Abstract
Telomeres are highly conserved tandem nucleotide repeats that include proximal double-stranded and distal single-stranded regions that in complex with shelterin proteins afford protection at chromosomal ends to maintain genomic integrity. Due to the inherent limitations of DNA replication and telomerase suppression in most somatic cells, telomeres undergo age-dependent incremental attrition. Short or dysfunctional telomeres are recognized as DNA double-stranded breaks, triggering cells to undergo replicative senescence. Telomere shortening, therefore, acts as a counting mechanism that drives replicative senescence by limiting the mitotic potential of cells. Telomere length, a complex hereditary trait, is associated with aging and age-related diseases. Epidemiological data, in general, support an association with varying magnitudes between constitutive telomere length and several disorders, including cancers. Telomere attrition is also influenced by oxidative damage and replicative stress caused by genetic, epigenetic, and environmental factors. Several single nucleotide polymorphisms at different loci, identified through genome-wide association studies, influence inter-individual variation in telomere length. In addition to genetic factors, environmental factors also influence telomere length during growth and development. Telomeres hold potential as biomarkers that reflect the genetic predisposition together with the impact of environmental conditions and as targets for anti-cancer therapies.
1. Introduction
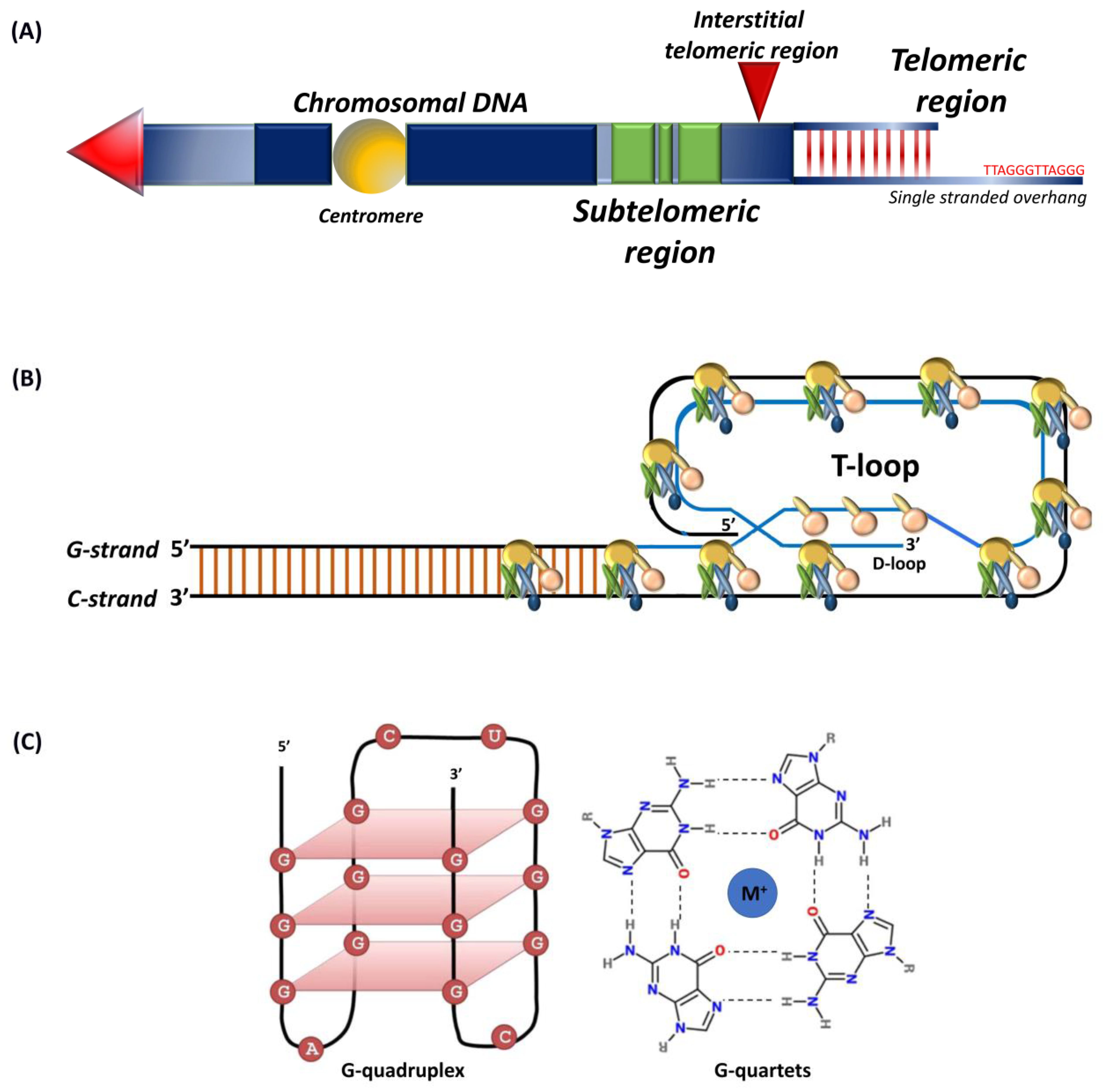
Telomeres are conserved tandem repeats at chromosomal ends that differ in length in diverse species [1,2,3,4,5]. Initially discovered in the extrachromosomal ribosomal DNA of Tetrahymena thermophile, the protozoan telomeres contain 20–70 hexameric TTGGGG tandem repeats [6]. The telomeres in yeast comprise of GGTTACA repeat sequences that extend up to 300 bp [7,8]. In plants, TTTAGGG repeats typically range between 2 to 100 kb, and certain protozoan and fungi carry short telomeres ranging between 18 to 600 bp [9,10]. In vertebrates, chromosomal ends consist of TTAGGG repeats with the longest telomeres being in rats and some strains of Mus musculus that extend up to 150 kb [5,7]. Human telomeres typically range between 10 to 15 kb [7,11,12]. Telomeres include proximal double-stranded and distal single-stranded regions (Figure 1A) with subtelomeres and interstitial sections separating repeats from the rest of the chromosome [13,14]. Telomeres, intrinsically unstable fragile sites, are stabilized through binding with so-called shelterin complex proteins [12,15,16].
Single-stranded 50–300 nucleotide guanine rich telomeric G-tail folds back into the duplex DNA to form a t-loop (Figure 1B) that resembles a large “lariat-like” structure [1,17,18]. The G-tail can also fold into a four-stranded helical structure known as the G-quadruplex (Figure 1C) that involves stacking of G-quartets and intra-molecular folding by overcoming kinetic barriers, with each quartet formed by the association of four guanines into a cyclic Hoogsten hydrogen-bonding arrangement [19,20]. Those compact and stable structures, besides forming a telomeric cap, inhibit access to telomerase [21]. Although the G-quadruplex structure in vivo has been observed by nuclear magnetic resonance, its biological function remains unknown [20,22].
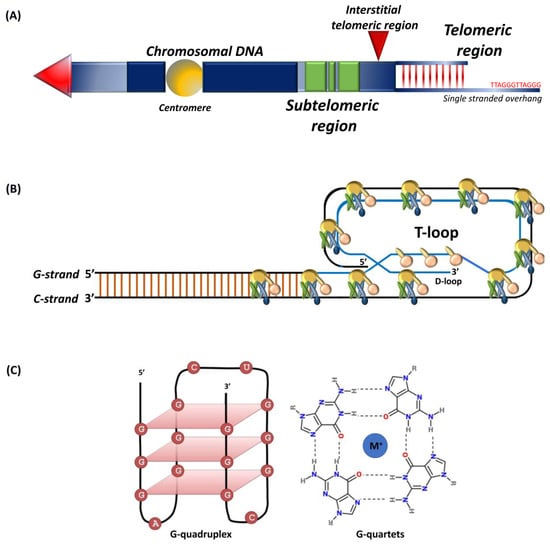
Figure 1.
Schematic representation of (A) telomeres and subtelomeric regions, tandem nucleotide repeats at chromosomal ends that include a double-stranded region and a 50–300 nucleotide single-stranded guanine rich G-tail. Subtelomers (green) represent regions of genes interspersed within repeat elements and interstitial telomeric sequence (red arrow); (B) shelterin complex, the G-tail folds back into the duplex DNA to form the t-loop; (C) G-quadruplex structure, intramolecular G-quadruplex (left) built from G-quartets that are formed through cyclic Hoogsten hydrogen-bonding arrangement of four guanines with each other with G-tetrad structure on the right. Adapted from [23,24].
Figure 1.
Schematic representation of (A) telomeres and subtelomeric regions, tandem nucleotide repeats at chromosomal ends that include a double-stranded region and a 50–300 nucleotide single-stranded guanine rich G-tail. Subtelomers (green) represent regions of genes interspersed within repeat elements and interstitial telomeric sequence (red arrow); (B) shelterin complex, the G-tail folds back into the duplex DNA to form the t-loop; (C) G-quadruplex structure, intramolecular G-quadruplex (left) built from G-quartets that are formed through cyclic Hoogsten hydrogen-bonding arrangement of four guanines with each other with G-tetrad structure on the right. Adapted from [23,24].
2. Telomere-Associated Proteins
Telomeres are, in general, associated with three types of proteins that include nucleosomes, shelterin complex, and chromosomal transcription factors [13,25,26].
2.1. Nucleosomes
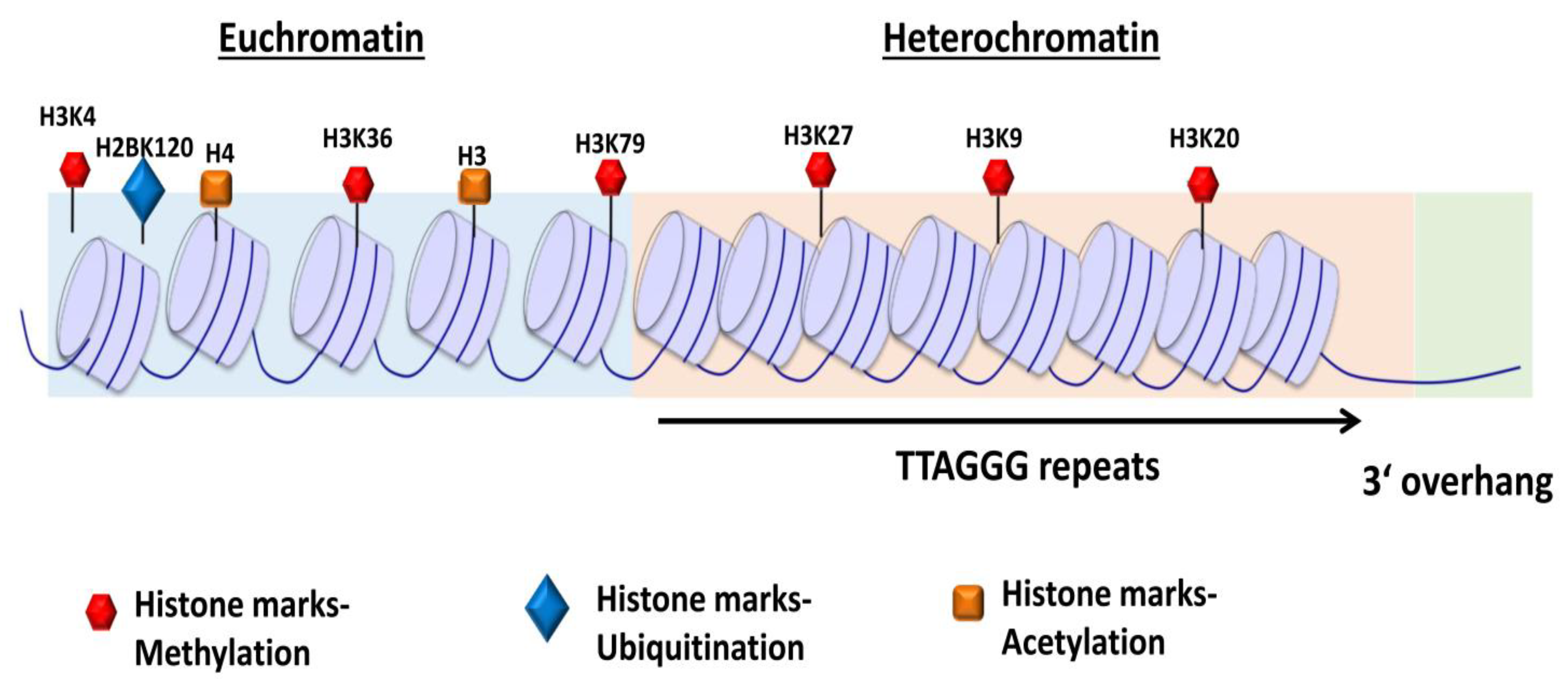
The telomeres, organized within tightly packed histone octamer composed nucleosomes (Figure 2), are stabilized through specific protein–protein and protein–DNA interactions between shelterin subunits and tandem repeat sequences [25,27]. Telomeres in higher eukaryotes are mainly heterochromatins enriched with histone 3 trimethylated at lysine 9 (H3K9me3) and histone 4 trimethylated at lysine 20 (H4K20me3) and heterochromatin protein (HP) isoforms [28,29,30]. The histone methyltransferases, SUV39H1 and SUV39H2, promote the methylation of H3K9 residues [31]. H3K9me3 recruits HP1 proteins, which are important for chromatin compaction through a high binding affinity site [29,32]. The heterochromatic region maintains telomeric structural integrity [29]. The loss of heterochromatic marks results in an open chromatin conformation, defective telomere function, aberrantly increased telomere length, and chromosomal instability [33].
