A ”Clickable” Probe for Active MGMT in Glioblastoma Demonstrates Two Discrete Populations of MGMT
Abstract
:1. Introduction
1.1. Design of Fluorogenic Clickable MGMT Probes
1.2. Synthesis of Fluorogenic Probes
2. Results
2.1. Measuring Levels of Active MGMT in GBM Cells
2.2. Labelling of MGMT in GBM Cells and Reduction in Labeling by Incubation with TMZ
2.3. MGMT Levels Increase Following TMZ Treatment, but Active MGMT Levels Fall
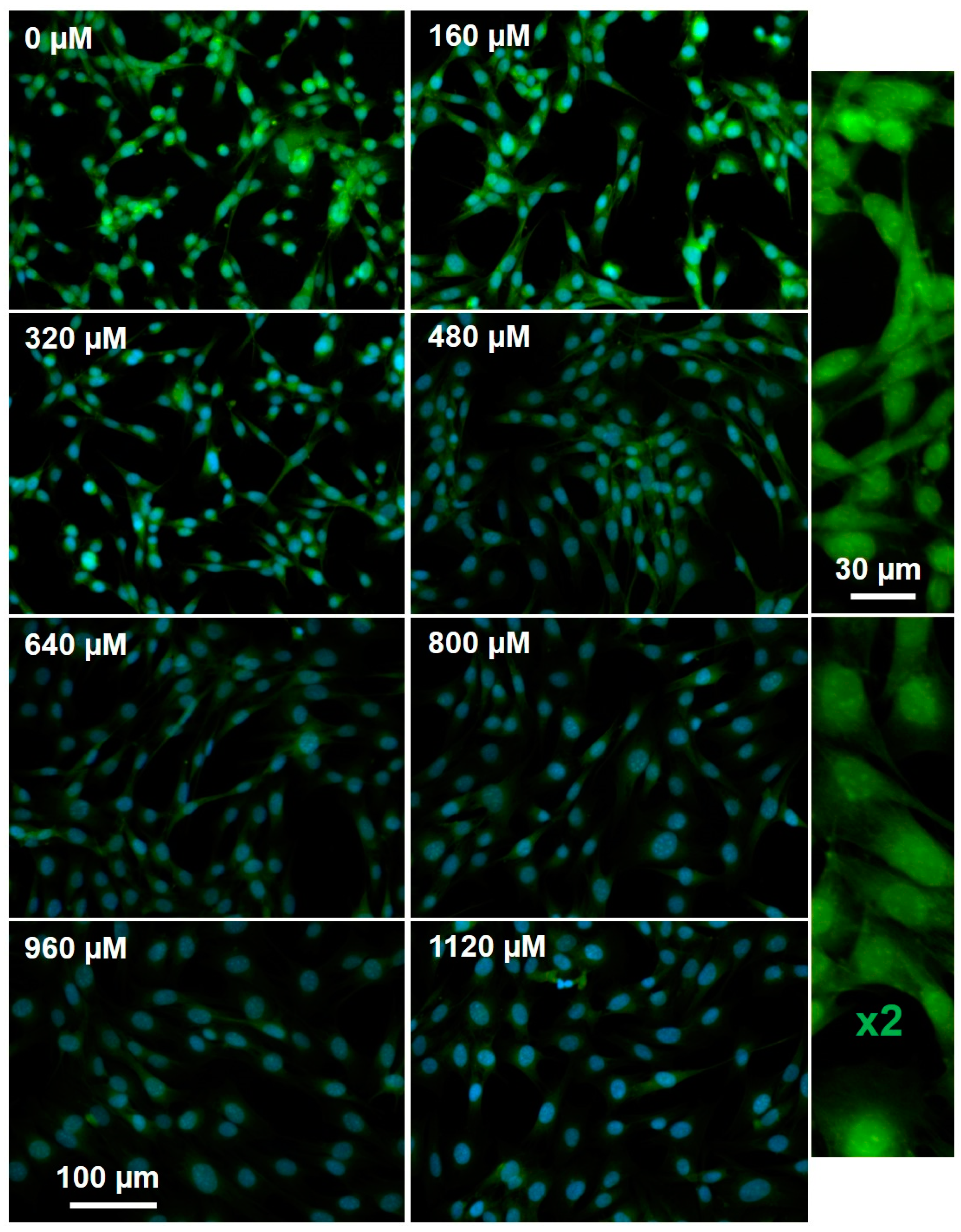
2.4. Active MGMT Levels Rapidly Increase at Higher Concentrations of Reactive Oxygen Species
2.5. Levels of PCNA and Phosphorylated MGMT Rise in Response to Oxidative stress, but Fall in Response to TMZ Induced DNA Damage
2.6. Staining of GBM Microarray Demonstrates Variability in Active and Total MGMT Levels
3. Discussion
Active MGMT Levels are Finely Tuned and Responsive to Levels of DNA Damage
4. Materials and Methods
4.1. Fluorescence Microscopy
4.2. Measurement of Active MGMT Levels
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gerson, S.L. MGMT: Its Role in Cancer Aetiology and Cancer Therapeutics. Nature Rev. Cancer 2004, 4, 296. [Google Scholar] [CrossRef]
- Zhang, J.; Stevens, M.F.G.; Bradshaw, T.D. Temozolomide: Mechanisms of Action, Repair and Resistance. Curr. Mol. Pharmacol. 2012, 5, 102–114. [Google Scholar] [CrossRef]
- Warren, J.J.; Forsberg, L.J.; Beese, L.S. The structural basis for the mutagenicity of O6-methyl-guanine lesions. Proc. Natl. Acad. Sci. USA 2006, 103, 19701–19706. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Batchelor, T.T.; Mulholland, P.; Neyns, B.; Nabors, L.B.; Campone, M.; Wick, A.; Mason, W.; Mikkelsen, T.; Phuphanich, S.; Ashby, L.S.; et al. Phase III randomized trial comparing the efficacy of cediranib as monotherapy, and in combination with lomustine, versus lomustine alone in patients with recurrent glioblastoma. J. Clin. Oncol. 2013, 31, 3212–3218. [Google Scholar] [CrossRef] [Green Version]
- Taal, W.; Oosterkamp, H.M.; Walenkamp, A.M.; Dubbink, H.J.; Beerepoot, L.V.; Hanse, M.C.; Buter, J.; Honkoop, A.H.; Boerman, D.; de Vos, F.Y.; et al. Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): A randomised controlled phase 2 trial. Lancet. Oncol. 2014, 15, 943–953. [Google Scholar] [CrossRef]
- Sharpe, M.A.; Raghavan, S.; Baskin, D.S. PAM-OBG: A monoamine oxidase B specific prodrug that inhibits MGMT and generates DNA interstrand crosslinks, potentiating temozolomide and chemoradiation therapy in intracranial glioblastoma. Oncotarget 2018, 9, 23923–23943. [Google Scholar] [CrossRef] [Green Version]
- Cabrini, G.; Fabbri, E.; Lo Nigro, C.; Dechecchi, M.C.; Gambari, R. Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (Review). Int. J. Oncol. 2015, 47, 417–428. [Google Scholar] [CrossRef] [Green Version]
- Hazra, T.K.; Roy, R.; Biswas, T.; Grabowski, D.T.; Pegg, A.E.; Mitra, S. Specific recognition of O6-methylguanine in DNA by active site mutants of human O6-methylguanine-DNA methyltransferase. Biochemistry 1997, 36, 5769–5776. [Google Scholar] [CrossRef]
