Molecular Profiling of Keratinocyte Skin Tumors Links Staphylococcus aureus Overabundance and Increased Human β-Defensin-2 Expression to Growth Promotion of Squamous Cell Carcinoma
Abstract
1. Introduction
2. Results
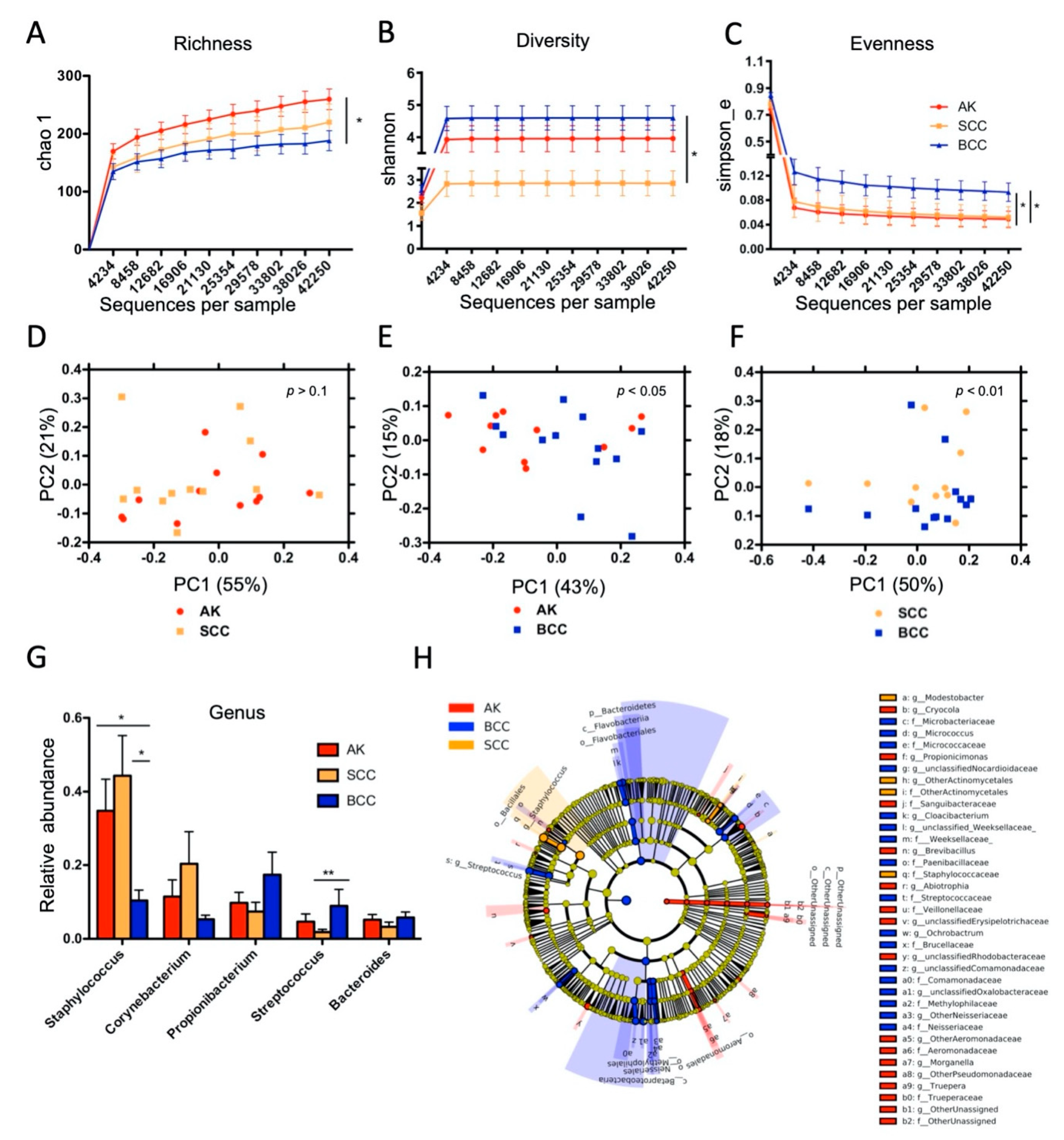
2.1. Microbial Colonization of Keratinocyte Skin Tumors
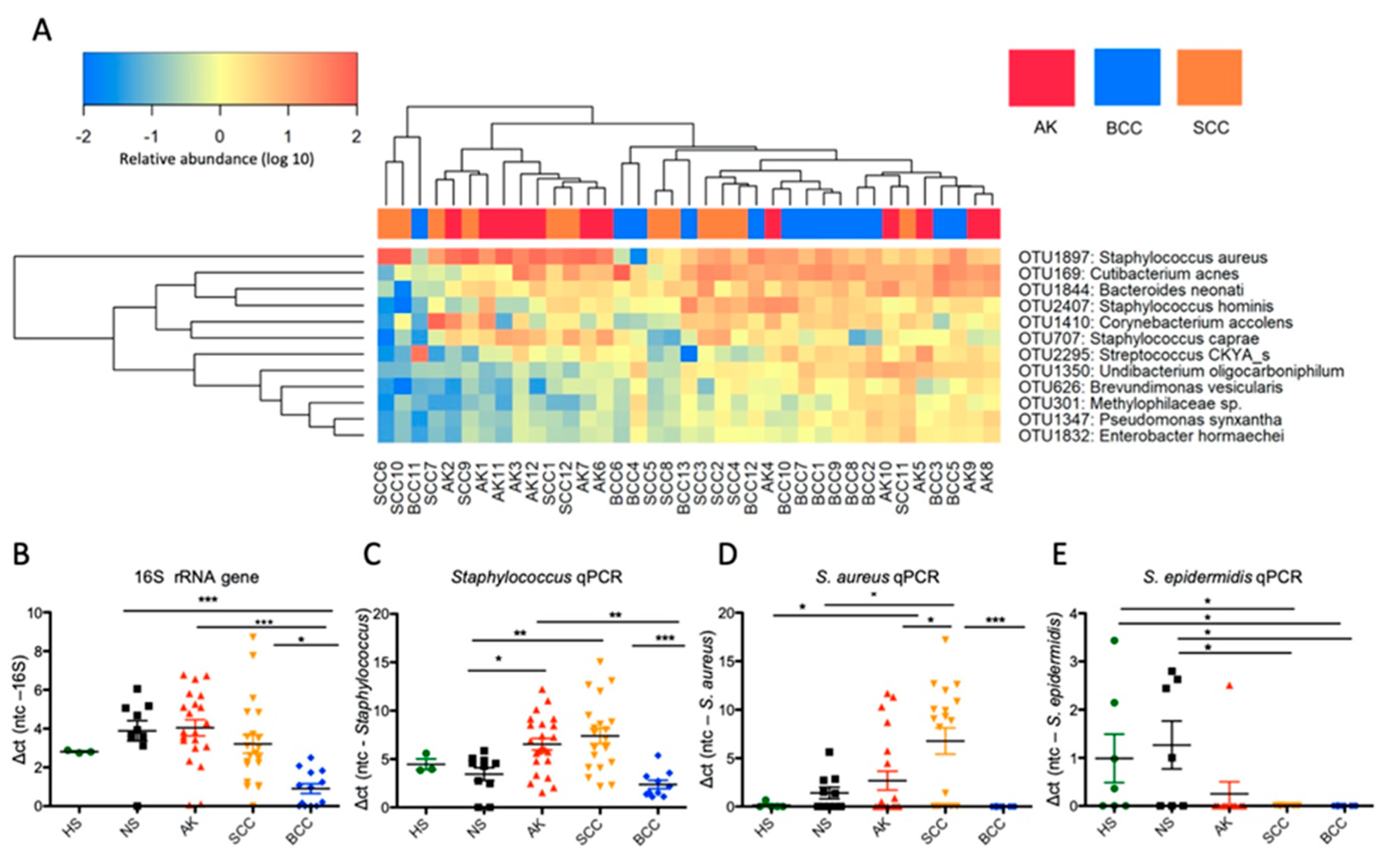
2.2. Distinct Microbial Community Types Are Prevalent in Keratinocyte Skin Tumors, wherein AK and SCC Are Characterized by Staphylococcus Overabundance
2.3. S. aureus Is Associated with Hyperkeratotic Regions in AK and SCC and Is Found in Areas with Invasive Tissue
2.4. Antimicrobial Peptide Expression Is Changed in Keratinocyte Skin Tumors, and hBD-2 Expression Correlates Significantly with S. aureus Loads in SCC
2.5. S. aureus Challenge Induces hBD-2 mRNA Expression and Stimulates Growth of Cutaneous SCC Cells
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Specimens, Histology, and Scoring
4.3. DNA Isolation and Quantification
4.4. 16S rRNA Gene PCR, Library Preparation and Sequencing
4.5. Microbiota and Correlation Analysis
4.6. Quantification of Bacterial Load, Staphylococcus, S. aureus, and S. epidermidis Abundance via qPCR
4.7. Reverse Transcription Quantitative PCR (RT-qPCR)
4.8. Fluorescent In Situ Hybridization (FISH)
4.9. Cell Culture, Infection Assay, and Flow Cytometry
4.10. CCK-8 Assay
4.11. XCELLigenceTM Real-Time Cell Proliferation Assay
4.12. Statistical Analysis
4.13. Availability of Data and Material
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wassberg, C.; Thorn, M.; Johansson, A.M.; Bergstrom, R.; Berne, B.; Ringborg, U. Increasing incidence rates of squamous cell carcinoma of the skin in Sweden. Acta Derm. Venereol. 2001, 81, 268–272. [Google Scholar] [CrossRef]
- Brand, D.; Ackerman, A.B. Squamous cell carcinoma, not basal cell carcinoma, is the most common cancer in humans. J. Am. Acad. Dermatol. 2000, 42, 523–526. [Google Scholar] [CrossRef]
- Organisation, W.H. Skin Cancers. Available online: www.who.int/uv/faq/skincancer/en/index1.html (accessed on 22 December 2018).
- Forslund, O.; Iftner, T.; Andersson, K.; Lindelof, B.; Hradil, E.; Nordin, P.; Stenquist, B.; Kirnbauer, R.; Dillner, J.; de Villiers, E.M.; et al. Cutaneous human papillomaviruses found in sun-exposed skin: Beta-papillomavirus species 2 predominates in squamous cell carcinoma. J. Infect. Dis. 2007, 196, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Asgari, M.M.; Kiviat, N.B.; Critchlow, C.W.; Stern, J.E.; Argenyi, Z.B.; Raugi, G.J.; Berg, D.; Odland, P.B.; Hawes, S.E.; de Villiers, E.M. Detection of human papillomavirus DNA in cutaneous squamous cell carcinoma among immunocompetent individuals. J. Investig. Dermatol. 2008, 128, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Program, N.C.S.; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef]