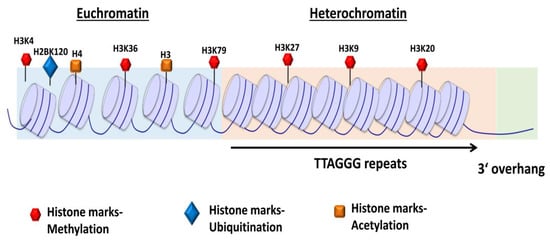
Figure 2.
Schematic representation of chromatin structure and distribution of histone marks on telomeres. The telomeres are tightly packed into nucleosomes, the structural and functional units of chromatin. The euchromatin-associated and heterochromatin-associated histone marks are indicated. The euchromatin-associated marks include H4ac, H4K20me1, H3ac, H3K4me1/2/3, H3K36me2/3, H3K27ac, H3K79me3, and H2BK120ub. The heterochromatin-associated marks include H4K20me3, H3K9me3, and H3K27me3. Adapted from [34].
Besides the routine post-translational modifications, histone proteins function in telomere capping, telomere transcription, homologous recombination at telomeres, cellular differentiation, and nuclear reprogramming [29,34]. The heterochromatin structure transcriptionally silences nearby genes, a phenomenon attributed to the telomere position effect (TPE) [34]. TPE mainly involves the shelterin protein, repressor and activator protein 1 (RAP1), and histone acetylase, SIRT6, a homolog of the yeast protein silent information regulator 2 (Sir2). RAP1 recruits SIRT6 protein, which on telomeres interact and promote hypo-acetylation of histone marks for active transcriptional repression of nearby genes [35].
2.2. Shelterin Complex
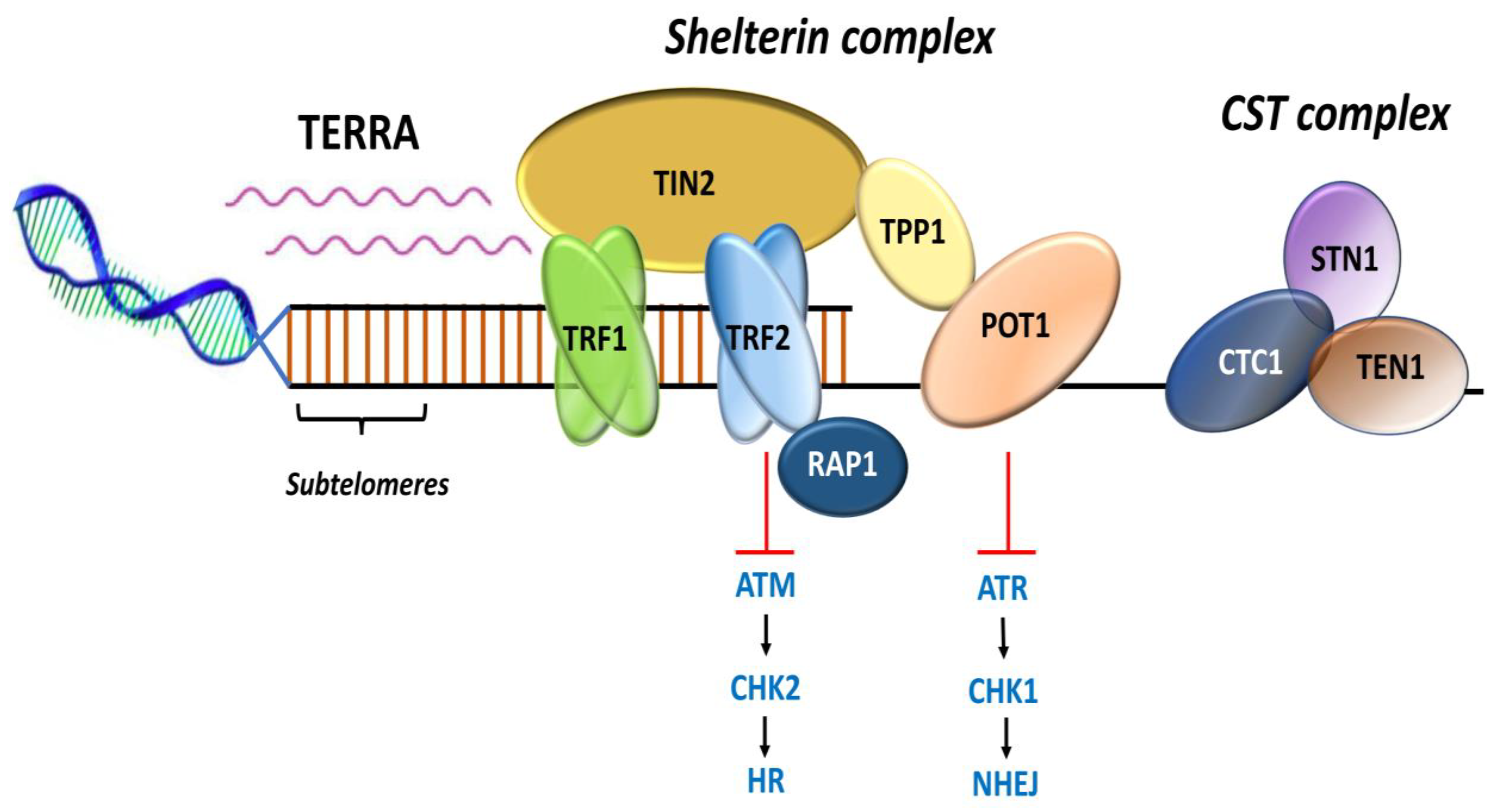
Shelterin complex comprises of six protein subunits [13]. Telomeric-repeat-binding factor 1 and 2 (TRF1 and TRF2) and protection of telomeres 1 (POT1) bind to DNA, and TRF1-interacting nuclear protein 2 (TIN2), TIN2-interacting protein (TPP1), and RAP1 act as adaptors (Figure 3) and mediate interactions among the constituents [5,36]. The shelterin complex functions as a dynamic unit in regulating telomere length, protects the chromosomal ends from being recognized as DNA damage, and represses DNA damage response (DDR) signals [13,37,38].
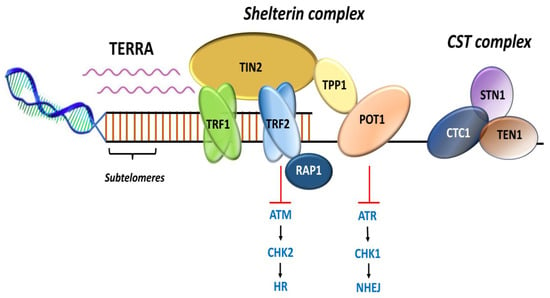
Figure 3.
Representation of shelterin complex, heterotrimeric complex CST, and telomeric repeat containing RNA (TERRA). Shelterin complex comprises of six distinct protein subunits: telomeric-repeat-binding factor 1 and 2 (TRF1 and TRF2), TRF1-interacting nuclear protein 2 (TIN2), protection of telomeres 1 (POT1), POT1 and TIN2-interacting protein (TPP1), and repressor and activator protein 1 (RAP1). TRF1 and TRF2 bind the double-stranded DNA; POT1 binds the single-stranded 3′ G-overhang. TIN2 bridges TRF1 and TRF2 by binding to both the proteins simultaneously through independent domains and recruits TPP1–POT1 complex. RAP1 interacts with TRF2 to localize at the telomeres. CST complex is a heterotrimeric protein consisting of conserved telomere protection component 1 (CTC1), suppressor of cdc13 a (STN1), and telomeric pathway with STN1 (TEN1), which specifically localize to the single-stranded 3′ overhang and protect the telomeres by mediating DNA replication and telomerase regulation, independent of shelterin complex. Telomeric repeat containing RNA (TERRA) transcription initiates within subtelomeres in the direction of telomeres. TERRA is involved in regulating telomere capping and the maintenance of telomeres. Adapted from [13].
TRF1 and TRF2 that exist as homodimers bind to the double-stranded DNA, and POT1 binds to the single-stranded 3′ G-overhang [39,40]. TRF1 and TRF2 contain a TRF homology (TRFH) domain that mediates homodimerization and a Myb-type domain that specifically binds to the telomere duplex [40,41,42]. TRF1 and TRF2 both negatively regulate telomere length and promote efficient telomere replication [42]. The TRFH domain of TRF2 regulates the formation of the t-loop, whose assembly and disassembly is coordinated during the cell cycle by a phospho-switch [18,43,44]. TRF1 and TRF2 also suppress non-homologous end joining (NHEJ) and ataxia telangiectasia mutated (ATM)-dependent DNA damage signaling [39]. TIN2 bridges TRF1 and TRF2 by binding to both the proteins simultaneously through independent domains. The TRFH domain of TRF1 mediates the TIN2–TRF1 interaction and the TIN2–TRF2 interaction is mediated by a short motif in the hinge domain of TRF2 [42,45]. TIN2 further recruits TPP1 forming a triple complex—TIN2-TPP1-TRF2 [46]. This interaction provides a structural basis for shelterin bridge assembly [47].
POT1, that binds to single-stranded DNA with high specificity, contains two N-terminal oligonucleotide/oligosaccharide binding (OB) folds [13]. The first OB fold binds to the hexamer repeat at the beginning of the strand while the second OB fold binds and protects the 3′ G-overhang [48,49]. POT1 represses the ATM- and RAD3- related protein (ATR)-dependent signaling pathway and protects the telomere ends from fusion [50]. TPP1 binding remains essential for recruiting POT1 to the telomeres as those form heterodimers, which enhances the function of POT1 at the single-stranded 3′ end of telomeres [51,52]. While POT1 directly binds to single-stranded DNA, it indirectly interacts with the double-stranded DNA through association with TPP1 [46]. A biological role for TIN2 dependent on TPP1-POT1 has been suggested where its binding stabilizes the complex and promotes telomere processivity [53]. Accordingly, TIN2, along with TPP1-POT1, forms as a specialized telomeric single-stranded DNA binding sub-complex within the shelterin complex [41,53].
