RAD52: Viral Friend or Foe?
Abstract
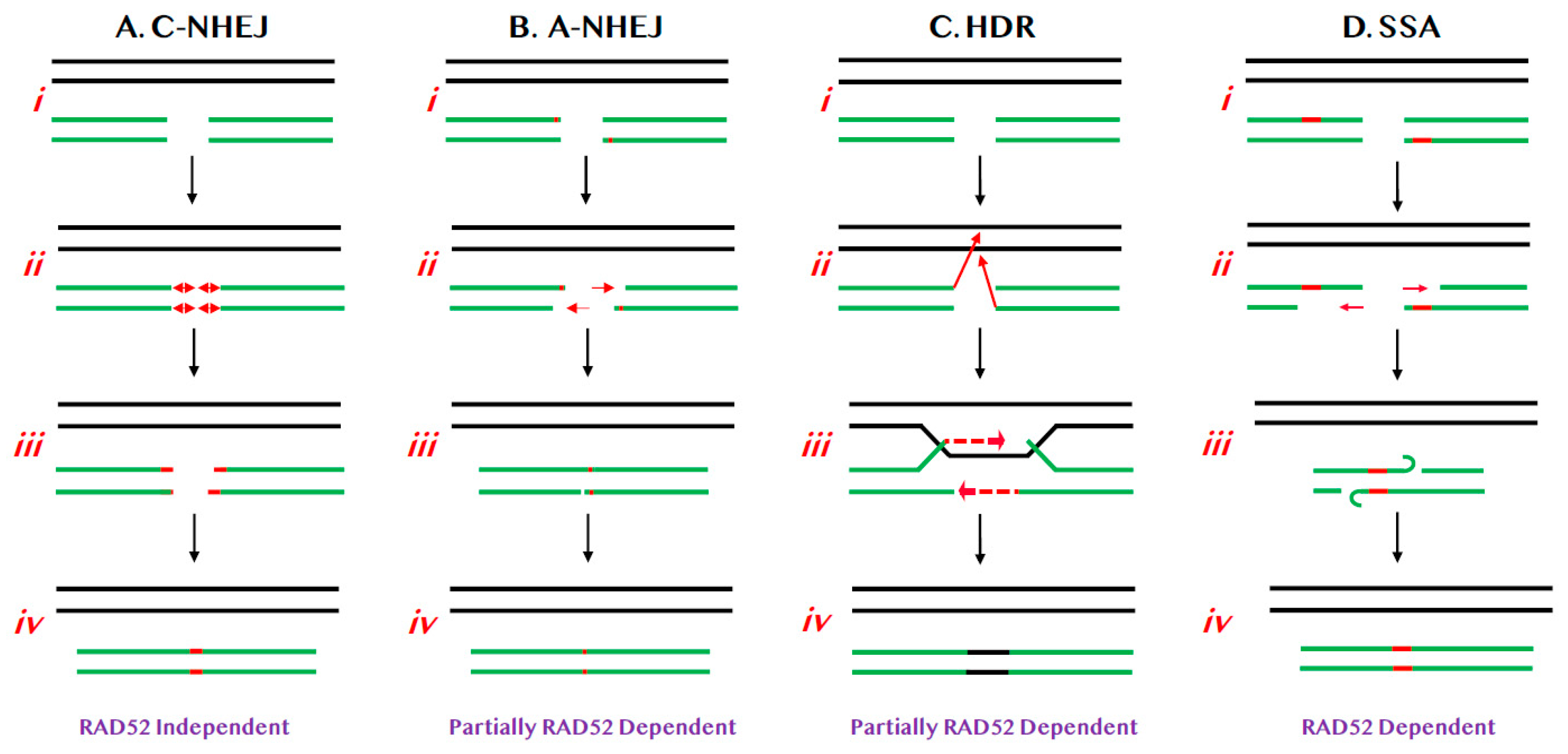
:1. Pathways of DNA Double-Stranded Break Repair
1.1. NHEJ-Mediated DSB Repair
1.2. HDR-Mediated DSB Repair
2. RAD52 and BIR
3. AAV Replication and Integration
4. AAV Replication, Integration and RAD52
4.1. AAV Replication and RAD52
4.2. AAV Integration and RAD52
5. Other Viruses and RAD52
5.1. Human Immunodeficiency Virus (HIV) and RAD52
5.2. Herpes Simplex Virus (HSV) Replication
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Bennett, C.B.; Lewis, A.L.; Baldwin, K.K.; Resnick, M.A. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA 1993, 90, 5613–5617. [Google Scholar] [CrossRef] [Green Version]
- Woodbine, L.; Gennery, A.R.; Jeggo, P.A. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair (Amsterdam) 2014, 16, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Jasin, M.; Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013, 5, a012740. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.J.; Chen, B.P.; Chen, D.J. DNA-PK: A dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amsterdam) 2014, 17, 21–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.E.; Mitchell, R.A.; Cheng, A.; Hendrickson, E.A. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 1997, 17, 1425–1433. [Google Scholar] [CrossRef] [Green Version]
- Betermier, M.; Bertrand, P.; Lopez, B.S. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014, 10, e1004086. [Google Scholar] [CrossRef] [Green Version]
- Krajewska, M.; Fehrmann, R.S.; de Vries, E.G.; van Vugt, M.A. Regulators of homologous recombination repair as novel targets for cancer treatment. Front. Genet. 2015, 6, 96. [Google Scholar] [CrossRef] [Green Version]
- Ward, A.; Khanna, K.K.; Wiegmans, A.P. Targeting homologous recombination, new pre-clinical and clinical therapeutic combinations inhibiting RAD51. Cancer Treat. Rev. 2015, 41, 35–45. [Google Scholar] [CrossRef]
- Huang, A.; Garraway, L.A.; Ashworth, A.; Weber, B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2019. [Google Scholar] [CrossRef] [PubMed]
- Toma, M.; Sullivan-Reed, K.; Sliwinski, T.; Skorski, T. RAD52 as a potential target for synthetic lethality-based anticancer therapies. Cancers 2019, 11, 1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalan, M.; Olsen, K.S.; Powell, S.N. Emerging roles of RAD52 in genome maintenance. Cancers 2019, 11, 1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iliakis, G.; Murmann, T.; Soni, A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2015, 793, 166–175. [Google Scholar] [CrossRef]
- Bhargava, R.; Sandhu, M.; Muk, S.; Lee, G.; Vaidehi, N.; Stark, J.M. C-NHEJ without indels is robust and requires synergistic function of distinct XLF domains. Nat. Commun. 2018, 9, 2484. [Google Scholar] [CrossRef]
- Kan, Y.; Batada, N.N.; Hendrickson, E.A. Human somatic cells deficient for RAD52 are impaired for viral integration and compromised for most aspects of homology-directed repair. DNA Repair (Amsterdam) 2017, 55, 64–75. [Google Scholar] [CrossRef]
- Xue, C.; Greene, E.C. New roles for RAD52 in DNA repair. Cell Res. 2018, 28, 1127–1128. [Google Scholar] [CrossRef]
- Hunter, N. Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 2015, 7. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, M.; de Lange, T. 53BP1: Prochoice in DNA repair. Trends Cell Biol. 2014, 24, 108–117. [Google Scholar] [CrossRef] [Green Version]
