The Impact of Focused Ultrasound in Two Tumor Models: Temporal Alterations in the Natural History on Tumor Microenvironment and Immune Cell Response
Abstract
1. Introduction
2. Results
2.1. Natural History Progress of B16 or 4T1 Tumors TME
2.1.1. Proteomic Profiling During the Natural History Growth of Naïve Tumors
2.1.2. Flow Cytometry of Naïve Tumors
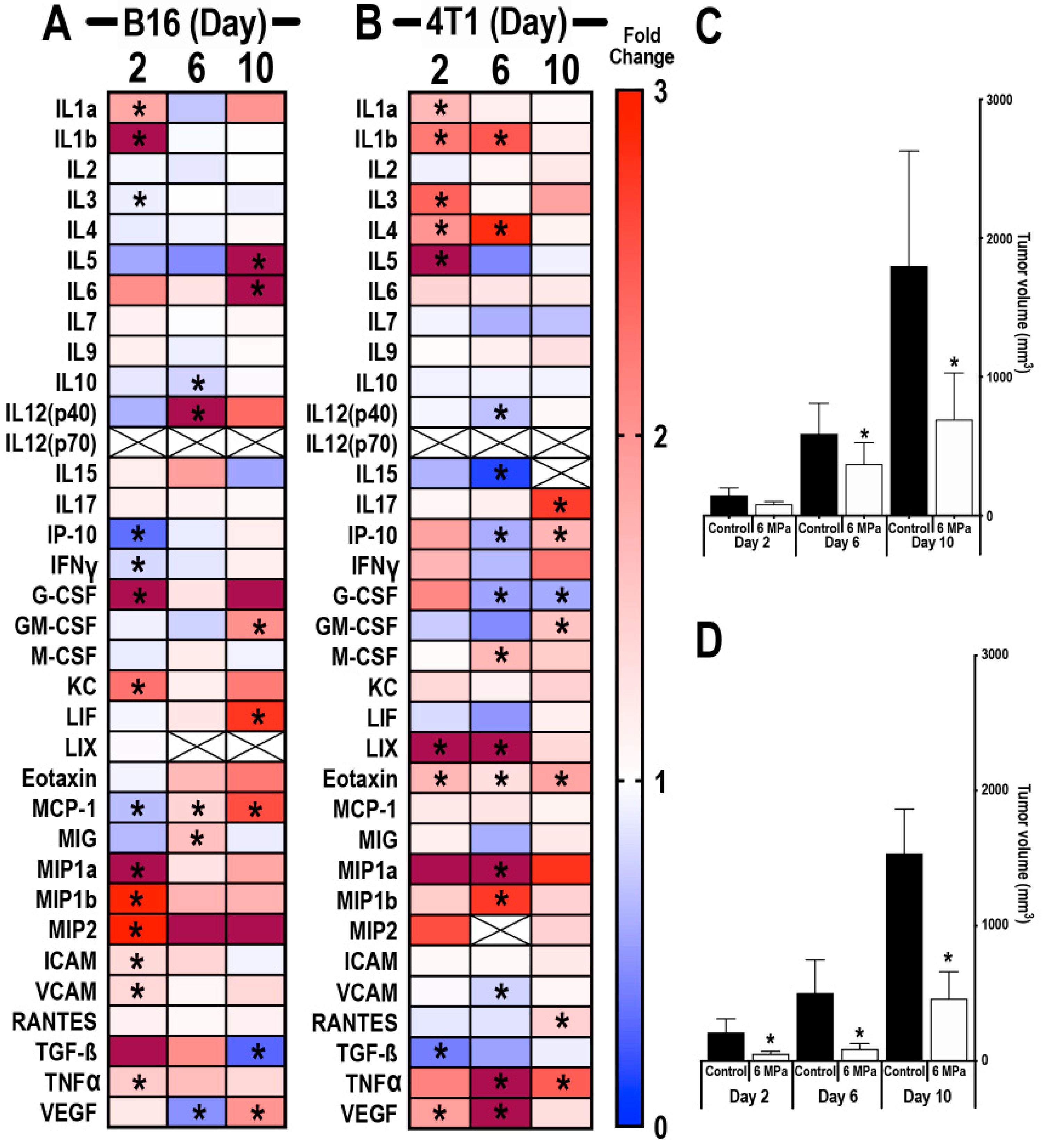
2.2. pFUS-Immunomodulatory Effects in Relation to Different Growth Stages of B16 and 4T1 Tumors
2.2.1. Proteomic Response to pFUS on Different Days with Changes in TME of B16 and 4T1 Tumors
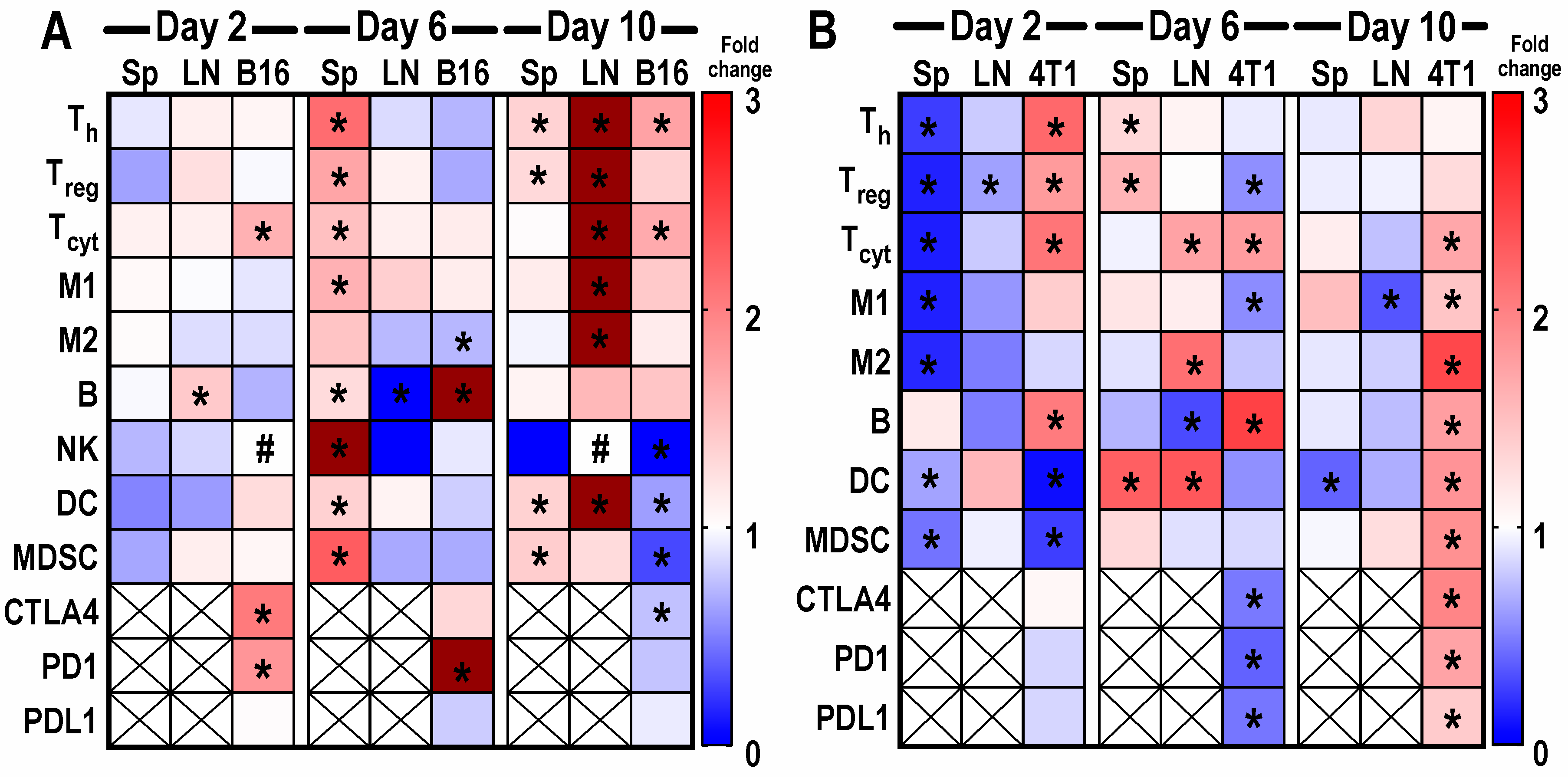
2.2.2. Flow Cytometry Analysis of Sonicated Tumors on Different Days Following pFUS
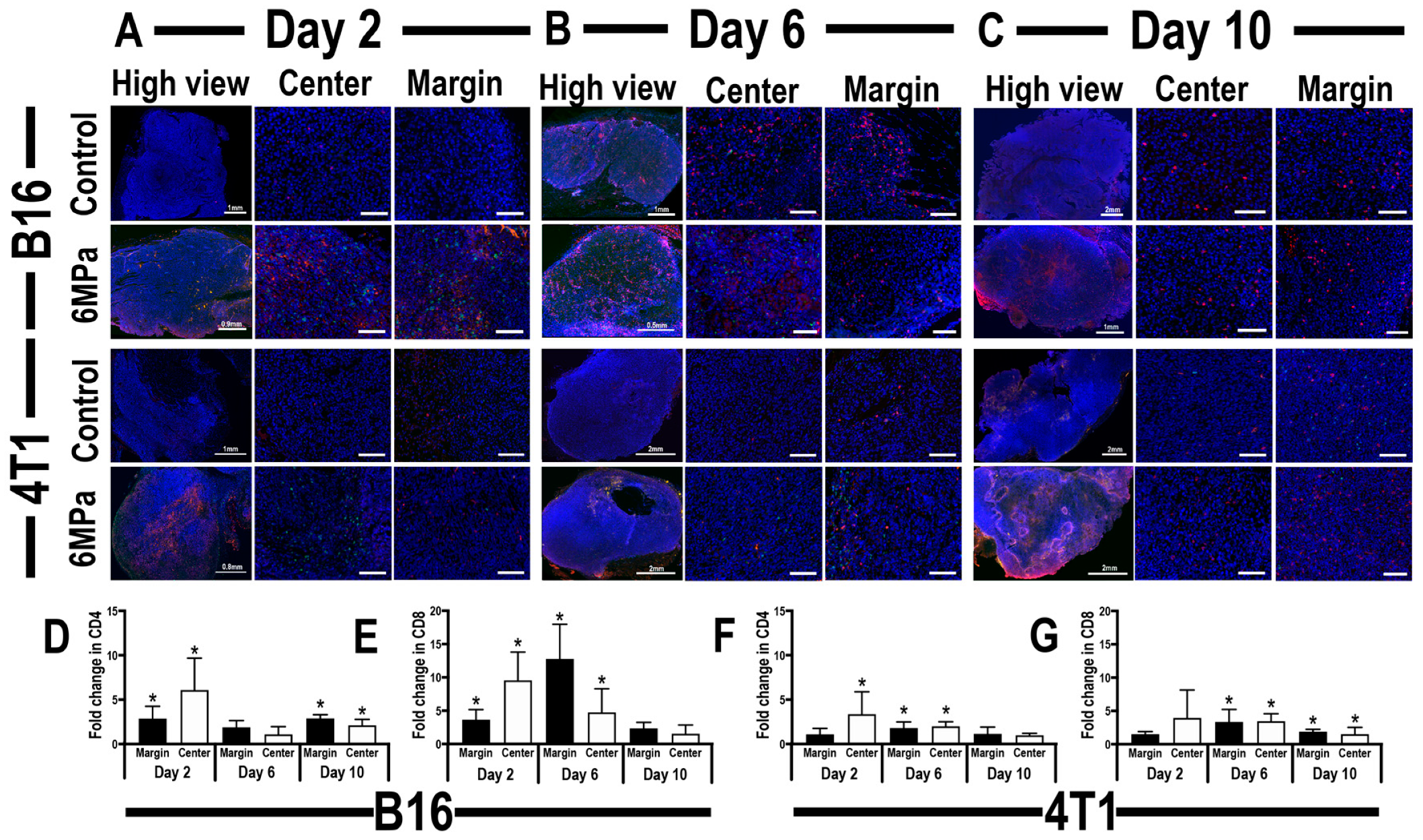
2.2.3. Histological Evaluation of Tumor Infiltrating CD4+ Th and CD8+ T-cells Localization and Relative DNA Damage in pFUS-Treated Tumors
3. Discussion
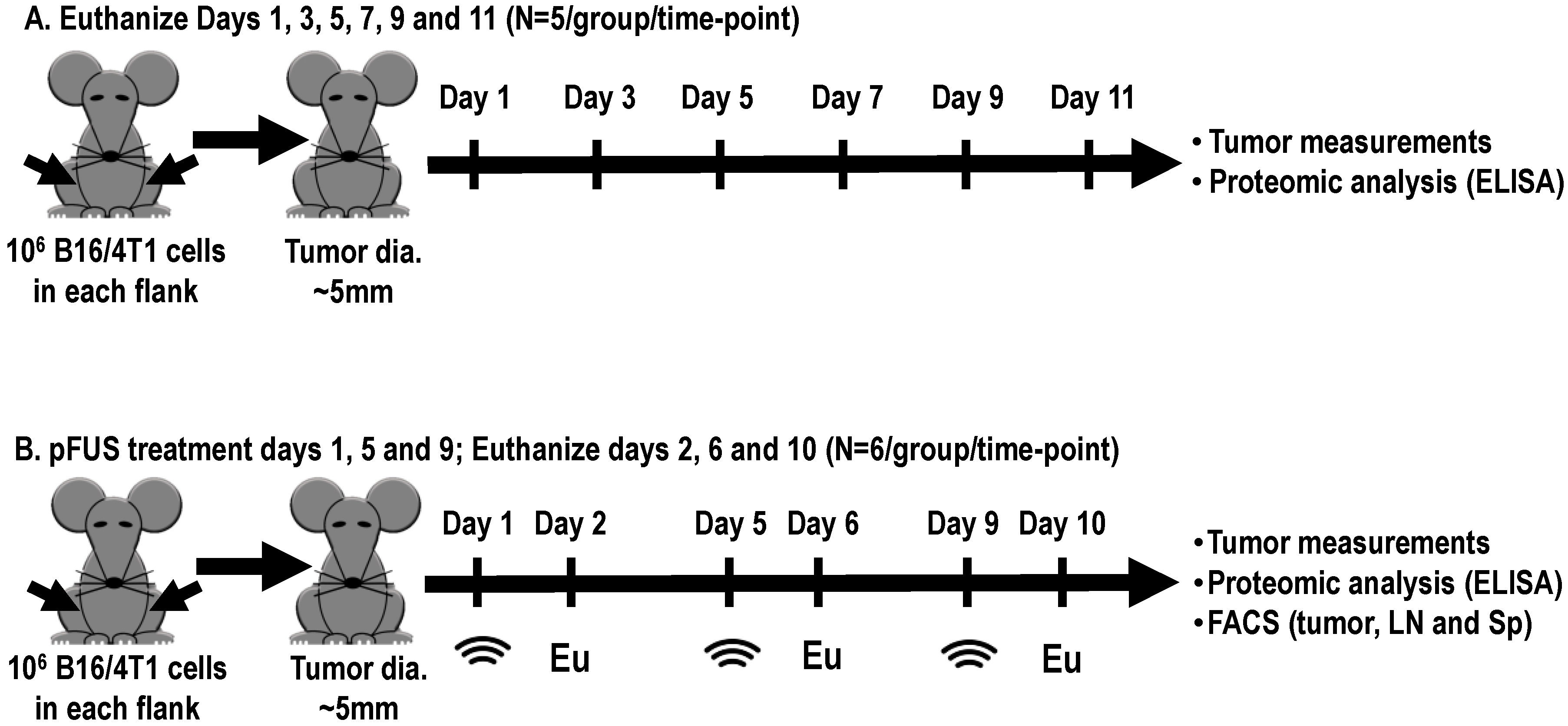
4. Materials and Methods
4.1. Cell Culture
4.2. Mouse Xenograft B16 and 4T1 Tumor Models
4.3. pFUS Treatment
4.4. Proteomic Analyses
4.5. Flow Cytometric Analyses
4.6. Histology and Immunohistochemistry
4.7. DNA Damage Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tang, H.; Qiao, J.; Fu, Y.X. Immunotherapy and tumor microenvironment. Cancer Lett. 2016, 370, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Klemm, F.; Joyce, J.A. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015, 25, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Devaud, C.; John, L.B.; Westwood, J.A.; Darcy, P.K.; Kershaw, M.H. Immune modulation of the tumor microenvironment for enhancing cancer immunotherapy. Oncoimmunology 2013, 2, e25961. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; de Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501, 346–354. [Google Scholar] [CrossRef]
- Lee, S.; Margolin, K. Cytokines in cancer immunotherapy. Cancers (Basel) 2011, 3, 3856–3893. [Google Scholar] [CrossRef]
- Makkouk, A.; Weiner, G.J. Cancer immunotherapy and breaking immune tolerance: New approaches to an old challenge. Cancer Res. 2015, 75, 5–10. [Google Scholar] [CrossRef]