RAP1 does not bind directly to the DNA, but rather forms a complex with TRF2 and its Myb domain binds to the primary domain of TRF2 for suppressing telomeric homologous recombination [54,55]. The RAP1-TRF2 complex represses the localization of proteins such as the poly (ADP-ribose) polymerase 1 and SLX4 (SLX4 structure-specific endonuclease subunit) to the telomeres [55].
2.3. Other Telomere-Interacting Complexes
Several protein complexes, apart from the shelterin, contribute to telomere regulation and maintenance [56,57]. Those are either directly recruited to the telomeres or through interactions with the shelterin components [56]. CST, a heterotrimeric protein complex (Figure 3) consisting of conserved telomere protection component 1 (CTC1), suppressor of cdc13a (STN1), and telomeric pathway with STN1 (TEN1), localizes at single strand and functions in telomere capping and length regulation [58,59,60]. The CST complex interacts with DNA Polα-primase during telomere replication [58]. In vitro biochemical analysis has shown that CST unfolds G-quadruplex structures to facilitate replication through telomeres [61,62]. The complex has also been shown to localize with Polα at DNA damage sites and fill in double-stranded breaks through interaction with the shieldin complex, a 53BP1 effector complex involved in DDR [63]. The STN1-TEN1 subunit of CST complex functions in resolving replication fork during replication stress and regulates telomerase-mediated extension of the 3′ G-overhang [64,65].
Some of the proteins associated with telomeres are also involved in the DDR mechanism [57]. RecQ-family DNA helicases, Werner (WRN) and bloom (BLM), are recruited to the telomeres through TRF1 and TRF2 [66,67]. RecQ helicase proteins are involved in unwinding of G-quadruplex structure and initiation of DNA replication [68]. In addition, excision repair cross complementing associated with xeroderma pigmentosum group F (ERCC/XPF) mediates the 3′ overhang process; the recombination protein RAD51 and the helicase regulator of telomere length 1 (RTEL1) are involved in the replication and recombination of telomeric DNA [69,70].
2.4. Subtelomeres
Subtelomeres are transcriptionally active chromatin regions (Figure 1A) between main chromosomal sequences and telomeres [71]. The subtelomeric region constitutes two major zones: polymorphic patchworks of inter-chromosomal segmental duplication region and a chromosome specific non-duplicated region [72,73]. Segmental duplicated regions constitute about 5% of the human genome and cover 5 to 300 kb of terminal chromosome sequences [72]. Subtelomeres are packed into constitutive heterochromatin that mainly contains H3K9me3 heterochromatin marks and also harbors transcriptional start sites for telomeric repeat-containing RNAs (TERRA) [74,75,76]. TERRA transcription initiates from within subtelomeres (Figure 3) towards telomeres [76,77]. TERRAs, associated with heterochromatin marks such as HP1 and H3K9me3, actively participate in telomere maintenance/end protection and heterochromatin formation [78,79,80]. Transcription factors such as SNAIL1, involved in the epithelial-to-mesenchymal transition, control telomere transcription, and integrity by negatively regulating TERRA [81]. The segmental duplicated region of subtelomeres contains protein coding genes that vary in copy number and is located on different chromosomes, such as WASH at 9p, 2p, Xq/Yq, 1p, 15q, and 16p; immunoglobulin heavy chain genes at 14q; and olfactory receptor genes at 1p, 6p, 8p, 11p, 15q, 19p, and 3q [82]. The subtelomeres function in the process of chromosome recognition and pairing during meiosis and are also involved in maintaining chromosomal stability and regulation of gene expression [83,84,85]. The subtelomeric homologous sequences prevent heterochromatin spreading into neighboring gene-rich regions to prevent suppression of the genes within those segments [84].
3. Telomere End Replication Problem
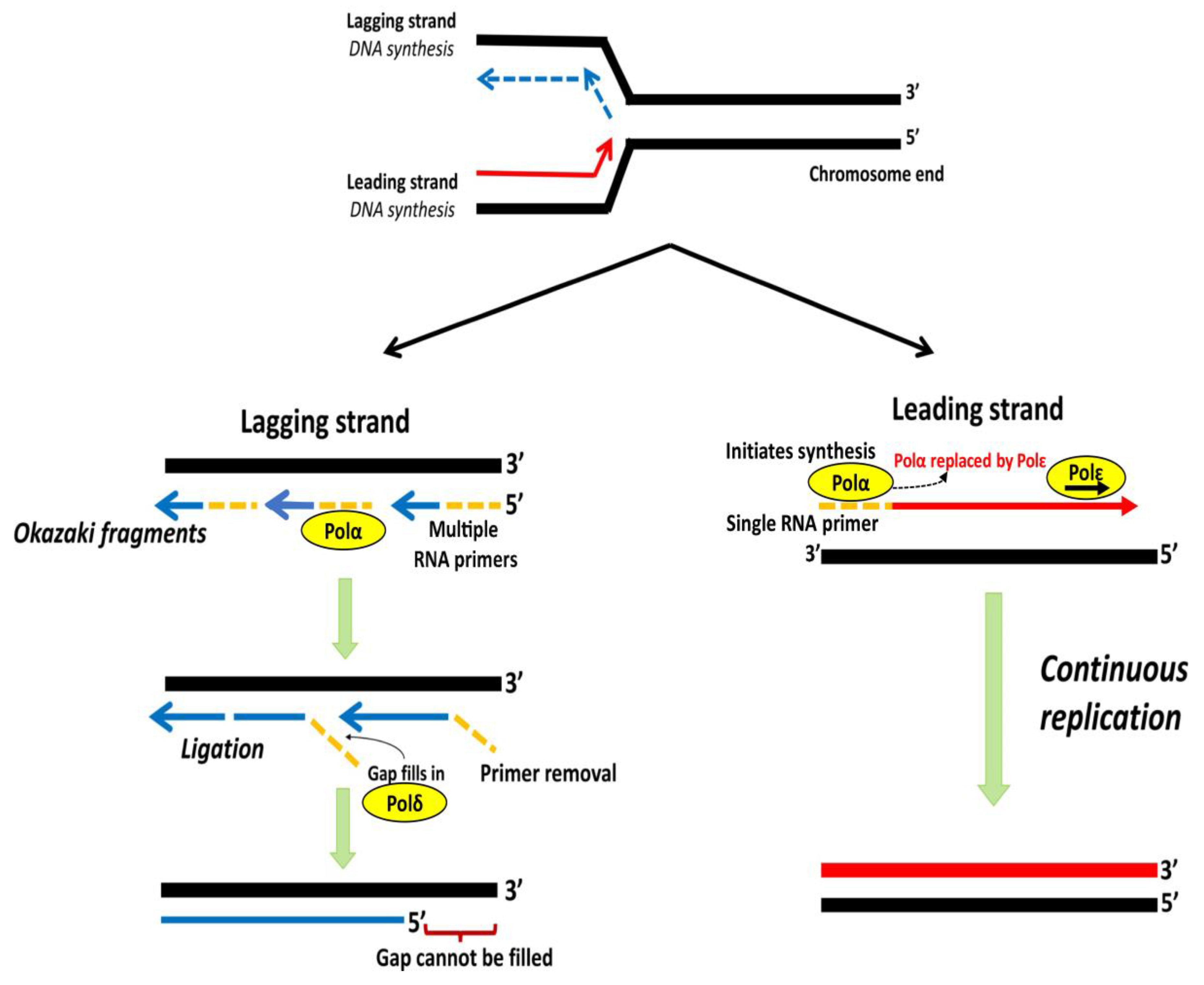
Incomplete replication at chromosomal ends by DNA polymerase results in progressive shortening of telomeres with each successive cell division and is termed as the “end replication problem” [1]. During DNA replication, a semi-conservative process, each DNA strand of a double helix acts as a template for the generation of a new complementary strand [7]. DNA polymerase Polα with a single RNA primer initiates the synthesis of a new strand in 5′ to 3′ direction towards replication fork, which is subsequently replaced by Polε for further elongation, forming the “leading strand” [86,87]. The synthesis of the “lagging strand” in the 5′ to 3′ direction requires annealing of multiple primers that elongate into short Okazaki fragments opposite to the replication fork and occurs less efficiently than the leading strand [88,89]. On completion of replication, the primer degradation results in internal gaps, filled by the polymerase, Polδ, and ligated to form a continuous strand. The gap left by the primer degradation at the terminal end remains unfilled, which results in the loss of a short segment of DNA at the 5′ end of the lagging strand [89]. The lagging strand synthesis fails to replicate an average length of ~250 nucleotides at the end of linear templates, which is hypothesized due to an inability of DNA Polα-primase to initiate lagging strand synthesis from the very end of linear DNA [90]. The loss of nucleotides at the chromosomal end leads to the G-rich single strand (Figure 4) at the end of the telomeres and, according to one hypothesis, the size of the 3′overhang is determinant of the rate of telomere shortening [91].
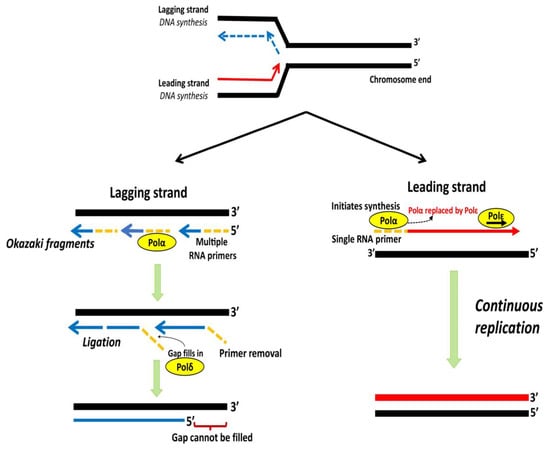
Figure 4.
Schematic representation of lagging and leading strand replication. DNA polymerase Polα with a single RNA primer initiates synthesis of leading strand, which is subsequently replaced by Polε for further elongation. The lagging strand is copied through discontinuous Okazaki fragments from multiple primers. RNA primers are degraded and the gaps filled by Polδ followed by ligation of discontinuous fragments. The gap at 5′ end remains unfilled, leading to a non-replicated terminal region. Adapted from [7].
Normal human cells in a culture stop dividing after 40 to 60 passages, a phenomenon first observed by Leonard Hayflick and eponymously called the Hayflick limit [89,92]. Incomplete replication with a gradual shortening of telomeres acts as a counting mechanism that eventually leads to the replicative senescence [93]. On average, a single human telomere contains enough repeats to buttress the effect of telomere erosion in the absence of a maintenance mechanism, with an estimated loss of about 50 to 250 bp per mitosis [12,90,94]. Telomere shortening, to an extent, in proliferating cells of self-renewal tissues, such as hematopoietic cells, cells of the skin, and cells from gastrointestinal epithelium, is mitigated by holoenzyme telomerase [5,12,95]. Most of the adult stem cells and somatic tissues do not contain sufficient telomerase to maintain telomere length infinitely and therefore undergo age-related telomere shortening [96].
4. Mechanisms of Telomere Maintenance
The ribonucleic protein, telomerase, counteracts the replication-related telomere attrition. Telomerase is upregulated in tumors from over 90% of cancers; in 10% to 15% of tumors, telomeres are elongated through a homologous recombination-based alternative lengthening of telomeres (ALT) [97].