- Bouslimani, A.; Porto, C.; Rath, C.M.; Wang, M.; Guo, Y.; Gonzalez, A.; Berg-Lyon, D.; Ackermann, G.; Moeller Christensen, G.J.; Nakatsuji, T.; et al. Molecular cartography of the human skin surface in 3D. Proc. Natl. Acad. Sci. USA 2015, 112, E2120–E2129. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Butcher, A.M.; Trzoss, L.L.; Nam, S.J.; Shirakawa, K.T.; Zhou, W.; Oh, J.; Otto, M.; Fenical, W.; et al. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci. Adv. 2018, 4, eaao4502. [Google Scholar] [CrossRef]
- Abed, J.; Emgard, J.E.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-galnac. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Bottomley, M.J.; Thomson, J.; Harwood, C.; Leigh, I. The role of the immune system in cutaneous squamous cell carcinoma. Int. J. Mol. Sci. 2019, 20, 2009. [Google Scholar] [CrossRef]
- Hoste, E.; Arwert, E.N.; Lal, R.; South, A.P.; Salas-Alanis, J.C.; Murrell, D.F.; Donati, G.; Watt, F.M. Innate sensing of microbial products promotes wound-induced skin cancer. Nat. Commun. 2015, 6, 5932. [Google Scholar] [CrossRef] [PubMed]
- Niyonsaba, F.; Ushio, H.; Nakano, N.; Ng, W.; Sayama, K.; Hashimoto, K.; Nagaoka, I.; Okumura, K.; Ogawa, H. Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J. Investig. Dermatol. 2007, 127, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Mburu, Y.K.; Abe, K.; Ferris, L.K.; Sarkar, S.N.; Ferris, R.L. Human beta-defensin 3 promotes NF-kappaB-mediated CCR7 expression and anti-apoptotic signals in squamous cell carcinoma of the head and neck. Carcinogenesis 2011, 32, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Brandwein, M.; Bentwich, Z.; Steinberg, D. Endogenous antimicrobial peptide expression in response to bacterial epidermal colonization. Front. Immunol. 2017, 8, 1637. [Google Scholar] [CrossRef]
- Dinulos, J.G.; Mentele, L.; Fredericks, L.P.; Dale, B.A.; Darmstadt, G.L. Keratinocyte expression of human beta defensin 2 following bacterial infection: Role in cutaneous host defense. Clin. Diagn. Lab. Immunol. 2003, 10, 161–166. [Google Scholar] [CrossRef]
- Schauber, J.; Gallo, R.L. Expanding the roles of antimicrobial peptides in skin: Alarming and arming keratinocytes. J. Investig. Dermatol. 2007, 127, 510–512. [Google Scholar] [CrossRef]
- Bardan, A.; Nizet, V.; Gallo, R.L. Antimicrobial peptides and the skin. Expert Opin. Biol. Ther. 2004, 4, 543–549. [Google Scholar] [CrossRef]
- Braff, M.H.; Gallo, R.L. Antimicrobial peptides: An essential component of the skin defensive barrier. Curr. Top. Microbiol. Immunol. 2006, 306, 91–110. [Google Scholar]