- Balkwill, F. Cancer and the chemokine network. Nat. Rev. Cancer 2004, 4, 540–550. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhu, X.Q.; Zhang, J.; Xu, Z.L.; Lu, P.; Wu, F. Changes in circulating immunosuppressive cytokine levels of cancer patients after high intensity focused ultrasound treatment. Ultrasound. Med. Biol. 2008, 34, 81–87. [Google Scholar] [CrossRef]
- Zou, W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat. Rev. Cancer 2005, 5, 263–274. [Google Scholar] [CrossRef]
- Taylor, A.; Verhagen, J.; Blaser, K.; Akdis, M.; Akdis, C.A. Mechanisms of immune suppression by interleukin-10 and transforming growth factor-beta: The role of T regulatory cells. Immunology 2006, 117, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Barker, H.E.; Paget, J.T.; Khan, A.A.; Harrington, K.J. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat. Rev. Cancer 2015, 15, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Restifo, N.P.; Dudley, M.E.; Rosenberg, S.A. Adoptive immunotherapy for cancer: Harnessing the T cell response. Nat. Rev. Immunol. 2012, 12, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A. Decade in review-cancer immunotherapy: Entering the mainstream of cancer treatment. Nat. Rev. Clin. Oncol. 2014, 11, 630–632. [Google Scholar] [CrossRef]
- Quezada, S.A.; Peggs, K.S.; Simpson, T.R.; Allison, J.P. Shifting the equilibrium in cancer immunoediting: From tumor tolerance to eradication. Immunol. Rev. 2011, 241, 104–118. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef]
- Twyman-Saint Victor, C.; Rech, A.J.; Maity, A.; Rengan, R.; Pauken, K.E.; Stelekati, E.; Benci, J.L.; Xu, B.; Dada, H.; Odorizzi, P.M.; et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2015, 520, 373–377. [Google Scholar] [CrossRef]
- Hamanishi, J.; Mandai, M.; Ikeda, T.; Minami, M.; Kawaguchi, A.; Murayama, T.; Kanai, M.; Mori, Y.; Matsumoto, S.; Chikuma, S.; et al. Safety and antitumor activity of anti-PD-1 antibody, Nivolumab, in patients with platinum-resistant ovarian cancer. J. Clin. Oncol. 2015, 33, 4015–4022. [Google Scholar] [CrossRef]