4.1. Telomerase Structure and Biogenesis
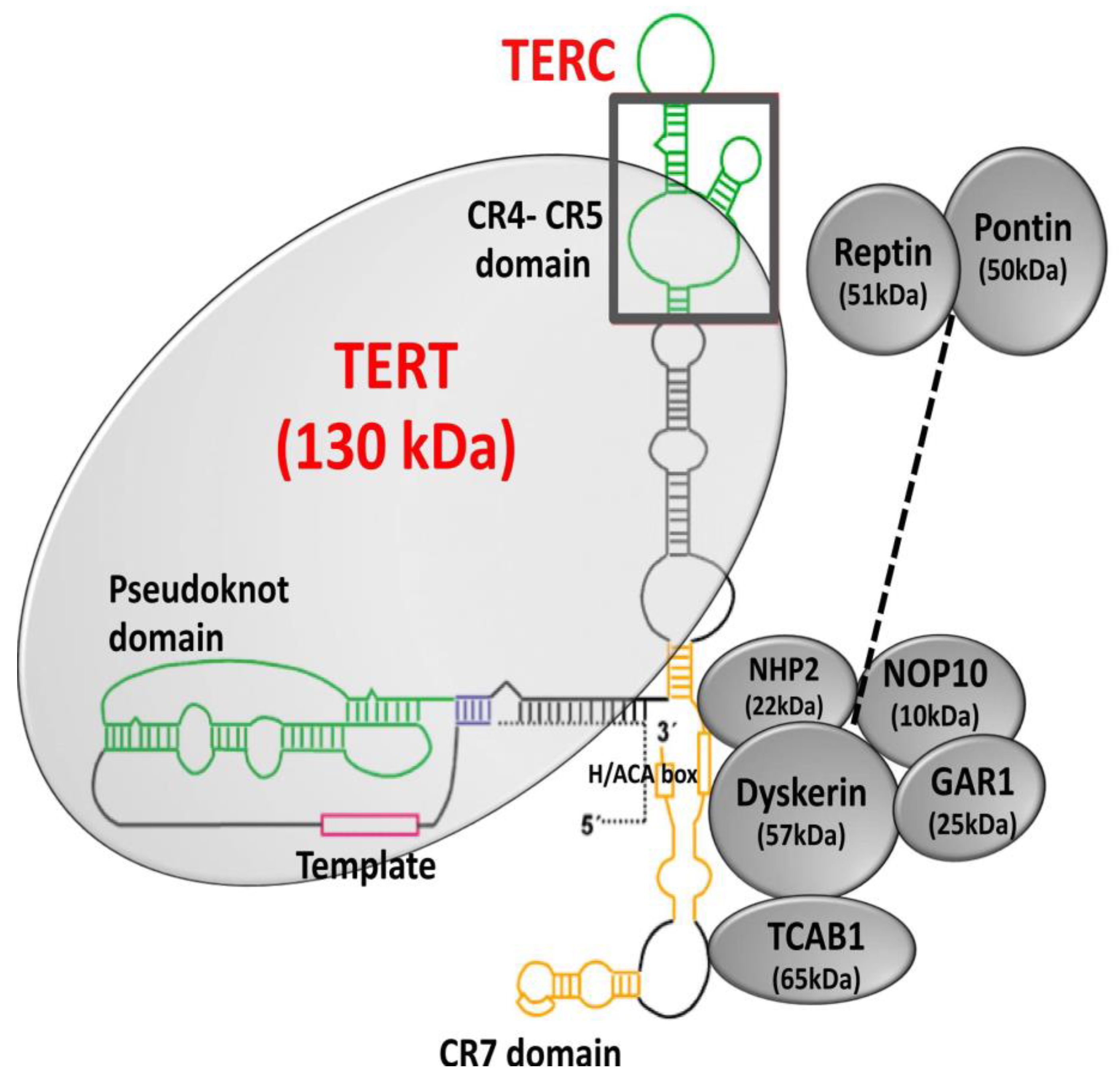
Telomerase consists of a catalytic subunit, telomerase reverse transcriptase (TERT), and an RNA component (TERC), which acts as a template for the extension of telomeric nucleotide repeats [6,98,99]. A number of accessory molecules regulate telomerase biogenesis, subcellular localization, and function [100,101,102,103]. The 3′ end of TERC contains a conserved H/ACA domain (Figure 5) that binds the protein complex formed by dyskerin (DKC1), nucleolar protein 10 (NOP10), non-histone protein 2 (NHP2), and encoding H/ACA ribonucleoprotein complex subunit 1 (GAR1) [17,103,104]. NOP10 and GAR1 bind to dyskerin, and NHP2 binds to the RNA directly [105]. TERC in the nucleolus assembles with TERT to form a mature telomerase complex, followed by recognition of the Cajal body (CAB) box by telomerase and telomerase cajal body protein 1 (TCAB1), which in turn recruits mature telomerase complex to Cajal body [106]. During the S-phase of the cell cycle, Cajal bodies facilitate the recruitment of the mature telomerase complex to the telomeres [107]. Further, auxiliary proteins, such as ATPases reptin and pontin, have shown to be involved in telomerase assembly by interacting with TERT and dyskerin [108]. Pontin and reptin facilitate the assembly of TERT with TERC and dyskerin or remodel the mature telomerase complex. Through their interaction with dyskerin, pontin and reptin are involved in assembling and stabilizing TERC [108].
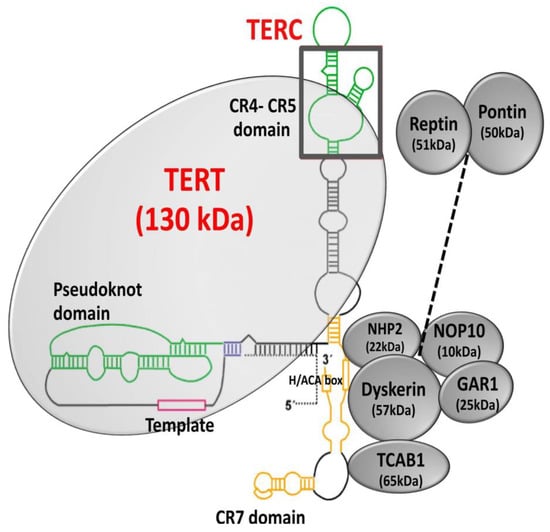
Figure 5.
Structure of telomerase. Telomerase is a holoenzyme composed of the catalytic subunit, TERT (telomerase reverse transcriptase), and the RNA component, TERC (telomerase RNA component). Dyskerin and other associated proteins, GAR1, NHP2, and NOP10 interact with TERC by binding to the H/ACA box and regulate telomerase biogenesis, subcellular localization, and function. Adapted from [109,110].
Telomerase activity remains tightly controlled at multiple levels- from transcriptional regulation of components for biogenesis to recruitment to the telomeres [111,112]. The model of repeat-addition processivity involves the addition of telomere repeats by the holoenzyme in successive steps without primer dissociation and requires several elements [12]. The number of repeats added by telomerase remains a controlled phenomenon with a set equilibrium and any disruption becomes causal for different telomere related diseases [12]. A number of proposed models have explained telomere length homeostasis [113]. The protein counting model predicated on telomere-bound proteins acting to block telomerase from a distance, with large numbers exerting a larger repressive effect and preferential elongation of shorter telomeres [114,115]. Another probabilistic model suggested the telomere length homeostasis via a switch between telomerase-extendible and telomerase non-extendible states, with a preferential shift towards the former state in short telomeres [116,117]. The replication fork model accounts for both negative regulation and preferential elongation of short telomeres with bound proteins exerting a negative effect that there would be increase in the probability of telomerase dissociation from the replication fork on short telomeres to reach the end for catalytic elongation [113].
4.2. Telomerase Reactivation
Telomerase reactivation occurs in tumors via multiple genetic and epigenetic mechanisms that include TERT and TERC amplification, genomic rearrangement of TERT, somatic mutations within the TERT promoter, and epigenetic modifications through TERT promoter methylation [97,118].
4.2.1. Gene Amplification of TERT and TERC and Rearrangement of TERT
The regions containing the TERT gene at chromosome 5p15.33 and the TERC gene at chromosome 3q26.3 (Figure 6A) are frequently amplified in cancers [119]. TERT expression based on correlation with the gene dosage has been shown to be haploinsufficient for telomere maintenance [119,120,121,122]. In a systematic analysis of TERT gene amplification based on 31 tumor types from 6835 patients, TERT amplifications were observed in 4% of tumors [118]. TERT amplifications were frequent in ovarian cancer, adrenocorticol carcinoma, esophageal cancer, lung adenocarcinoma, and squamous cell carcinoma. Overall, only in 3% of tumors, increased TERT expression was attributed to the amplifications; other tumors involved diverse mechanisms [118]. Increased TERT gene copy number was associated with upregulation of the gene expression and correlated with worse clinical outcomes in breast, lung adenocarcinoma, Merkel cell carcinoma, and thyroid carcinoma [123,124,125,126]. In systematic analyses, TERC amplifications leading to an increased expression occur in about 4% of the tumors, which included lung squamous cell carcinoma, esophageal cancer, and ovarian cancer [118].
Another mechanism of TERT upregulation, observed in neuroblastoma, comprises genomic rearrangements (Figure 6B) affecting the TERT locus at 5p15.33 [127,128]. The rearrangements mainly cluster in a region 50 kb upstream of the TERT transcriptional site, leading to the juxtaposition of active super-enhancers in close proximity to the TERT locus that causes chromatin remodeling and consequent increased expression [127,128]. The TERT rearrangements occur mainly in high-risk neuroblastoma in mutually exclusiveness with MYCN amplifications and ATRX mutations [127,128].
4.2.2. TERT Promoter Mutations
TERT promoter mutations represent frequent somatic genetic alterations that drive TERT expression and telomerase reactivation [12,129]. The recurrent somatic mutations within the TERT promoter mainly at −124 and −146 bp from the ATG start site generate de novo binding sites for E-twenty-six/ternary complex (ETS/TCF) transcription factors [130,131]. Other somatic TERT promoter mutations that create identical binding sites for ETS/TCF transcription factors include that detected at −57 bp, originally discovered as the causal germline mutation in a melanoma pedigree, and at −124/−125 bp and −138/−139 bp as CC > TT tandem mutations that occur mainly in skin cancers [130,132,133,134]. In glioblastoma, liver cancer and bladder cancer cell lines, GA binding protein transcription factor subunit alpha (GABPA) as in a heteromeric complex with GABPB1, binds to the de novo E-twenty-six (ETS) binding sites created by the TERT promoter mutations (Figure 6C) in cooperation with in-proximity native sites [135]. TERT promoter mutations occur mainly in cancers arising from tissues with low-rates of self-renewal that include glioblastoma, melanoma, urothelial carcinoma, squamous cell carcinoma, medulloblastomas, and aggressive thyroid carcinoma subtypes [12,131,136,137,138,139,140,141,142,143]. TERT promoter mutations contribute to tumorigenesis in a two-step mechanism. Those mutations during the initial phase, instead of preventing bulk telomere shortening, extend the cellular lifespan by stabilizing the shortest telomeres. In the second phase, the critically short telomeres lead to genomic instability and telomerase is further upregulated to sustain cell proliferation [144].
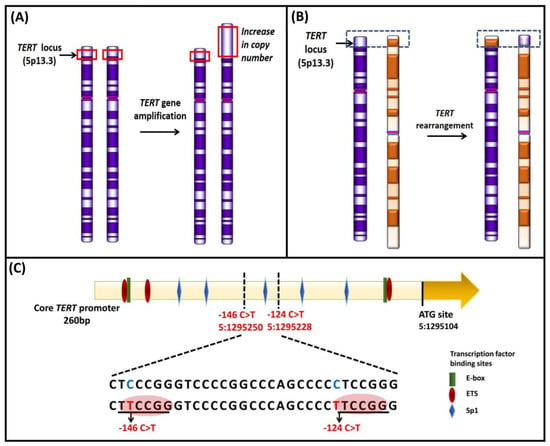
Figure 6.
