015. [Google Scholar] [CrossRef]
- Jung, S.H.; Lee, S.E.; Lee, M.; Kim, S.H.; Yim, S.H.; Kim, T.W.; Min, C.K.; Chung, Y.J. Circulating microRNA expressions can predict the outcome of lenalidomide plus low-dose dexamethasone treatment in patients with refractory/relapsed multiple myeloma. Haematologica 2017, 102, e456–e459. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Kumar, R.; Seth, T.; Garg, B.; Sati, H.C.; Sharma, A. Clinical significance of circulatory microRNA-203 in serum as novel potential diagnostic marker for multiple myeloma. J. Cancer Res. Clin. Oncol. 2019, 145, 1601–1611. [Google Scholar] [CrossRef]
- Manier, S.; Liu, C.J.; Avet-Loiseau, H.; Park, J.; Shi, J.; Campigotto, F.; Salem, K.Z.; Huynh, D.; Glavey, S.V.; Rivotto, B.; et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017, 129, 2429–2436. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Li, Y.C.; Geng, C.Y.; Zhou, H.X.; Gao, W.; Chen, W.M. Serum exosomal microRNAs as novel biomarkers for multiple myeloma. Hematol. Oncol. 2019, 37, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Bouyssou, J.M.; Liu, C.J.; Bustoros, M.; Sklavenitis-Pistofidis, R.; Aljawai, Y.; Manier, S.; Yosef, A.; Sacco, A.; Kokubun, K.; Tsukamoto, S.; et al. Profiling of circulating exosomal miRNAs in patients with Waldenström Macroglobulinemia. PLoS ONE 2018, 13, e0204589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isin, M.; Ozgur, E.; Cetin, G.; Erten, N.; Aktan, M.; Gezer, U.; Dalay, N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin. Chim. Acta 2014, 431, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Chen, H.; Shen, X.; Wang, X.; Ju, S.; Lu, M.; Cong, H. Serum level of long noncoding RNA H19 as a diagnostic biomarker of multiple myeloma. Clin. Chim Acta 2018, 480, 199–205. [Google Scholar] [CrossRef]

| PC Disorder | Bone Marrow PCs or Lymphoplasmacytic Cells, % | MC Serum/24 h FLC Urine | CRAB Features (Y/N) * | Best Therapeutic Options (First Line) |
|---|---|---|---|---|
| Symptomatic Multiple Myeloma | >10% PCs | > 3g/dL 500 mg | Y | PIs/ImiDs +/− MoAbs ** based regimens |
| Smoldering Multiple Myeloma | >10%<60% PCs | >or <3 g/dL/ 500 mg | N | No therapy−strict follow up |
| Plasma Cell Leukemia | >20% circulating PC in peripheral blood | > or <3 g/dL 500 mg | Y | PIs/ImiDs based regimens |
| MGUS | <10% PCs | <3 g/dL 500 mg | N | No therapy-follow up |
| Primary Amyloidosis | <10% PCs | <3 g/dL 500 mg | N | PIs/ImiDs +/− MoAbs ** based regimens |
| Solitary Plasmacytoma | <10% PCs | <3 g/dL 500 mg | Y *** | Radiotherapy |
| Smoldering Waldenström Macroglobulinemia | Usually <30% LPCs | <3g/dL | N | No therapy−strict follow up |
| Waldenström Macroglobulinemia | Usually >30% LPCs | >3 g/dL | N | PI based regimens+anti CD20 monoclonal; BTK inhibitors if MYD88mut |
| POEMS | >10% (in the case of an underlying MM) | >3 g/ dL (in the case of an underlying MM) | Y (in the case of an underlying MM) | MM regimens (****) |
| Name | Class | Disease | Role in Tumorigenesis | Mechanisms sncRNAs→targets lncRNAs→pathways | References |
|---|---|---|---|---|---|
| Let-7b | miRNA | MM | Tumor-suppressor | MYC | [174] |
| miR-15a/16-1 | miRNA | MM | Tumor-suppressor | MAP3KIP3, BCL2, AKT3, RPS6, VEGFA, IL17A, CABIN1 | [105,175,176] |