- Santegoets, S.J.; Stam, A.G.; Lougheed, S.M.; Gall, H.; Scholten, P.E.; Reijm, M.; Jooss, K.; Sacks, N.; Hege, K.; Lowy, I.; et al. T cell profiling reveals high CD4+CTLA-4 + T cell frequency as dominant predictor for survival after prostate GVAX/ipilimumab treatment. Cancer Immunol. Immunother. 2013, 62, 245–256. [Google Scholar] [CrossRef]
- Dodd, G.D., 3rd; Soulen, M.C.; Kane, R.A.; Livraghi, T.; Lees, W.R.; Yamashita, Y.; Gillams, A.R.; Karahan, O.I.; Rhim, H. Minimally invasive treatment of malignant hepatic tumors: At the threshold of a major breakthrough. Radiographics 2000, 20, 9–27. [Google Scholar] [CrossRef]
- Showalter, A.; Limaye, A.; Oyer, J.L.; Igarashi, R.; Kittipatarin, C.; Copik, A.J.; Khaled, A.R. Cytokines in immunogenic cell death: Applications for cancer immunotherapy. Cytokine 2017, 97, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Webb, H.; Lubner, M.G.; Hinshaw, J.L. Thermal ablation. Semin Roentgenol 2011, 46, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Van den Bijgaart, R.J.; Eikelenboom, D.C.; Hoogenboom, M.; Futterer, J.J.; den Brok, M.H.; Adema, G.J. Thermal and mechanical high-intensity focused ultrasound: Perspectives on tumor ablation, immune effects and combination strategies. Cancer Immunol Immunother 2017, 66, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Matsutani, N.; Nakayama, T.; Dejima, H.; Uehara, H.; Kawamura, M. Immunological effect of local ablation combined with immunotherapy on solid malignancies. Chin. J. Cancer 2017, 36, 49. [Google Scholar] [CrossRef] [PubMed]
- Den Brok, M.H.; Sutmuller, R.P.; van der Voort, R.; Bennink, E.J.; Figdor, C.G.; Ruers, T.J.; Adema, G.J. In situ tumor ablation creates an antigen source for the generation of antitumor immunity. Cancer Res 2004, 64, 4024–4029. [Google Scholar] [CrossRef] [PubMed]
- Prise, K.M.; O’Sullivan, J.M. Radiation-induced bystander signalling in cancer therapy. Nat Rev Cancer 2009, 9, 351–360. [Google Scholar] [CrossRef]
- Whiteside, T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef]