Schematic representation of genetic mechanisms of telomerase activation. (A) TERT gene amplification leading to an increase in TERT copy number at the 5p15.33 locus; (B) genomic rearrangement in TERT result in inter-chromosomal translocation; (C) Mutations at two hotspots in the TERT promoter, −124 and −146 bp from ATG start site create de novo binding sites for ETS transcription factors (red circles). Various transcriptional binding elements are represented in the core promoter region, E-box (green), ETS (red), and Sp1 (blue). Adapted from [12,97,145].
Figure 6.
Schematic representation of genetic mechanisms of telomerase activation. (A) TERT gene amplification leading to an increase in TERT copy number at the 5p15.33 locus; (B) genomic rearrangement in TERT result in inter-chromosomal translocation; (C) Mutations at two hotspots in the TERT promoter, −124 and −146 bp from ATG start site create de novo binding sites for ETS transcription factors (red circles). Various transcriptional binding elements are represented in the core promoter region, E-box (green), ETS (red), and Sp1 (blue). Adapted from [12,97,145].
4.2.3. Epigenetic Mechanisms
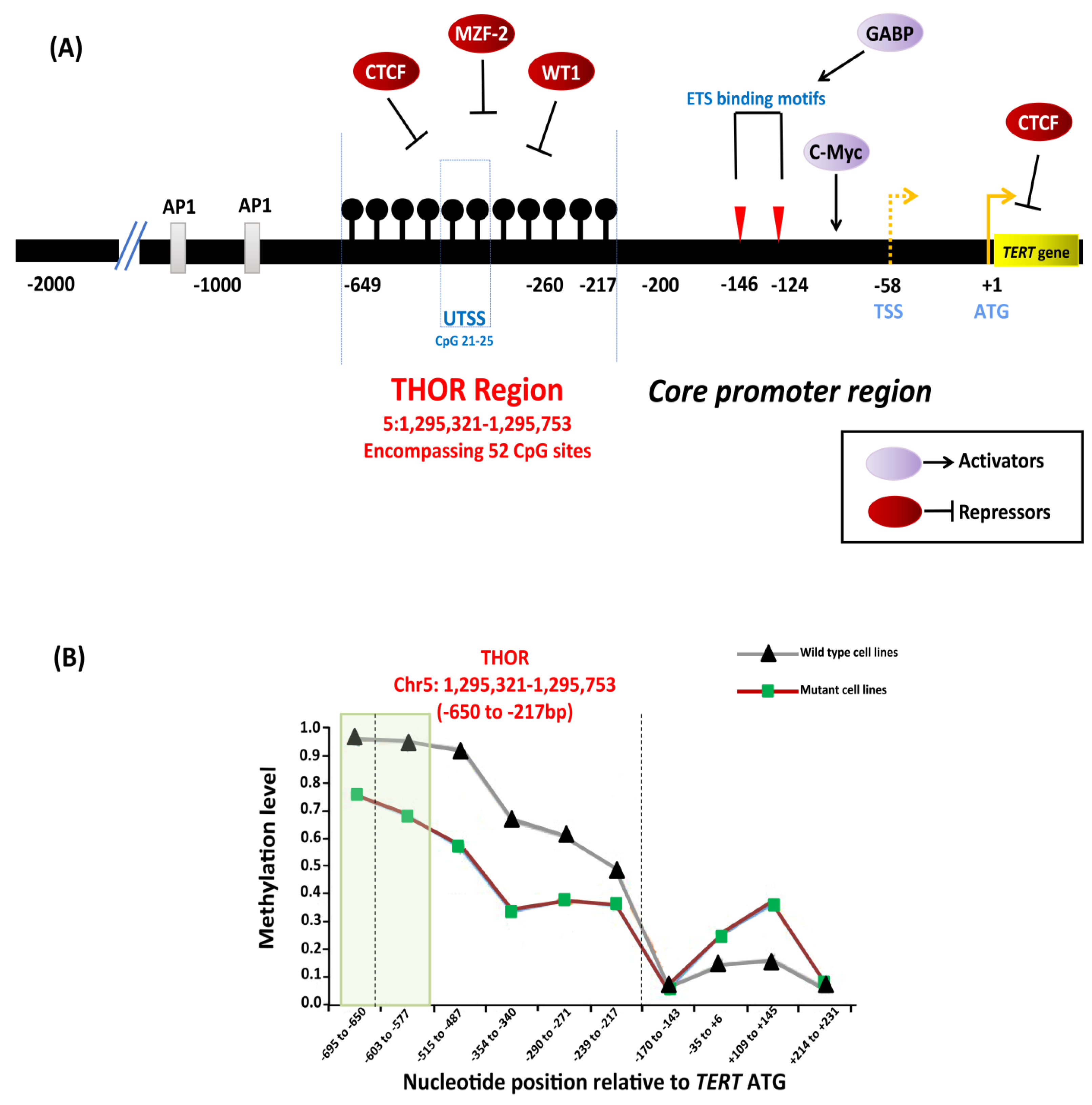
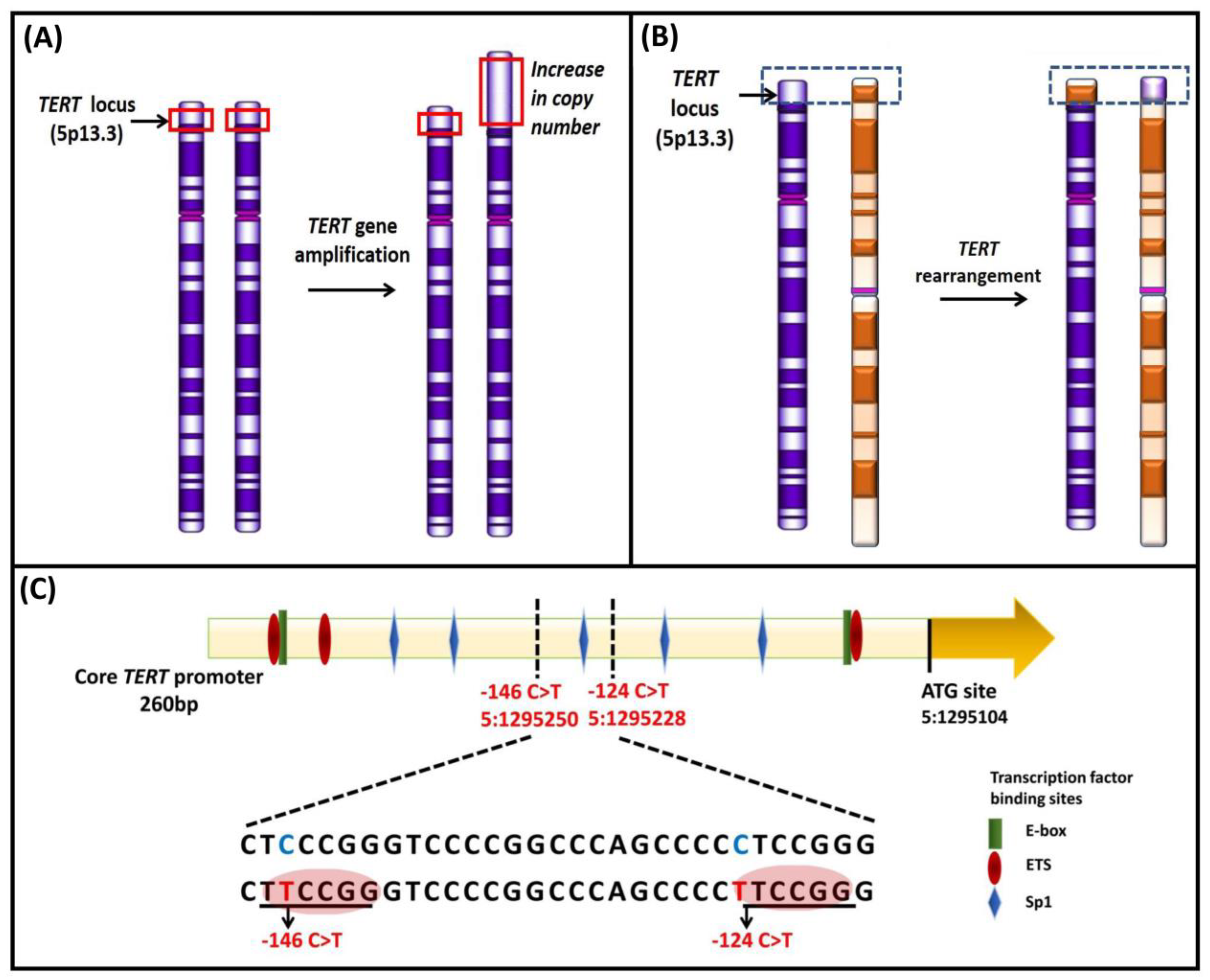
The TERT gene contains a CpG island that extends from −838 bp from the ATG start site to a position near the end of exon 2 (Chr 5: 1,296,000–1,293,450) [146]. Conventional and next generation sequencing studies characterized a 433 bp genomic region within the promoter, extending from −650 to −217 bp from the ATG start site (GRCh37/hg19, Chr5: 1,295,321–1,295,753), that encompasses 52 CpG sites known as the TERT hypermethylated oncological region (THOR) [147]. THOR is hypermethylated in malignant tumors and hypomethylated in normal tissues and stem cells. DNA methylation controls the binding of transcriptional activators, c-Myc, and repressors CCCTC-binding factor (CTCF), myeloid zinc finger protein-2 (MZF-2), and Wilms tumor 1 (WT1) to the TERT promoter (Figure 7A). Hypermethylation prevents binding of the repressors to the promoter that leads to TERT upregulation and telomerase activation [123,147,148]. THOR methylation has been reported to have a diagnostic and prognostic role in pediatric brain tumors and prostate cancer [149,150].
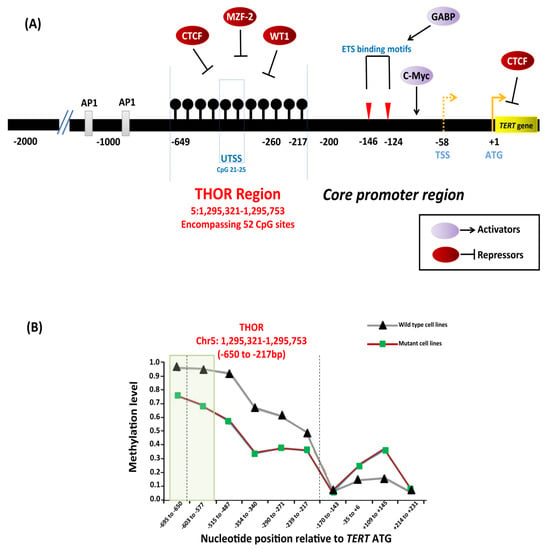
Figure 7.
Epigenetic regulation of TERT in cancers. (A) Depiction of transcription factors along with binding sites, TERT promoter mutations −124C > T and −146C > T, hypermethylated region upstream of transcription start site (THOR). Binding of transcriptional activators, c-Myc, and repressors, CCCTC-binding factor (CTCF), myeloid zinc finger protein-2 (MZF-2), and Wilms tumor 1 (WT1) to the TERT promoter is controlled by DNA methylation as methylated CpGs prevent the binding to the target sites leading to TERT activation. The black lollipops represent methylated CpG sites. (B) Relative DNA methylation in tumor-derived cell lines with and without TERT promoter mutations. The green box represents a specific region in THOR (−668 to −577 bp relative to ATG) that is shown to be less methylated in cell lines with TERT promoter mutations than in cell lines without mutations. Adapted from [123,146,147].