- Srivenugopal, K.S.; Yuan, X.H.; Friedman, H.S.; Ali-Osman, F. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chloroethyl)-1-nitrosourea. Biochemistry 1996, 35, 1328–1334. [Google Scholar] [CrossRef]
- Fan, C.H.; Liu, W.L.; Cao, H.; Wen, C.; Chen, L.; Jiang, G. O6-methylguanine DNA methyltransferase as a promising target for the treatment of temozolomide-resistant gliomas. Cell Death Dis. 2013, 4, e876. [Google Scholar] [CrossRef] [Green Version]
- Everhard, S.; Tost, J.; Abdalaoui, H.E.; Crinière, E.; Busato, F.; Marie, Y.; Gut, I.G.; Sanson, M.; Mokhtari, K.; Laigle-Donadey, F.; et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro-Oncology 2009, 11, 348–356. [Google Scholar] [CrossRef] [Green Version]
- Weller, M.; Stupp, R.; Reifenberger, G.; Brandes, A.A.; van den Bent, M.J.; Wick, W.; Hegi, M.E. MGMT promoter methylation in malignant gliomas: Ready for personalized medicine? Nat. Rev. Neurol. 2009, 6, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, G.; Brenner, W.; Johnsson, K.; Kaina, B. DNA repair protein O6-methylguanine-DNA methyltransferase in testis and testicular tumors as determined by a novel nonradioactive assay. Anal. Biochem. 2003, 321, 38–43. [Google Scholar] [CrossRef]
- Yeager, N.D.; Dolan, M.E.; Gastier, J.M.; Gross, T.G.; Delaney, S.; Frick, J.; Ruymann, F.B.; Ewesuedo, R. O6-methylguanine-DNA methyltransferase activity and promoter methylation status in pediatric rhabdomyosarcoma. J. Pediat. Hematol. Oncol. 2003, 25, 941–947. [Google Scholar] [CrossRef]
- Encell, L.P.; Loeb, L.A. Redesigning the substrate specificity of human O6-alkylguanine-DNA alkyltransferase. Mutants with enhanced repair of O(4)-methylthymine. Biochemistry 1999, 38, 12097–12103. [Google Scholar] [CrossRef]
- Kreklau, E.L.; Limp-Foster, M.; Liu, N.; Xu, Y.; Kelley, M.R.; Erickson, L.C. A novel fluorometric oligonucleotide assay to measure O-6-methylguanine DNA methyltransferase, methylpurine DNA glycosylase, 8-oxoguanine DNA glycosylase and abasic endonuclease activities: DNA repair status in human breast carcinoma cells overexpressing methylpurine DNA glycosylase. Nucleic Acids Res. 2001, 29, 2558–2566. [Google Scholar] [CrossRef] [Green Version]
- Beharry, A.A.; Nagel, Z.D.; Samson, L.D.; Kool, E.T. Fluorogenic Real-Time Reporters of DNA Repair by MGMT, a Clinical Predictor of Antitumor Drug Response. PLoS ONE 2016, 11, e0152684. [Google Scholar] [CrossRef] [Green Version]
- Tintoré, M.; Aviñó, A.; Ruiz, F.M.; Eritja, R.; Fàbrega, C. Development of a Novel Fluorescence Assay Based on the Use of the Thrombin-Binding Aptamer for the Detection of O6-Alkylguanine-DNA Alkyltransferase Activity. J. Nucl. Acids 2010, 2010, 632041. [Google Scholar] [CrossRef] [Green Version]
- Robinson, C.; Palomo, J.; Vogelbaum, M.A. Thin layer chromatography-based assay of O6-methylguanine-DNA methyltransferase activity in tissue. Anal. Biochem. 2010, 405, 263–265. [Google Scholar] [CrossRef]