- Becker, J.C.; Andersen, M.H.; Schrama, D.; Thor Straten, P. Immune-suppressive properties of the tumor microenvironment. Cancer Immunol. Immunother. 2013, 62, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Kuper, H.; Adami, H.O.; Trichopoulos, D. Infections as a major preventable cause of human cancer. J. Int. Med. 2000, 248, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.H.; Parsonnet, J. Role of bacteria in oncogenesis. Clin. Microbiol. Rev. 2010, 23, 837–857. [Google Scholar] [CrossRef] [PubMed]
- Uemura, N.; Okamoto, S.; Yamamoto, S.; Matsumura, N.; Yamaguchi, S.; Yamakido, M.; Taniyama, K.; Sasaki, N.; Schlemper, R.J. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001, 345, 784–789. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Kluytmans, J.; van Belkum, A.; Verbrugh, H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef]
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota-host interactions. Nature 2018, 553, 427–436. [Google Scholar] [CrossRef]
- Kobayashi, T.; Glatz, M.; Horiuchi, K.; Kawasaki, H.; Akiyama, H.; Kaplan, D.H.; Kong, H.H.; Amagai, M.; Nagao, K. Dysbiosis and staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 2015, 42, 756–766. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Program, N.C.S.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef]
- Kooistra-Smid, M.; Nieuwenhuis, M.; van Belkum, A.; Verbrugh, H. The role of nasal carriage in Staphylococcus aureus burn wound colonization. FEMS Immunol. Med. Microbiol. 2009, 57, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kullander, J.; Forslund, O.; Dillner, J. Staphylococcus aureus and squamous cell carcinoma of the skin. Cancer Epidemiol. Biomarkers Prev. 2009, 18, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.L.A.; Lachner, N.; Tan, J.M.; Tang, S.; Angel, N.; Laino, A.; Linedale, R.; Le Cao, K.A.; Morrison, M.; Frazer, I.H.; et al. A natural history of actinic keratosis and cutaneous squamous cell carcinoma microbiomes. MBio 2018, 9, e01432-18. [Google Scholar] [CrossRef] [PubMed]
- Fahlen, A.; Engstrand, L.; Baker, B.S.; Powles, A.; Fry, L. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch. Dermatol. Res. 2012, 304, 15–22. [Google Scholar] [CrossRef]
- Thio, H.B. The microbiome in psoriasis and psoriatic arthritis: The skin perspective. J. Rheumatol. Suppl. 2018, 94, 30–31. [Google Scholar] [CrossRef]
- Gao, Z.; Tseng, C.H.; Strober, B.E.; Pei, Z.; Blaser, M.J. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE 2008, 3, e2719. [Google Scholar] [CrossRef]
- Prohic, A. Identification of Malassezia species isolated from scalp skin of patients with psoriasis and healthy subjects. Acta Dermatovenerol. Croat. 2003, 11, 10–16. [Google Scholar]
- Ayala-Fontanez, N.; Soler, D.C.; McCormick, T.S. Current knowledge on psoriasis and autoimmune diseases. Psoriasis 2016, 6, 7–32. [Google Scholar] [CrossRef]