- Bunting, S.F.; Nussenzweig, A. End-joining, translocations and cancer. Nat. Rev. Cancer 2013, 13, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Daley, J.M.; Sung, P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 2014, 34, 1380–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kass, E.M.; Moynahan, M.E.; Jasin, M. When genome maintenance goes badly awry. Mol. Cell 2016, 62, 777–787. [Google Scholar] [CrossRef] [Green Version]
- Bhargava, R.; Onyango, D.O.; Stark, J.M. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016, 32, 566–575. [Google Scholar] [CrossRef] [Green Version]
- Verma, P.; Greenberg, R.A. Noncanonical views of homology-directed DNA repair. Genes Dev. 2016, 30, 1138–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortensen, U.H.; Bendixen, C.; Sunjevaric, I.; Rothstein, R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 1996, 93, 10729–10734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, T.; New, J.H.; Kowalczykowski, S.C. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA 1998, 95, 6049–6054. [Google Scholar] [CrossRef] [Green Version]
- Van Dyck, E.; Stasiak, A.Z.; Stasiak, A.; West, S.C. Visualization of recombination intermediates produced by RAD52-mediated single-strand annealing. EMBO Rep. 2001, 2, 905–909. [Google Scholar] [CrossRef] [Green Version]
- Wray, J.; Liu, J.; Nickoloff, J.A.; Shen, Z. Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Res. 2008, 68, 2699–2707. [Google Scholar] [CrossRef] [Green Version]
- Grimme, J.M.; Honda, M.; Wright, R.; Okuno, Y.; Rothenberg, E.; Mazin, A.V.; Ha, T.; Spies, M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010, 38, 2917–2930. [Google Scholar] [CrossRef] [Green Version]
- Sugawara, N.; Ira, G.; Haber, J.E. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000, 20, 5300–5309. [Google Scholar] [CrossRef] [Green Version]
- Lisby, M.; Barlow, J.H.; Burgess, R.C.; Rothstein, R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 2004, 118, 699–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi-Iwai, Y.; Sonoda, E.; Buerstedde, J.M.; Bezzubova, O.; Morrison, C.; Takata, M.; Shinohara, A.; Takeda, S. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol. 1998, 18, 6430–6435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rijkers, T.; Van Den Ouweland, J.; Morolli, B.; Rolink, A.G.; Baarends, W.M.; Van Sloun, P.P.; Lohman, P.H.; Pastink, A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 1998, 18, 6423–6429. [Google Scholar] [CrossRef] [Green Version]
- de Vries, F.A.; Zonneveld, J.B.; van Duijn-Goedhart, A.; Roodbergen, M.; Boei, J.; van Buul, P.P.; Essers, J.; van Steeg, H.; van Zeeland, A.A.; van Benthem, J.; et al. Inactivation of RAD52 aggravates RAD54 defects in mice but not in Schizosaccharomyces pombe. DNA Repair (Amsterdam) 2005, 4, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.P.; Lovett, S.T.; Haber, J.E. Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakofsky, C.J.; Malkova, A. Break induced replication in eukaryotes: Mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 395–413. [Google Scholar] [CrossRef] [PubMed]
- Donnianni, R.A.; Zhou, Z.X.; Lujan, S.A.; Al-Zain, A.; Garcia, V.; Glancy, E.; Burkholder, A.B.; Kunkel, T.A.; Symington, L.S. DNA polymerase delta synthesizes both strands during break-induced replication. Mol. Cell 2019, 76, 371–381. [Google Scholar] [CrossRef]