- Alteber, Z.; Azulay, M.; Cafri, G.; Vadai, E.; Tzehoval, E.; Eisenbach, L. Cryoimmunotherapy with local co-administration of ex vivo generated dendritic cells and CpG-ODN immune adjuvant, elicits a specific antitumor immunity. Cancer Immunol. Immunother. 2014, 63, 369–380. [Google Scholar] [CrossRef]
- Den Brok, M.H.; Nierkens, S.; Wagenaars, J.A.; Ruers, T.J.; Schrier, C.C.; Rijke, E.O.; Adema, G.J. Saponin-based adjuvants create a highly effective anti-tumor vaccine when combined with in situ tumor destruction. Vaccine 2012, 30, 737–744. [Google Scholar] [CrossRef]
- Chiang, P.H.; Liu, Y.Y. Comparisons of oncological and functional outcomes among radical retropubic prostatectomy, high dose rate brachytherapy, cryoablation and high-intensity focused ultrasound for localized prostate cancer. Springerplus 2016, 5, 1905. [Google Scholar] [CrossRef]
- Mauri, G.; Sconfienza, L.M.; Pescatori, L.C.; Fedeli, M.P.; Ali, M.; Di Leo, G.; Sardanelli, F. Technical success, technique efficacy and complications of minimally-invasive imaging-guided percutaneous ablation procedures of breast cancer: A systematic review and meta-analysis. Eur. Radiol. 2017, 27, 3199–3210. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.E. High-intensity focused ultrasound in the treatment of solid tumours. Nat. Rev. Cancer 2005, 5, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Merckel, L.G.; Knuttel, F.M.; Deckers, R.; van Dalen, T.; Schubert, G.; Peters, N.H.; Weits, T.; van Diest, P.J.; Mali, W.P.; Vaessen, P.H.; et al. First clinical experience with a dedicated MRI-guided high-intensity focused ultrasound system for breast cancer ablation. Eur. Radiol. 2016, 26, 4037–4046. [Google Scholar] [CrossRef] [PubMed]
- Mauri, G.; Nicosia, L.; Xu, Z.; Di Pietro, S.; Monfardini, L.; Bonomo, G.; Varano, G.M.; Prada, F.; Della Vigna, P.; Orsi, F. Focused ultrasound: Tumour ablation and its potential to enhance immunological therapy to cancer. Br. J. Radiol. 2018, 91, 20170641. [Google Scholar] [CrossRef] [PubMed]