- Benhadou, F.; Mintoff, D.; Schnebert, B.; Thio, H.B. Psoriasis and microbiota: A systematic review. Diseases 2018, 6, 47. [Google Scholar] [CrossRef]
- Chang, H.W.; Yan, D.; Singh, R.; Liu, J.; Lu, X.; Ucmak, D.; Lee, K.; Afifi, L.; Fadrosh, D.; Leech, J.; et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome 2018, 6, 154. [Google Scholar] [CrossRef]
- Todd, J.K. Staphylococcal infections. Pediatr. Rev. 2005, 26, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Gribbon, E.M.; Cunliffe, W.J.; Holland, K.T. Interaction of Propionibacterium acnes with skin lipids In Vitro. J. Gen. Microbiol. 1993, 139, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Deming, C.; Cassidy, S.K.B.; Harrison, O.J.; Ng, W.I.; Conlan, S.; Program, N.C.S.; Belkaid, Y.; Segre, J.A.; Kong, H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543. [Google Scholar] [CrossRef] [PubMed]
- Scola, N.; Gambichler, T.; Saklaoui, H.; Bechara, F.G.; Georgas, D.; Stucker, M.; Glaser, R.; Kreuter, A. The expression of antimicrobial peptides is significantly altered in cutaneous squamous cell carcinoma and precursor lesions. Br. J. Dermatol. 2012, 167, 591–597. [Google Scholar] [CrossRef]
- Harder, J.; Bartels, J.; Christophers, E.; Schroder, J.M. A peptide antibiotic from human skin. Nature 1997, 387, 861. [Google Scholar] [CrossRef]
- Menzies, B.E.; Kenoyer, A. Signal transduction and nuclear responses in Staphylococcus aureus-induced expression of human beta-defensin 3 in skin keratinocytes. Infect. Immun. 2006, 74, 6847–6854. [Google Scholar] [CrossRef]
- Cho, J.W.; Cho, S.Y.; Lee, K.S. Roles of SEA-expressing Staphylococcus aureus, isolated from an atopic dermatitis patient, on expressions of human beta-defensin-2 and inflammatory cytokines in HaCaT cells. Int. J. Mol. Med. 2009, 23, 331–335. [Google Scholar] [CrossRef][Green Version]
- Menzies, B.E.; Kenoyer, A. Staphylococcus aureus infection of epidermal keratinocytes promotes expression of innate antimicrobial peptides. Infect. Immun. 2005, 73, 5241–5244. [Google Scholar] [CrossRef]
- Midorikawa, K.; Ouhara, K.; Komatsuzawa, H.; Kawai, T.; Yamada, S.; Fujiwara, T.; Yamazaki, K.; Sayama, K.; Taubman, M.A.; Kurihara, H.; et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect. Immun. 2003, 71, 3730–3739. [Google Scholar] [CrossRef] [PubMed]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Gotz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Investig. Dermatol. 2011, 131, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.R.; Mohamed, R.; Bian, L.; Routh, A.F.; Kokai-Kun, J.F.; Mond, J.J.; Tarkowski, A.; Foster, S.J. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 2007, 1, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.K.; McCormick, T.S.; Weinberg, A. Human beta defensins and cancer: Contradictions and common ground. Front. Oncol. 2019, 9, 341. [Google Scholar] [CrossRef]