| miR-17-92 | miRNA | MM | Tumor-promoting | BCL2l11, TP53, PTEN, CDKN1A, SOCS1 | [104,129] |
| miR-21 | miRNA | MM | Tumor-promoting | PTEN, PIAS3 | [137,138] |
| miR-22 | miRNA | MM | Tumor-suppressor | LIG3 | [177] |
| miR-29b | miRNA | MM | Tumor-suppressor | MCL1, CDK6, SP1, DNMT3A DNMT3B, FOS, MMP2 | [116,122,123,136,139,142] |
| miR-34a | miRNA | MM | Tumor-suppressor | BCL2, CDK6, NOTCH1 | [110] |
| miR-125a | miRNA | MM | Tumor-promoting | TP53 | [178] |
| miR-125b | miRNA | MM | Tumor-suppressor | IRF4, PRDM1 | [112] |
| miR-155 | miRNA | MM | Tumor-suppressor | PSMβ5 | [119] |
| miR-155 | miRNA | WM | Tumor-promoting | CEBPB, SMAD5, SOCS1, MAFB, SHANK2, SH3PXD2A | [113] |
| miR-181a/b | miRNA | MM | Tumor-suppressor | KAT2B | [104] |
| miR-194-2-192 | miRNA | MM | Tumor-suppressor | MDM2, IGF1 | [126] |
| miR-199a-5p | miRNA | MM | Tumor-suppressor | HIF1A, VEGFA, CXCL8, FGF | [140] |
| miR-203 | miRNA | MM | Tumor-suppressor | CREB-1 | [179] |
| miR-215-194-1 | miRNA | MM | Tumor-suppressor | MDM2, IGF1R | [126] |
| miR-214 | miRNA | MM | Tumor-suppressor | PSMD10, ASF1B | [180] |
| miR-215 | miRNA | MM | Tumor-suppressor | RUNX1 | [181] |
| miR-221/222 | miRNA | MM | Tumor-promoting | CDKN1B, CDKN1C, BBC3, PTEN | [106,107,109] |
| piRNA-823 | piRNA | MM | Tumor-promoting | DNMTA, DNMT3B | [133] |
| ACA11 | snoRNA | MM | Tumor-promoting | DHX9, ILF3, NCL, ADAR, HNRNPU | [25] |
| MALAT1 | lncRNA | MM | Tumor-promoting | Transcriptional regulation of proteasome machinery; Activation of A-NHEJ DNA repair | [151,152] |
| NEAT1 | lncRNA | MM | Tumor-promoting | Activation of HR DNA repair. | [154] |
| H19 | lncRNA | MM | Tumor-promoting | Activation of NF-kB pathway; ceRNA of miR-29b-3p resulting in positive regulation of MCL1; | [157,158] |
| CCAT1 | lncRNA | MM | Tumor-promoting | ceRNA of miR-181a-5p resulting in positive regulation of HOXA | [156] |
| circ_00001190 | circRNA | MM | Tumor-suppressor | ceRNA of miR-767-5p resulting in upregulation of MAPK4 | [162] |
| circ_SMARCA5 | circRNA | MM | Tumor-suppressor | ceRNA of miR-767-5p | [163] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morelli, E.; Gullà, A.; Rocca, R.; Federico, C.; Raimondi, L.; Malvestiti, S.; Agosti, V.; Rossi, M.; Costa, G.; Giavaresi, G.; et al. The Non-Coding RNA Landscape of Plasma Cell Dyscrasias. Cancers 2020, 12, 320. https://doi.org/10.3390/cancers12020320
Morelli E, Gullà A, Rocca R, Federico C, Raimondi L, Malvestiti S, Agosti V, Rossi M, Costa G, Giavaresi G, et al. The Non-Coding RNA Landscape of Plasma Cell Dyscrasias. Cancers. 2020; 12(2):320. https://doi.org/10.3390/cancers12020320
Chicago/Turabian StyleMorelli, Eugenio, Annamaria Gullà, Roberta Rocca, Cinzia Federico, Lavinia Raimondi, Stefano Malvestiti, Valter Agosti, Marco Rossi, Giosuè Costa, Gianluca Giavaresi, and et al. 2020. "The Non-Coding RNA Landscape of Plasma Cell Dyscrasias" Cancers 12, no. 2: 320. https://doi.org/10.3390/cancers12020320
APA StyleMorelli, E., Gullà, A., Rocca, R., Federico, C., Raimondi, L., Malvestiti, S., Agosti, V., Rossi, M., Costa, G., Giavaresi, G., Azab, A. K., Cagnetta, A., Cea, M., Tagliaferri, P., Neri, A., Munshi, N. C., Viglietto, G., Tassone, P., & Amodio, N. (2020). The Non-Coding RNA Landscape of Plasma Cell Dyscrasias. Cancers, 12(2), 320. https://doi.org/10.3390/cancers12020320