- Eranki, A.; Srinivasan, P.; Ries, M.; Kim, A.; Lazarski, C.A.; Rossi, C.T.; Khokhlova, T.D.; Wilson, E.; Knoblach, S.; Sharma, K.V.; et al. High intensity focused ultrasound (HIFU) triggers immune sensitization of refractory murine neuroblastoma to checkpoint inhibitor therapy. Clin. Cancer Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Burks, S.R.; Nguyen, B.A.; Tebebi, P.A.; Kim, S.J.; Bresler, M.N.; Ziadloo, A.; Street, J.M.; Yuen, P.S.; Star, R.A.; Frank, J.A. Pulsed focused ultrasound pretreatment improves mesenchymal stromal cell efficacy in preventing and rescuing established acute kidney injury in mice. Stem Cells 2015, 33, 1241–1253. [Google Scholar] [CrossRef] [PubMed]
- Burks, S.R.; Ziadloo, A.; Hancock, H.A.; Chaudhry, A.; Dean, D.D.; Lewis, B.K.; Frenkel, V.; Frank, J.A. Investigation of cellular and molecular responses to pulsed focused ultrasound in a mouse model. PLoS ONE 2011, 6, e24730. [Google Scholar] [CrossRef]
- Jang, K.W.; Tu, T.W.; Nagle, M.E.; Lewis, B.K.; Burks, S.R.; Frank, J.A. Molecular and histological effects of MR-guided pulsed focused ultrasound to the rat heart. J. Transl. Med. 2017, 15, 252. [Google Scholar] [CrossRef]
- Kovacs, Z.I.; Kim, S.; Jikaria, N.; Qureshi, F.; Milo, B.; Lewis, B.K.; Bresler, M.; Burks, S.R.; Frank, J.A. Disrupting the blood-brain barrier by focused ultrasound induces sterile inflammation. Proc. Natl. Acad. Sci. USA 2017, 114, E75–E84. [Google Scholar] [CrossRef]
- Cohen-Inbar, O.; Xu, Z.; Sheehan, J.P. Focused ultrasound-aided immunomodulation in glioblastoma multiforme: A therapeutic concept. J. Ther. Ultrasound. 2016, 4, 2. [Google Scholar] [CrossRef]
- Aydin, O.; Chandran, P.; Lorsung, R.R.; Cohen, G.; Burks, S.R.; Frank, J.A. The proteomic effects of pulsed focused ultrasound on tumor microenvironments of murine melanoma and breast cancer models. Ultrasound. Med. Biol. 2019, 45, 3232–3245. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.W.; Karin, M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Investig. 2007, 117, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Ben-Baruch, A. Inflammation-associated immune suppression in cancer: The roles played by cytokines, chemokines and additional mediators. Semin. Cancer Biol. 2006, 16, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Latacz, E.; Caspani, E.; Barnhill, R.; Lugassy, C.; Verhoef, C.; Grunhagen, D.; Van Laere, S.; Fernandez Moro, C.; Gerling, M.; Dirix, M.; et al. Pathological features of vessel co-option versus sprouting angiogenesis. Angiogenesis 2019. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B. IFNgamma: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef]
- Jiang, W.; Chan, C.K.; Weissman, I.L.; Kim, B.Y.S.; Hahn, S.M. Immune priming of the tumor microenvironment by radiation. Trends Cancer 2016, 2, 638–645. [Google Scholar] [CrossRef]
- Chew, V.; Toh, H.C.; Abastado, J.P. Immune microenvironment in tumor progression: Characteristics and challenges for therapy. J. Oncol. 2012, 2012, 608406. [Google Scholar] [CrossRef]
- Vasilevska, J.; De Souza, G.A.; Stensland, M.; Skrastina, D.; Zhulenvovs, D.; Paplausks, R.; Kurena, B.; Kozlovska, T.; Zajakina, A. Comparative protein profiling of B16 mouse melanoma cells susceptible and non-susceptible to alphavirus infection: Effect of the tumor microenvironment. Cancer Biol. Ther. 2016, 17, 1035–1050. [Google Scholar] [CrossRef][Green Version]
- Villanueva, J.; Herlyn, M. Melanoma and the tumor microenvironment. Curr. Oncol. Rep. 2008, 10, 439–446. [Google Scholar] [CrossRef]
- Figenschau, S.L.; Knutsen, E.; Urbarova, I.; Fenton, C.; Elston, B.; Perander, M.; Mortensen, E.S.; Fenton, K.A. ICAM1 expression is induced by proinflammatory cytokines and associated with TLS formation in aggressive breast cancer subtypes. Sci. Rep. 2018, 8, 11720. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Ariztia, E.V.; Lee, C.J.; Gogoi, R.; Fishman, D.A. The tumor microenvironment: Key to early detection. Crit. Rev. Clin. Lab. Sci. 2006, 43, 393–425. [Google Scholar] [CrossRef]
- Roma-Rodrigues, C.; Raposo, L.R.; Cabral, R.; Paradinha, F.; Baptista, P.V.; Fernandes, A.R. Tumor microenvironment modulation via gold nanoparticles targeting malicious exosomes: Implications for cancer diagnostics and therapy. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, Z.; Xu, X.; Yu, Z.; Mi, J. The influence of microenvironment on tumor immunotherapy. FEBS J. 2019, 286, 4160–4175. [Google Scholar] [CrossRef]