- Winter, J.; Pantelis, A.; Reich, R.; Martini, M.; Kraus, D.; Jepsen, S.; Allam, J.P.; Novak, N.; Wenghoefer, M. Human beta-defensin-1, -2, and -3 exhibit opposite effects on oral squamous cell carcinoma cell proliferation. Cancer Investig. 2011, 29, 196–201. [Google Scholar] [CrossRef]
- Shi, N.; Jin, F.; Zhang, X.; Clinton, S.K.; Pan, Z.; Chen, T. Overexpression of human beta-defensin 2 promotes growth and invasion during esophageal carcinogenesis. Oncotarget 2014, 5, 11333–11344. [Google Scholar] [CrossRef]
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef]
- Nomura, I.; Goleva, E.; Howell, M.D.; Hamid, Q.A.; Ong, P.Y.; Hall, C.F.; Darst, M.A.; Gao, B.; Boguniewicz, M.; Travers, J.B.; et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J. Immunol. 2003, 171, 3262–3269. [Google Scholar] [CrossRef]
- Marcinkiewicz, M.; Majewski, S. The role of antimicrobial peptides in chronic inflammatory skin diseases. Postepy Dermatol. Alergol. 2016, 33, 6–12. [Google Scholar] [CrossRef]
- Glaser, R.; Meyer-Hoffert, U.; Harder, J.; Cordes, J.; Wittersheim, M.; Kobliakova, J.; Folster-Holst, R.; Proksch, E.; Schroder, J.M.; Schwarz, T. The antimicrobial protein psoriasin (S100A7) is upregulated in atopic dermatitis and after experimental skin barrier disruption. J. Investig. Dermatol. 2009, 129, 641–649. [Google Scholar] [CrossRef]
- Chieosilapatham, P.; Ogawa, H.; Niyonsaba, F. Current insights into the role of human beta-defensins in atopic dermatitis. Clin. Exp. Immunol. 2017, 190, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Cohen, D.B.; Ravel, J.; Abdo, Z.; Forney, L.J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE 2012, 7, e33865. [Google Scholar] [CrossRef] [PubMed]
- Klymiuk, I.; Bambach, I.; Patra, V.; Trajanoski, S.; Wolf, P. 16S based microbiome analysis from healthy subjects’ skin swabs stored for different storage periods reveal phylum to genus level changes. Front. Microbiol. 2016, 7, 2012. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef]
- Huse, S.M.; Welch, D.M.; Morrison, H.G.; Sogin, M.L. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 2010, 12, 1889–1898. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Roshan Moniri, M.; Young, A.; Reinheimer, K.; Rayat, J.; Dai, L.J.; Warnock, G.L. Dynamic assessment of cell viability, proliferation and migration using real time cell analyzer system (RTCA). Cytotechnology 2015, 67, 379–386. [Google Scholar] [CrossRef]
- Revelle, W.R.P. Psych: Procedures for Personality and Psychological Research; Northwestern University: Evanston, IL, USA, 2017. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Moissl, C.; Rudolph, C.; Huber, R. Natural communities of novel archaea and bacteria with a string-of-pearls-like morphology: Molecular analysis of the bacterial partners. Appl. Environ. Microbiol. 2002, 68, 933–937. [Google Scholar] [CrossRef]