Reduced methylation in the TERT promoter occurs in cancers that harbor TERT promoter mutations [146,151,152]. A specific region within the THOR, from −668 to −577 bp from the ATG start site (Chr5: 1,295,681–1,295,772), was shown to be hypomethylated in tumor-derived cell lines (Figure 7B) with TERT promoter mutations compared to those without mutations [146]. In the cell lines with TERT promoter mutations, the methylation was shown to be allele-specific, and H3K27me3 and H3K9me3 histone marks of inactivation promote the methylation [146]. The binding of the GABPA/B1 complex to the de novo sites on the mutant alleles causes an epigenetic change from an inactive H3K27me3 to an active chromatin mark H3K4me2/3, resulting in monoallelic expression [146,153]. The enzyme enhancer of zeste homolog 2 (EZH2), catalytic subunit of polycomb repressive complex 2 (PRC2), is responsible for the deposition of H3K27me3. The causal relationship between DNA and histone methylation was further supported by a strong binding preference for PRC2 at the methylated TERT promoter in vitro [146].
4.3. Alternative Lengthening of Telomeres
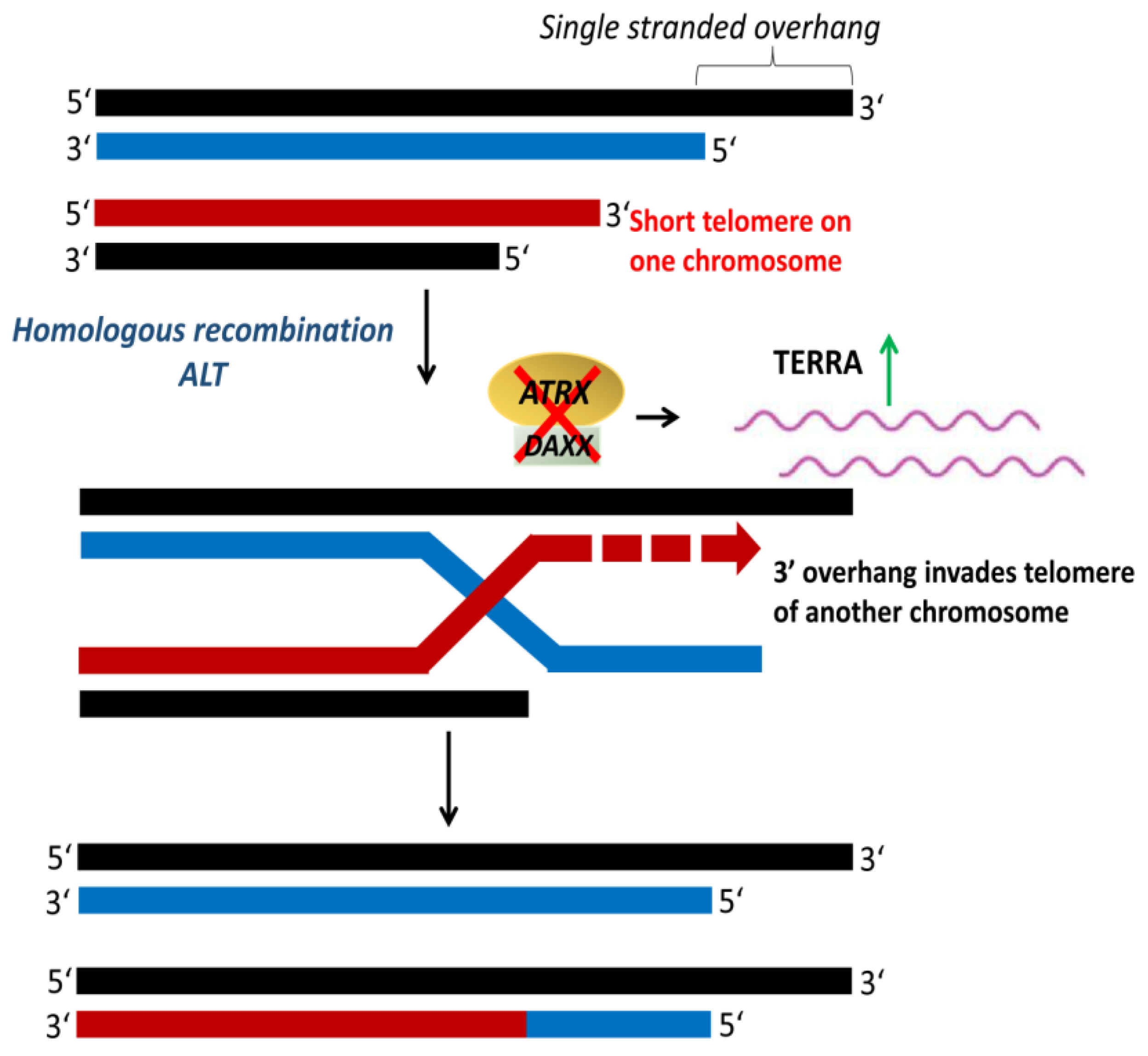
Cancer cells that maintain their telomeres by ALT (Figure 8) are characterized by heterogeneous telomere length with extremely long (>50 kb) and short (<5 kb) telomeres [154,155]. Telomeres in ALT cells cluster around promyelocytic leukemia (PML) nuclear bodies, referred to as ALT-associated PML bodies (APB) [156]. ALT is usually detected by telomere-specific fluorescence in situ hybridization, APB immunofluorescence, and ALT-associated molecule detection assays [157,158]. Mutations in the genes encoding for the α-thalassemia/mental retardation syndrome X-linked protein (ATRX) and the death domain-associated protein (DAXX) have been associated with ALT-positive tumors [159]. ATRX, together with DAXX, function as a chromatin remodeling complex that facilitates the deposition of histone variant H3.3 at the telomeres [160]. The loss of ATRX and DAXX due to mutations leads to a repressed heterochromatin state that activates recombination and initiation of ALT [154,161]. ATRX loss compromises the cell cycle regulation of TERRA and leads to the persistent association of replication protein A (RPA) with telomeres, resulting in a recombinant nucleoprotein structure [162]. ALT is observed at a high frequency in tumors of the central nervous system, peripheral nervous system, and sarcoma, but rare in carcinomas [159,163].
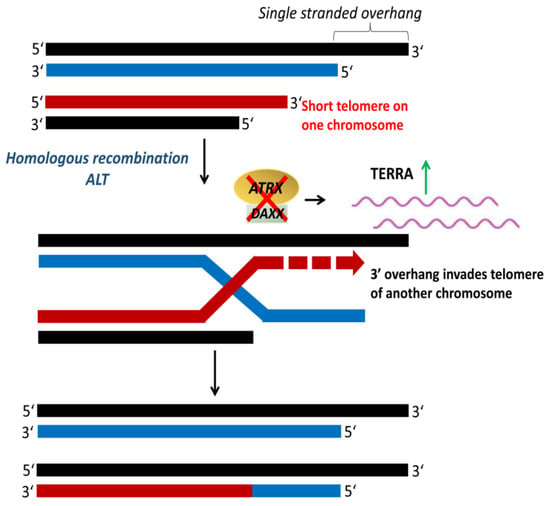
Figure 8.
Alternative lengthening of telomeres (ALT). ALT is a telomerase-independent mechanism that occurs via homologous recombination to maintain telomere length. The inactivation of α-thalassemia/mental retardation syndrome X-linked protein (ATRX) and death domain-associated protein (DAXX) upregulates telomeric repeat containing RNA (TERRA), which activates telomeric recombination and initiation of ALT. Adapted from [97].
5. Telomere Length Heritability
Epidemiological studies have shown telomere length as a complex heritable trait with estimated heritability derived from twin studies from 36% to 82% compared to 34% to 50% from familial studies [164,165,166]. The predominant environmental factors shared between twin-pairs impact the telomere length during initial growth and development [164]. The two potential sources of heritability are inherited genetic variations that influence telomere maintenance and variability in telomere length per se [165,167,168].
The variability in telomere length in parental gametes is directly expressed in the offspring zygotes, but a correlation between offspring and paternal telomere length or offspring and maternal telomere length is not clear [165,169]. In a meta-analysis involving six different populations with 19,713 subjects, a high heritability estimate of 70% and a statistically significant correlation between maternal and offspring telomere was reported, which was attributed to an X-linked mechanism and mitochondrial DNA [170,171]. The effect of paternal age at conception on offspring telomere length has been widely reported in several studies, with evidence suggesting that newborns with older fathers had statistically significant long telomeres [170,171,172,173,174].
Telomere length inter-individual variation arises early in life due to an interplay between genetic and environmental factors [175,176]. Several genetic variants associated with telomere length have been identified through genome-wide association studies (GWAS), which to some extent account for inter-individual variation in telomere length in the general population [177,178,179,180,181,182,183]. In addition, the impact of environmental factors influencing telomere length during growth and development is also relevant to telomere heritability estimates [169].
5.1. Genetic Factors Associated with Telomere Length
A number of telomere length associated genetic loci associated have been identified through linkage analysis and GWAS [178,179,180,181,182,183,184,185,186]. In a study conducted on 383 adult subjects from 173 families, comprising of 258 sibling pairs, the first locus associated with mean telomere length was mapped to chromosome 12p11.2 [187]. In another linkage study with 1025 dizygotic twin pairs, chromosome 14q23.2 and two additional suggestive loci at 10q26.13 and 3p26.1 associated with telomere length were identified [188]. In a linkage analysis carried out on 4289 individuals from 586 families, with evidence for longevity, two additional loci were mapped at 17q23.2 and 10q11.21 [184].
Twelve GWAS (Table 1) on telomere length conducted on different study populations have been reported so far [178,179,180,181,182,183,184,186,189,190,191,192]. In a first GWAS conducted on 1625 women from the UK adult twin registry, two single-nucleotide polymorphisms (SNPs) represented by rs2162440 and rs7235755 on chromosome 18q12.2 were shown to be associated with telomere length, which could not be replicated in additional 1165 men and women from the UK twin registry [189]. First, statistically significant associated SNPs with telomere length were at 3q26, represented by rs12696304 and rs16847897, identified through GWAS on 1487 individuals with coronary heart disease and 1430 healthy controls with association replicated in independent cohorts [179,186]. In a meta-analysis on 3417 individuals from four populations, telomere length associated SNPs were identified within the segments with genes OBFC1 and chemokine C-X-C motif receptor 4 (CXCR4) [181]. Following the initial discovery of SNPs in the TERC and OBFC1 loci, several GWAS have identified additional variants associated with telomere length in loci containing TERT, CTC1, NAF1, and RTEL1 [178,179,180,181,182,183,186].
Through a large-scale GWAS based on 26,089 healthy controls from breast, ovarian, and prostate cancer, four loci with telomere length associated SNPs were identified, including 3q26.2 (TERC), 5p15.33 (TERT) and 10q24.3 (OBFC1), and at chromosome 3p14.1 with the PXK gene [178]. So far, through GWAS, nine different loci telomere length associated variants have been identified. The individual SNPs in those genes exert only a small effect on telomere length; the combined effect of numerous such polymorphisms can be substantial [193].

Table 1.
Telomere length-associated single nucleotide polymorphisms.
Table 1.