- Griffin, R.J.; Arris, C.E.; Bleasdale, C.; Boyle, F.T.; Calvert, A.H.; Curtin, N.J.; Dalby, C.; Kanugula, S.; Lembicz, N.K.; Newell, D.R.; et al. Resistance-Modifying Agents. 8. Inhibition of O6-Alkylguanine-DNA Alkyltransferase by O6-Alkenyl-, O6-Cycloalkenyl-, and O6-(2-Oxoalkyl)guanines and Potentiation of Temozolomide Cytotoxicity in Vitro by O6-(1-Cyclopentenylmethyl)guanine. J. Med. Chem. 2000, 43, 4071–4083. [Google Scholar] [CrossRef] [PubMed]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.-L. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paranjpe, A.; Bailey, N.I.; Konduri, S.; Bobustuc, G.C.; Ali-Osman, F.; Yusuf, M.A.; Punganuru, S.R.; Madala, H.R.; Basak, D.; Mostofa, A.; et al. New insights into estrogenic regulation of O(6)-methylguanine DNA-methyltransferase (MGMT) in human breast cancer cells: Co-degradation of ER-alpha and MGMT proteins by fulvestrant or O(6)-benzylguanine indicates fresh avenues for therapy. J. Biomed. Res. 2016, 30, 393–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Lin, H.; Zhang, X.; Li, J. Resveratrol reverses temozolomide resistance by downregulation of MGMT in T98G glioblastoma cells by the NF-κB-dependent pathway. Oncol. Rep. 2012, 27, 2050–2056. [Google Scholar] [PubMed]
- Mullapudi, S.R.; Ali-Osman, F.; Shou, J.; Srivenugopal, K.S. DNA repair protein O6-alkylguanine-DNA alkyltransferase is phosphorylated by two distinct and novel protein kinases in human brain tumour cells. Biochem. J. 2000, 351 Pt 2, 393–402. [Google Scholar] [CrossRef]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radical Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [Green Version]
- Mostofa, A.; Punganuru, S.R.; Madala, H.R.; Srivenugopal, K.S. S-phase Specific Downregulation of Human O6-Methylguanine DNA Methyltransferase (MGMT) and its Serendipitous Interactions with PCNA and p21(cip1) Proteins in Glioma Cells. Neoplasia 2018, 20, 305–323. [Google Scholar] [CrossRef]
- Sharpe, M.A.; Baskin, D.S. Monoamine oxidase B levels are highly expressed in human gliomas and are correlated with the expression of HiF-1α and with transcription factors Sp1 and Sp3. Oncotarget 2016, 7, 3379–3393. [Google Scholar] [CrossRef] [Green Version]









© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raghavan, S.; Baskin, D.S.; Sharpe, M.A. A ”Clickable” Probe for Active MGMT in Glioblastoma Demonstrates Two Discrete Populations of MGMT. Cancers 2020, 12, 453. https://doi.org/10.3390/cancers12020453
Raghavan S, Baskin DS, Sharpe MA. A ”Clickable” Probe for Active MGMT in Glioblastoma Demonstrates Two Discrete Populations of MGMT. Cancers. 2020; 12(2):453. https://doi.org/10.3390/cancers12020453
Chicago/Turabian StyleRaghavan, Sudhir, David S. Baskin, and Martyn A. Sharpe. 2020. "A ”Clickable” Probe for Active MGMT in Glioblastoma Demonstrates Two Discrete Populations of MGMT" Cancers 12, no. 2: 453. https://doi.org/10.3390/cancers12020453