- Wagner, M.; Amann, R.; Kampfer, P.; Assmus, B.; Hartmann, A.; Hutzler, P.; Springer, N.; Schleifer, K.H. Identification and In Situ detection of gram-negative filamentous bacteria in activated-sludge. Syst. Appl. Microbiol. 1994, 17, 405–417. [Google Scholar] [CrossRef]
- Lawson, T.S.; Connally, R.E.; Iredell, J.R.; Vemulpad, S.; Piper, J.A. Detection of Staphylococcus aureus with a fluorescence in situ hybridization that does not require lysostaphin. J. Clin. Lab. Anal. 2011, 25, 142–147. [Google Scholar] [CrossRef]
- Probst, A.J.; Auerbach, A.K.; Moissl-Eichinger, C. Archaea on human skin. PLoS ONE 2013, 8, e65388. [Google Scholar] [CrossRef]
- Kempf, V.A.; Trebesius, K.; Autenrieth, I.B. Fluorescent In situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 2000, 38, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Aso, K. Establishment of a cell line of human skin squamous cell carcinoma In Vitro. Br. J. Dermatol. 1981, 105, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Boukamp, P.; Tilgen, W.; Dzarlieva, R.T.; Breitkreutz, D.; Haag, D.; Riehl, R.K.; Bohnert, A.; Fusenig, N.E. Phenotypic and genotypic characteristics of a cell line from a squamous cell carcinoma of human skin. J. Natl. Cancer Inst. 1982, 68, 415–427. [Google Scholar] [PubMed]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell. Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Montalban-Arques, A.; Wurm, P.; Trajanoski, S.; Schauer, S.; Kienesberger, S.; Halwachs, B.; Hogenauer, C.; Langner, C.; Gorkiewicz, G. Propionibacterium acnes overabundance and natural killer group 2 member D system activation in corpus-dominant lymphocytic gastritis. J. Pathol. 2016, 240, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Brakstad, O.G.; Aasbakk, K.; Maeland, J.A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 1992, 30, 1654–1660. [Google Scholar] [CrossRef]
- Li, W.; Han, L.; Yu, P.; Ma, C.; Wu, X.; Moore, J.E.; Xu, J. Molecular characterization of skin microbiota between cancer cachexia patients and healthy volunteers. Microb. Ecol. 2014, 67, 679–689. [Google Scholar] [CrossRef]
- Vordenbaumen, S.; Pilic, D.; Otte, J.M.; Schmitz, F.; Schmidt-Choudhury, A. Defensin-mRNA expression in the upper gastrointestinal tract is modulated in children with celiac disease and Helicobacter pylori-positive gastritis. J. Pediatr. Gastroenterol. Nutr. 2010, 50, 596–600. [Google Scholar] [CrossRef]
- Zanger, P.; Holzer, J.; Schleucher, R.; Steffen, H.; Schittek, B.; Gabrysch, S. Constitutive expression of the antimicrobial peptide RNase 7 is associated with Staphylococcus aureus infection of the skin. J. Infect. Dis. 2009, 200, 1907–1915. [Google Scholar] [CrossRef]