- Deem, A.; Keszthelyi, A.; Blackgrove, T.; Vayl, A.; Coffey, B.; Mathur, R.; Chabes, A.; Malkova, A. Break-induced replication is highly inaccurate. PLoS Biol. 2011, 9, e1000594. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.C.; Harris, S.T.; Dinter, T.; Shah, K.A.; Mirkin, S.M. The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat. Struct. Mol. Biol. 2017, 24, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Symington, L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002, 66, 630–670. [Google Scholar] [CrossRef] [Green Version]
- Sotiriou, S.K.; Kamileri, I.; Lugli, N.; Evangelou, K.; Da-Re, C.; Huber, F.; Padayachy, L.; Tardy, S.; Nicati, N.L.; Barriot, S.; et al. Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Mol. Cell 2016, 64, 1127–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479, 131–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charman, M.; Herrmann, C.; Weitzman, M.D. Viral and cellular interactions during adenovirus DNA replication. FEBS Lett. 2019, 593, 3531–3550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muzyczka, N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 1992, 158, 97–129. [Google Scholar] [CrossRef]
- Khan, I.F.; Hirata, R.K.; Russell, D.W. AAV-mediated gene targeting methods for human cells. Nat. Protoc. 2011, 6, 482–501. [Google Scholar] [CrossRef] [Green Version]
- Atchison, R.W.; Casto, B.C.; Hammon, W.M. Adenovirus-associated defective virus particles. Science 1965, 149, 754–756. [Google Scholar] [CrossRef]
- Blacklow, N.R.; Hoggan, M.D.; Rowe, W.P. Serologic evidence for human infection with adenovirus-associated viruses. J. Natl. Cancer Inst. 1968, 40, 319–327. [Google Scholar]
- Hendrickson, E.A. Gene targeting in human somatic cells. In Sourcebook of Models for Biomedical Research; Conn, P.M., Ed.; Humana Press: Totowa, NJ, USA, 2008; pp. 509–525. [Google Scholar] [CrossRef]
- Hori, T.; Fukutome, M.; Koike, C. Adeno associated virus (AAV) as a tool for clinical and experimental delivery of target genes into the mammalian retina. Biol. Pharm. Bull. 2019, 42, 343–347. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, T.; Ishikawa, K.; Hinkel, R.; Kupatt, C. Translational aspects of adeno-associated virus-mediated cardiac gene therapy. Hum. Gene Ther. 2018, 29, 1341–1351. [Google Scholar] [CrossRef]
- Rose, J.A.; Berns, K.I.; Hoggan, M.D.; Koczot, F.J. Evidence for a single-stranded adenovirus-associated virus genome: Formation of a DNA density hybrid on release of viral DNA. Proc. Natl. Acad. Sci. USA 1969, 64, 863–869. [Google Scholar] [CrossRef] [Green Version]
- Samulski, R.J.; Chang, L.S.; Shenk, T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J. Virol. 1987, 61, 3096–3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lusby, E.; Fife, K.H.; Berns, K.I. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J. Virol. 1980, 34, 402–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Xiao, W.; Li, J.; Samulski, R.J. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J. Virol. 1997, 71, 941–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotin, R.M.; Siniscalco, M.; Samulski, R.J.; Zhu, X.D.; Hunter, L.; Laughlin, C.A.; McLaughlin, S.; Muzyczka, N.; Rocchi, M.; Berns, K.I. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 1990, 87, 2211–2215. [Google Scholar] [CrossRef] [Green Version]