- Karakasilioti, I.; Kamileri, I.; Chatzinikolaou, G.; Kosteas, T.; Vergadi, E.; Robinson, A.R.; Tsamardinos, I.; Rozgaja, T.A.; Siakouli, S.; Tsatsanis, C.; et al. DNA damage triggers a chronic autoinflammatory response, leading to fat depletion in NER progeria. Cell Metab. 2013, 18, 403–415. [Google Scholar] [CrossRef]
- Borges, H.L.; Linden, R.; Wang, J.Y. DNA damage-induced cell death: Lessons from the central nervous system. Cell Res. 2008, 18, 17–26. [Google Scholar] [CrossRef]
- Honda, H.; Kondo, T.; Zhao, Q.L.; Feril, L.B., Jr.; Kitagawa, H. Role of intracellular calcium ions and reactive oxygen species in apoptosis induced by ultrasound. Ultrasound. Med. Biol. 2004, 30, 683–692. [Google Scholar] [CrossRef]
- Burks, S.R.; Lorsung, R.M.; Nagle, M.E.; Tu, T.W.; Frank, J.A. Focused ultrasound activates voltage-gated calcium channels through depolarizing TRPC1 sodium currents in kidney and skeletal muscle. Theranostics 2019, 9, 5517–5531. [Google Scholar] [CrossRef]
- Fan, Z.; Kumon, R.E.; Park, J.; Deng, C.X. Intracellular delivery and calcium transients generated in sonoporation facilitated by microbubbles. J. Control Release 2010, 142, 31–39. [Google Scholar] [CrossRef]
- Khalil, D.N.; Smith, E.L.; Brentjens, R.J.; Wolchok, J.D. The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016, 13, 394. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Fridman, W.H.; Pages, F. The adaptive immunologic microenvironment in colorectal cancer: A novel perspective. Cancer Res. 2007, 67, 1883–1886. [Google Scholar] [CrossRef] [PubMed]
- Angell, H.; Galon, J. From the immune contexture to the immunoscore: The role of prognostic and predictive immune markers in cancer. Curr. Opin. Immunol. 2013, 25, 261–267. [Google Scholar] [CrossRef]
- Galluzzi, L.; Senovilla, L.; Vacchelli, E.; Eggermont, A.; Fridman, W.H.; Galon, J.; Sautes-Fridman, C.; Tartour, E.; Zitvogel, L.; Kroemer, G. Trial watch: Dendritic cell-based interventions for cancer therapy. Oncoimmunology 2012, 1, 1111–1134. [Google Scholar] [CrossRef]