Telomere length-associated single nucleotide polymorphisms.
| Genes (Locus) | SNP a | Genomic Position (GR37/hg19) b | ΔTL c | p-Value d | Reference |
|---|---|---|---|---|---|
| TERC (3q26.2) | rs1317082 rs10936601 rs12696304 rs16847897 rs10936599 rs3772190 | 3:169497585 3:169528449 3:169481271 3:169568116 3:169492101 3:169500487 | (−) 77 bp NA (−) 75 bp (−) 33 bp NA NA | 1 × 10−8 4 × 10−15 4 × 10−14 1 × 10−5 3 × 10−31 2 × 10−1 | [178] [178] [179] [179] [180] [186] |
| OBFC1 (10q24) | rs2487999 rs9420907 rs4387287 rs9419958 | 10:105659826 10:105676465 10:105677897 10:105675946 | (+) 100 bp NA (−) 230 bp NA | 4 × 10−14 7 × 10−11 2 × 10−11 9 × 10−11 | [178] [180] [181] [182] |
| TERT (5p15.3) | rs7726159 rs2736100 rs2736108 rs7705526 rs2853669 | 5:1282319 5:1286516 5:1297488 5:1285974 5:1295349 | (+) 73 bp (−) 94 bp NA (+) 90 bp (+) 60 bp | 5 × 10−17 4 × 10−6 5 × 10−5 1 × 10−15 - | [178] [180] [194] [194] |
| PXK (3p14.4) | rs6772228 | 3:58376019 | (−) 120 bp | 4 × 10−10 | [178] |
| ZNF311 (6p22.1) | rs9257445 | 6:28949206 | (−) 38 bp | 1 × 10−7 | [178] |
| BCL2L1 (20q11.2) | rs6060627 | 20:30262159 | (+) 36 bp | 6 × 10−7 | [178] |
| GRIA4 (11q22.3) | rs610160 | 11:105696895 | (+) 30 bp | 7 × 10−6 | [179] |
| NAF1 (4q32.2) | rs7675998 | 4:164007820 | (−) 90bp | 4 × 10−16 | [180] |
| RTEL1 (20q13.3) | rs755017 | 20:62421622 | (−) 74 bp | 7 × 10−9 | [180] |
| ACYP2 (2p16.2) | rs11125529 | 2:54475866 | (−) 67 bp | 8 × 10−10 | [180] |
| ZNF208 (19p12) | rs8105767 | 19:22215441 | (−) 58 bp | 1 × 10−9 | [180] |
| MPHOSPH6 (16q23.3) | rs2967374 | 16:82209861 | NA | 3 × 10−7 | [180] |
| CTC1 (17p13.1) | rs3027234 | 17:8136092 | (−) 57 bp | 2 × 10−8 | [182] |
| ZNF676 (19p12) | rs412658 | 19:22359440 | (−) 49 bp | 1 × 10−8 | [182] |
| DCAF4 (14q24.2) | rs2535913 | 14: 73415233 | (−) 45 bp | 2 × 10−7 | [195] |
| DHX35 (20q11.23) | rs6028466 | 20:38129002 | NA | 3 × 10−7 | [183] |
| DKK2 (4q25) | rs7680468 | 4:108304199 | NA | 5 × 10−8 | [184] |
| CSNKA2 (16q21) | rs74019828 | 16:58209274 | (−) 38 bp | 5 × 10−8 | [190] |
a SNP, single nucleotide polymorphism. b Genomic position of SNP from GRCh37/hg19 reference genome. c Differences in telomere length estimates (given in base pairs, bp) for the variant allele of each SNP associated with telomere length, determined from GWAS. NA, data not available. d p-values from GWAS summary data showing genome-wide statistical significance.
Functionality of Telomere Length-Associated Single Nucleotide Polymorphisms
The functional impacts of the SNPs rs3027234 and rs2535913 at the loci 17p13.1 and 14q24.2 associated with telomere length were assessed from genome-wide expression data [182,195]. The minor allele (T-allele) of the SNP rs3027234, located in intron 11 (GRCh37/hg19 Chr17: 8,136,092) of the CTC1 gene, associated with low expression of the gene [182]. CTC1 is a component of the telomere-binding CST complex, which binds to the telomeric 3′ single strand and functions to promote replication by stimulating Polα-primase activity [180,196]. Reduced expression of CTC1 impairs complex formation with STN1 and TEN1 [197,198]. Depletion of the CST complex results in insufficient accumulation of Polα for efficient replication at the telomeres, leading to progressive telomere attrition [198].
The SNP rs2535913, located in intron 8 (GRCh37/hg19 Chr14: 73,415,233) of the DDB1 and CUL4-associated factor 4 (DCAF4) gene, leads to the reduced gene expression by affecting the binding of CTCF and Rad21. DCAF4 forms a complex with DDB1 and CUL4 that is involved in nucleotide excision repair [195]. Rad21, a component of the cohesion complex, and CTCF have been implicated in telomere maintenance [195,199]. The depletion of CTCF or Rad21 results in reduced binding of TRF1 and TRF2 to telomeres [200]. DCAF4 indirectly influences telomerase activity and telomere length through its interaction with DDB1. DDB1 functions as a binding partner for the transcription factor E2F1, a member of the E2F family of transcription factors that regulate cell proliferation and telomerase activity [201,202]. The exact function of DCAF4 on telomere regulation remains unclear [203]. In another functional study, the minor allele of the SNP rs2630578 located in intron 1 (GRCh37/hg 19 Chr12: 32,305,787) was shown to be associated with a reduced mRNA expression level of Bicaudal D Homolog 1 (BICD1), which functions in vacuolar traffic and regulates telomere length via telomerase and Ku-protein pathways [185,204]. The region surrounding the SNP exhibited the heterochromatin mark, H3K4me3, and the minor allele was shown to disrupt a putative binding sequence for Nuclear Factor Y (NF-Y) transcription factor, which is essential for TERC expression [185,205].
In addition, SNPs at chromosome 5p15.33 and 3q26.2, not associated with telomere length, were shown to affect TERT and TERC expression, respectively. The genomic region on chromosome 5 at 5p15.33, harboring TERT and cleft lip and palate associated transmembrane 1-like protein (CLPTM1L) genes, has been reported to contain several independent cancer susceptibility loci [194,206,207]. Fine-mapping analysis of the region in GWAS from four cancers identified an SNP, rs36115365, with a functional role in regulating TERT expression, which is located in-between the 5′end of TERT and 3′ end of CLPTM1L, with active histone modification marks and multiple transcription factor binding sites [208]. Transcriptional silencing of the regulatory region, encompassing the SNP, results in reduced telomerase activity and telomere length. The transcriptional regulator Zinc finger transcription factor 148 (ZNF148) preferentially binds to the minor allele of the variant that mediates increased TERT expression [208]. In a study based on 3912 individuals from the general population, the rs2293607 variant at 3q26.2, harboring the TERC gene was shown to alter the secondary structure of TERC mRNA, with the minor allele associating with an increase in the gene expression and telomere length [209].
The functional studies provide a framework for a genetic approach to investigate the causal role of telomere length in age-associated diseases [182,185,195]. However, to establish a causal link between the genetic variants associated with telomere length and disease risk is particularly challenging because other environmental and lifestyle factors also affect telomere length [169].
5.2. Environmental Factors Affecting Telomere Length
Several other factors that influence telomere length include oxidative stress, inflammation, lifestyle factors, physiological stress, and exposure to carcinogens [210,211,212]. The association between telomere length and environmental, occupational, and health risk factors has been reported in several cross-sectional epidemiological studies [210,211,212]. Oxidative stress is reportedly one of the most important causes of telomere shortening and reflects an imbalance between antioxidants and reactive oxygen species (ROS) [213,214]. Telomeres, due to high guanine content, are targets of oxidative damage through the formation of 8-hydroxy-2-deoxyguanosine (8-oxodG), an important marker of oxidative stress, which causes accelerated shortening [214,215]. Single-stranded breaks preferentially accumulate at telomeres in conditions of mild oxidative stress, which cause replication fork stalling and incomplete replication of chromosome ends, again leading to telomere shortening [216]. Environmental exposure to ultraviolet and ionizing radiation and exposure to carcinogens such as arsenic and lead cause DNA damage either directly or indirectly through the induction of oxidative damage or onset of chronic inflammation [192,215,217,218,219,220].
Other lifestyle factors like smoking, obesity, and lack of exercise increase the rate of telomere shortening. In a meta-analysis based on 84 studies, it was reported that smokers had shorter telomeres than non-smokers [221]. Various aspects of socio-economic status, particularly educational attainment and social support, have shown to influence telomere length [222,223]. In a study based on 84,996 non-Hispanic whites, individuals with low socio-economic status had short telomeres [224].
6. Telomere Length and Risk of Cancers
The association between telomere length and risk of cancers has been reported in several epidemiological studies [225,226,227,228,229]. Studies conducted in large cohorts have consistently demonstrated an association between increased telomere length and risk of various cancers, including melanoma, basal cell carcinoma, glioma, lung cancers, tumors of the urogenital system, and lymphoma [226,230,231,232]. The genetic basis for those observations is provided through Mendelian randomization and studies showing that various polymorphisms that modulate telomere length also affect the risk of different cancers, with alleles segregating with long telomeres associating with increased risk [226,230,231]. Within the cellular context, long telomeres afford increased proliferative potential until the replicative crisis and the telomere length acts as a deterministic factor in cancer development [144,233]. Different investigations over the years, in contrast, have suggested that short telomeres associate with poor patient survival in different cancers [234,235,236,237,238]. Extremely short telomeres, caused by defective components that either protect or elongate telomeres due to genetic mutations, result in various debilitating disorders, referred to as telomeropathies [239,240,241].
In a Mendelian randomization study on 22 primary cancers involving 4,20,081 cases and 10,93,105 controls, genetically increased telomere length was shown to be associated with increased risk of nine cancers, which included glioma, serous low-malignant potential ovarian cancer, lung adenocarcinoma, neuroblastoma, bladder cancer, melanoma, testicular germ-cell cancer, kidney and endometrial cancer [226]. Those findings were similar in direction and magnitude of risk estimates reported previously in observational and Mendelian randomization studies [142,193,225,227,228,229,230,231,242,243,244,245,246,247]. Although long telomeres have been consistently reported to show statistically significant association with increased risk of various cancers, some conspicuous exceptions to that generalization have been reported in different studies [226,230,231,248,249]. In a Mendelian randomization study based on 2374 pancreatic cancer cases and 4326 controls, genetically decreased telomeres were associated with increased risk of pancreatic cancer [248]. Exposure to carcinogens such as arsenic has been shown to modulate the direction of the effect of telomere length on cancer risk. In a study on basal cell carcinoma, a reversal of the effect was reported, where arsenic exposure and short telomeres were shown to synergistically increase the risk in a dose-dependent manner [249].
7. Telomeres as Potential Targets for Anti-Cancer Therapy
Telomeres and telomerase-based therapies are emerging as prospective cancer treatment strategies [250]. Telomerase inhibitors such as small molecule inhibitors, antisense nucleotides, G-quadruplex stabilizers, TERT-dependent anticancer immunotherapy, and chemical inhibition of telomerase are the most commonly studied anti-cancer treatment strategies [251,252]. Telomeres are also targeted using guanine-rich oligonucleotide (GRO) homologous to the 3′ single-stranded overhang, known as T-oligos, a specific 11-base oligonucleotide sequence, (5′ GTTAGGGTTAG), which accumulates in the nucleus and induces DDR with minimal or no functional effect on normal cells [250,253]. Treatment with T-oligos in vitro has been shown to be effective in reducing viability and tumor growth in several cancers, including melanoma, prostate, ovarian, lung, breast, and colorectal cancer [250,254,255,256,257,258]. T-oligos are hypothesized to interfere with normal telomeric structure and form G-quadruplexes, inducing genomic stress in addition to aberrant upregulation of DDR pathways, and TRF2 and POT1 have shown to be upregulated after T-oligo treatment [250,254,259]. T-oligos induce DDR mechanism via two potential modes, the shelterin dissociation model (SDM) and the exposed telomere mimicry model (ETM). The SDM model proposes that the T-oligos upon introduction into the nucleus compromise the integrity of telomeres through the displacement of shelterin proteins, leading to the unfolding of t-loops and induction of DDR response. The ETM model proposes that T-oligos accumulate in the nucleus and are recognized as damaged telomeres, initiating a DDR mechanism similar to those occurring during excessive telomere shortening [250].