- Samulski, R.J.; Zhu, X.; Xiao, X.; Brook, J.D.; Housman, D.E.; Epstein, N.; Hunter, L.A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991, 10, 3941–3950. [Google Scholar] [CrossRef]
- Kan, Y.; Ruis, B.; Lin, S.; Hendrickson, E.A. The mechanism of gene targeting in human somatic cells. PLoS Genet. 2014, 10, e1004251. [Google Scholar] [CrossRef] [Green Version]
- Zentilin, L.; Marcello, A.; Giacca, M. Involvement of cellular double-stranded DNA break binding proteins in processing of the recombinant adeno-associated virus genome. J. Virol. 2001, 75, 12279–12287. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, R.A.; Palacios, J.A.; Cassell, G.D.; Adam, S.; Giacca, M.; Weitzman, M.D. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 2007, 81, 12936–12945. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.G.; Trobridge, G.D.; Petek, L.M.; Jacobs, M.A.; Kaul, R.; Russell, D.W. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J. Virol. 2005, 79, 11434–11442. [Google Scholar] [CrossRef] [Green Version]
- Rutledge, E.A.; Russell, D.W. Adeno-associated virus vector integration junctions. J. Virol. 1997, 71, 8429–8436. [Google Scholar] [CrossRef] [Green Version]
- Ndung’u, T.; McCune, J.M.; Deeks, S.G. Why and where an HIV cure is needed and how it might be achieved. Nature 2019, 576, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Kanaar, R.; Jackson, S.P.; O’Connor, M.J. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 2004, 23, 3421–3429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Olvera, J.M.; Yoder, K.E.; Mitchell, R.S.; Butler, S.L.; Lieber, M.; Martin, S.L.; Bushman, F.D. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001, 20, 3272–3281. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.O. Integration. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Bernstein, D.I.; Bellamy, A.R.; Hook, E.W., III; Levin, M.J.; Wald, A.; Ewell, M.G.; Wolff, P.A.; Deal, C.D.; Heineman, T.C.; Dubin, G.; et al. Epidemiology, clinical presentation, and antibody response to primary infection with herpes simplex virus type 1 and type 2 in young women. Clin. Infect. Dis. 2013, 56, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Goins, W.F.; Huang, S.; Cohen, J.B.; Glorioso, J.C. Engineering HSV-1 vectors for gene therapy. Methods Mol. Biol. 2014, 1144, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.J.; Mohni, K.N.; Kan, Y.; Hendrickson, E.A.; Stark, J.M.; Weller, S.K. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 2012, 8, e1002862. [Google Scholar] [CrossRef]
- Weller, S.K.; Coen, D.M. Herpes simplex viruses: Mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 2012, 4, a013011. [Google Scholar] [CrossRef] [Green Version]
- Weller, S.K.; Sawitzke, J.A. Recombination promoted by DNA viruses: Phage lambda to herpes simplex virus. Annu. Rev. Microbiol. 2014, 68, 237–258. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hendrickson, E.A. RAD52: Viral Friend or Foe? Cancers 2020, 12, 399. https://doi.org/10.3390/cancers12020399
Hendrickson EA. RAD52: Viral Friend or Foe? Cancers. 2020; 12(2):399. https://doi.org/10.3390/cancers12020399
Chicago/Turabian StyleHendrickson, Eric A. 2020. "RAD52: Viral Friend or Foe?" Cancers 12, no. 2: 399. https://doi.org/10.3390/cancers12020399
APA StyleHendrickson, E. A. (2020). RAD52: Viral Friend or Foe? Cancers, 12(2), 399. https://doi.org/10.3390/cancers12020399