- Garreis, F.; Gottschalt, M.; Schlorf, T.; Glaser, R.; Harder, J.; Worlitzsch, D.; Paulsen, F.P. Expression and regulation of antimicrobial peptide psoriasin (S100A7) at the ocular surface and in the lacrimal apparatus. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4914–4922. [Google Scholar] [CrossRef]
- Hohnadel, M.; Felden, L.; Fijuljanin, D.; Jouette, S.; Chollet, R. A new ultrasonic high-throughput instrument for rapid DNA release from microorganisms. J. Microbiol. Methods 2014, 99, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Byrne, F.J.; Waters, S.M.; Waters, P.S.; Curtin, W.; Kerin, M. Development of a molecular methodology to quantify Staphylococcus epidermidis in surgical wash-out samples from prosthetic joint replacement surgery. Eur. J. Orthop. Surg. Traumatol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Furet, J.P.; Firmesse, O.; Gourmelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Dore, J.; Corthier, G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madhusudhan, N.; Pausan, M.R.; Halwachs, B.; Durdević, M.; Windisch, M.; Kehrmann, J.; Patra, V.; Wolf, P.; Boukamp, P.; Moissl-Eichinger, C.; et al. Molecular Profiling of Keratinocyte Skin Tumors Links Staphylococcus aureus Overabundance and Increased Human β-Defensin-2 Expression to Growth Promotion of Squamous Cell Carcinoma. Cancers 2020, 12, 541. https://doi.org/10.3390/cancers12030541
Madhusudhan N, Pausan MR, Halwachs B, Durdević M, Windisch M, Kehrmann J, Patra V, Wolf P, Boukamp P, Moissl-Eichinger C, et al. Molecular Profiling of Keratinocyte Skin Tumors Links Staphylococcus aureus Overabundance and Increased Human β-Defensin-2 Expression to Growth Promotion of Squamous Cell Carcinoma. Cancers. 2020; 12(3):541. https://doi.org/10.3390/cancers12030541
Chicago/Turabian StyleMadhusudhan, Nandhitha, Manuela R. Pausan, Bettina Halwachs, Marija Durdević, Markus Windisch, Jan Kehrmann, VijayKumar Patra, Peter Wolf, Petra Boukamp, Christine Moissl-Eichinger, and et al. 2020. "Molecular Profiling of Keratinocyte Skin Tumors Links Staphylococcus aureus Overabundance and Increased Human β-Defensin-2 Expression to Growth Promotion of Squamous Cell Carcinoma" Cancers 12, no. 3: 541. https://doi.org/10.3390/cancers12030541
APA StyleMadhusudhan, N., Pausan, M. R., Halwachs, B., Durdević, M., Windisch, M., Kehrmann, J., Patra, V., Wolf, P., Boukamp, P., Moissl-Eichinger, C., Cerroni, L., Becker, J. C., & Gorkiewicz, G. (2020). Molecular Profiling of Keratinocyte Skin Tumors Links Staphylococcus aureus Overabundance and Increased Human β-Defensin-2 Expression to Growth Promotion of Squamous Cell Carcinoma. Cancers, 12(3), 541. https://doi.org/10.3390/cancers12030541