- Galon, J.; Angell, H.K.; Bedognetti, D.; Marincola, F.M. The continuum of cancer immunosurveillance: Prognostic, predictive, and mechanistic signatures. Immunity 2013, 39, 11–26. [Google Scholar] [CrossRef]
- Taube, J.M.; Galon, J.; Sholl, L.M.; Rodig, S.J.; Cottrell, T.R.; Giraldo, N.A.; Baras, A.S.; Patel, S.S.; Anders, R.A.; Rimm, D.L.; et al. Implications of the tumor immune microenvironment for staging and therapeutics. Mod. Pathol. 2018, 31, 214–234. [Google Scholar] [CrossRef]
- National Research Council (U.S.); Committee for the Update of the Guide for the Care and Use of Laboratory Animals; Institute for Laboratory Animal Research (U.S.); National Academies Press (U.S.). Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011; p. xxv, 220. [Google Scholar]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohen, G.; Chandran, P.; Lorsung, R.M.; Tomlinson, L.E.; Sundby, M.; Burks, S.R.; Frank, J.A. The Impact of Focused Ultrasound in Two Tumor Models: Temporal Alterations in the Natural History on Tumor Microenvironment and Immune Cell Response. Cancers 2020, 12, 350. https://doi.org/10.3390/cancers12020350
Cohen G, Chandran P, Lorsung RM, Tomlinson LE, Sundby M, Burks SR, Frank JA. The Impact of Focused Ultrasound in Two Tumor Models: Temporal Alterations in the Natural History on Tumor Microenvironment and Immune Cell Response. Cancers. 2020; 12(2):350. https://doi.org/10.3390/cancers12020350
Chicago/Turabian StyleCohen, Gadi, Parwathy Chandran, Rebecca M. Lorsung, Lauren E. Tomlinson, Maggie Sundby, Scott R. Burks, and Joseph A. Frank. 2020. "The Impact of Focused Ultrasound in Two Tumor Models: Temporal Alterations in the Natural History on Tumor Microenvironment and Immune Cell Response" Cancers 12, no. 2: 350. https://doi.org/10.3390/cancers12020350
APA StyleCohen, G., Chandran, P., Lorsung, R. M., Tomlinson, L. E., Sundby, M., Burks, S. R., & Frank, J. A. (2020). The Impact of Focused Ultrasound in Two Tumor Models: Temporal Alterations in the Natural History on Tumor Microenvironment and Immune Cell Response. Cancers, 12(2), 350. https://doi.org/10.3390/cancers12020350