Telomere dysfunction mediated through telomerase substrate precursor, 6-thio-2′-deoxyguanosine (6-thio-dG), impairs cell viability and tumor growth [260]. 6-thio-dGTP, which is formed in cells from 6-thio-dG, gets recognized by telomerase and incorporated into telomeres leading to telomere dysfunction-induced foci in telomerase positive lung and colon cancer and BRAF-mutant melanoma cells [260,261]. Therapeutic inhibition of TRF1 binding to the telomeres using small molecules have been shown to suppress the growth of lung carcinomas and glioblastoma by inducing the DDR mechanism [262,263,264]. Imetelstat (GRN163L) directly targets telomerase by antagonistically binding to TERC; however, the long term effects are not known [265]. G-quadruplexes inhibit telomerase activity by blocking the binding of TERC [250]. The use of G-quadruplex stabilizers as treatment for progressive and malignant cancers gradually shortens the 3′ single-stranded ends of the telomeres, without reducing the overall length of the telomeres, thereby indirectly inhibiting telomerase activity [250,266]. Although the TERT-based therapeutic vaccination have limited anti-proliferation efficiency, the focus has shifted to personalized interventions specifically for patients with TERT promoter mutations and TERT genomic rearrangements, in combination with immune-checkpoint inhibitors [267]. Individuals with short telomeres are more prone to damage by irradiation compared to those with long telomeres. Telomerase inhibitors such as Imetelstat, coupled with radiotherapy, enhanced the cancer cell response to irradiation via telomere dysfunction [268]. Inhibitors directly targeting telomeres, such as T-oligos, G-quadruplexes, telomestatin, G-quadruplex ligand, and shelterin proteins-TRF2, TPP1, and POT1, are also shown to improve radiosensitivity [268,269].
8. Conclusions
Telomeres, the dynamic structures at chromosomal ends, are crucial for genomic integrity, and through age-dependent attrition, act as tumor suppressors [12]. Telomeres are protected from being recognized by DNA damage response by components of shelterin complex that also assists in the recruitment of telomerase for elongation of repeats [16]. Telomerase, a tightly regulated holoenzyme, while limited in most somatic cells, is upregulated as a major hallmark through different mechanisms in a majority of human cancers to impart unlimited replicative potential [5]. Telomere length per se, a hereditary trait, has been associated with different diseases, including various cancers. Extremely short telomeres, characteristics of various telomere diseases, are caused by genetic mutations in different components involved in telomerase function. Genetically-driven long telomeres, with some exceptions, in many studies, have been shown to increase the risk of different cancers. Telomeres not only represent functional segments in the human genome but also hold potential as targets for anti-cancer strategies.
Author contributions
R.K. developed the concept; N.S. gathered information from the relevant scientific literature for the review; and S.R. gathered information from the relevant scientific literature for the review. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Bundesministerium für Bildung und Forschung (BMBF) (01KT15511) and German Consortium for Translational Research (DKTK).
Conflicts of Interest
The authors declare no conflict of interest.
References
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.M.; Ryder, O.A.; Houck, M.L.; Charter, S.J.; Walker, W.; Forsyth, N.R.; Austad, S.N.; Venditti, C.; Pagel, M.; Shay, J.W.; et al. Comparative biology of mammalian telomeres: Hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell 2011, 10, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Toupance, S.; Villemonais, D.; Germain, D.; Gegout-Petit, A.; Albuisson, E.; Benetos, A. The individual’s signature of telomere length distribution. Sci. Rep. 2019, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Wright, W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Greider, C.W.; Szostak, J.W. Telomeres and telomerase: The path from maize, tetrahymena and yeast to human cancer and aging. Nat. Med. 2006, 12, 1133–1138. [Google Scholar] [CrossRef]
- Pfeiffer, V.; Lingner, J. Replication of telomeres and the regulation of telomerase. Cold Spring Harb. Perspect. Biol. 2013, 5, 010405. [Google Scholar] [CrossRef]
- Box, J.A.; Bunch, J.T.; Zappulla, D.C.; Glynn, E.F.; Baumann, P. A flexible template boundary element in the rna subunit of fission yeast telomerase. J. Biol. Chem. 2008, 283, 24224–24233. [Google Scholar] [CrossRef]
- Aksenova, A.Y.; Mirkin, S.M. At the beginning of the end and in the middle of the beginning: Structure and maintenance of telomeric DNA repeats and interstitial telomeric sequences. Genes 2019, 10, 118. [Google Scholar] [CrossRef]
- Fajkus, P.; Peska, V.; Zavodnik, M.; Fojtova, M.; Fulneckova, J.; Dobias, S.; Kilar, A.; Dvorackova, M.; Zachova, D.; Necasova, I.; et al. Telomerase rnas in land plants. Nucleic Acids Res. 2019, 47, 9842–9856. [Google Scholar] [CrossRef]
- Webb, C.J.; Wu, Y.; Zakian, V.A. DNA repair at telomeres: Keeping the ends intact. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, B.; Kumar, R. Tert promoter mutations in telomere biology. Mutat. Res. 2017, 771, 15–31. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.M. Telomeres and genomic evolution. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; de Lange, T. Mammalian telomeres resemble fragile sites and require trf1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef]
- Sfeir, A.; de Lange, T. Removal of shelterin reveals the telomere end-protection problem. Science 2012, 336, 593–597. [Google Scholar] [CrossRef]
- Nandakumar, J.; Cech, T.R. Finding the end: Recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013, 14, 69–82. [Google Scholar] [CrossRef]
- Doksani, Y.; Wu, J.Y.; de Lange, T.; Zhuang, X. Super-resolution fluorescence imaging of telomeres reveals trf2-dependent t-loop formation. Cell 2013, 155, 345–356. [Google Scholar] [CrossRef]
- Yang, S.Y.; Lejault, P.; Chevrier, S.; Boidot, R.; Robertson, A.G.; Wong, J.M.Y.; Monchaud, D. Transcriptome-wide identification of transient rna g-quadruplexes in human cells. Nat. Commun. 2018, 9, 4730. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]
- Lin, C.; Yang, D. Human telomeric g-quadruplex structures and g-quadruplex-interactive compounds. Methods Mol. Biol. 2017, 1587, 171–196. [Google Scholar] [PubMed]
- Zhang, Z.; Dai, J.; Veliath, E.; Jones, R.A.; Yang, D. Structure of a two-g-tetrad intramolecular g-quadruplex formed by a variant human telomeric sequence in k+ solution: Insights into the interconversion of human telomeric g-quadruplex structures. Nucleic Acids Res. 2010, 38, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Martinez, P.; Blasco, M.A. Heart-breaking telomeres. Circ. Res. 2018, 123, 787–802. [Google Scholar] [CrossRef]
- Frees, S.; Menendez, C.; Crum, M.; Bagga, P.S. Qgrs-conserve: A computational method for discovering evolutionarily conserved g-quadruplex motifs. Hum. Genom. 2014, 8, 8. [Google Scholar] [CrossRef]
- Tardat, M.; Dejardin, J. Telomere chromatin establishment and its maintenance during mammalian development. Chromosoma 2018, 127, 3–18. [Google Scholar] [CrossRef]
- Kar, A.; Willcox, S.; Griffith, J.D. Transcription of telomeric DNA leads to high levels of homologous recombination and t-loops. Nucleic Acids Res. 2016, 44, 9369–9380. [Google Scholar] [CrossRef][Green Version]
- Bandaria, J.N.; Qin, P.; Berk, V.; Chu, S.; Yildiz, A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell 2016, 164, 735–746. [Google Scholar] [CrossRef]
- Vaquero-Sedas, M.I.; Vega-Palas, M.A. On the chromatin structure of eukaryotic telomeres. Epigenetics 2011, 6, 1055–1058. [Google Scholar] [CrossRef]
- Chow, T.T.; Shi, X.; Wei, J.H.; Guan, J.; Stadler, G.; Huang, B.; Blackburn, E.H. Local enrichment of hp1alpha at telomeres alters their structure and regulation of telomere protection. Nat. Commun. 2018, 9, 3583. [Google Scholar] [CrossRef]
- Galati, A.; Micheli, E.; Cacchione, S. Chromatin structure in telomere dynamics. Front. Oncol. 2013, 3, 46. [Google Scholar] [CrossRef]
- Garcia-Cao, M.; O’Sullivan, R.; Peters, A.H.; Jenuwein, T.; Blasco, M.A. Epigenetic regulation of telomere length in mammalian cells by the suv39h1 and suv39h2 histone methyltransferases. Nat. Genet. 2004, 36, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Lachner, M.; O’Carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of histone h3 lysine 9 creates a binding site for hp1 proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. A ‘higher order’ of telomere regulation: Telomere heterochromatin and telomeric rnas. EMBO J. 2009, 28, 2323–2336. [Google Scholar] [CrossRef] [PubMed]
- Jezek, M.; Green, E.M. Histone modifications and the maintenance of telomere integrity. Cells 2019, 8, 199. [Google Scholar] [CrossRef]
- Tennen, R.I.; Bua, D.J.; Wright, W.E.; Chua, K.F. Sirt6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2011, 2, 433. [Google Scholar] [CrossRef] [PubMed]
- Martinez, P.; Blasco, M.A. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer 2011, 11, 161–176. [Google Scholar] [CrossRef]
- de Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef]
- Alexey Moskalev, A.M.V. Epigenetics of Aging and Longevity; Academic Press: Cambridge, MA, USA, 2017; Volume 4, p. 544. [Google Scholar]
- Schmutz, I.; de Lange, T. Shelterin. Curr. Biol. 2016, 26, R397–R399. [Google Scholar] [CrossRef]
- Erdel, F.; Kratz, K.; Willcox, S.; Griffith, J.D.; Greene, E.C.; de Lange, T. Telomere recognition and assembly mechanism of mammalian shelterin. Cell Rep. 2017, 18, 41–53. [Google Scholar] [CrossRef]
- Kim, H.; Li, F.; He, Q.; Deng, T.; Xu, J.; Jin, F.; Coarfa, C.; Putluri, N.; Liu, D.; Songyang, Z. Systematic analysis of human telomeric dysfunction using inducible telosome/shelterin crispr/cas9 knockout cells. Cell Discov. 2017, 3, 17034. [Google Scholar] [CrossRef]
- Lin, J.; Countryman, P.; Buncher, N.; Kaur, P.; Longjiang, E.; Zhang, Y.; Gibson, G.; You, C.; Watkins, S.C.; Piehler, J.; et al. Trf1 and trf2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014, 42, 2493–2504. [Google Scholar] [CrossRef] [PubMed]
- Van Ly, D.; Low, R.R.J.; Frolich, S.; Bartolec, T.K.; Kafer, G.R.; Pickett, H.A.; Gaus, K.; Cesare, A.J. Telomere loop dynamics in chromosome end protection. Mol. Cell 2018, 71, 510–525. [Google Scholar] [CrossRef] [PubMed]
- Sarek, G.; Kotsantis, P.; Ruis, P.; Van Ly, D.; Margalef, P.; Borel, V.; Zheng, X.F.; Flynn, H.R.; Snijders, A.P.; Chowdhury, D.; et al. Cdk phosphorylation of trf2 controls t-loop dynamics during the cell cycle. Nature 2019, 575, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Kalathiya, U.; Padariya, M.; Baginski, M. The structurally similar trfh domain of trf1 and trf2 dimers shows distinct behaviour towards tin2. Arch. Biochem. Biophys. 2018, 642, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Rai, R.; Huang, C.; Broton, C.; Long, J.; Xu, Y.; Xue, J.; Lei, M.; Chang, S.; Chen, Y. Structural and functional analyses of the mammalian tin2-tpp1-trf2 telomeric complex. Cell Res. 2017, 27, 1485–1502. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Liu, J.; Hu, X.; Yu, C.; Roskamp, K.; Sankaran, B.; Huang, L.; Komives, E.A.; Qiao, F. Structural basis for shelterin bridge assembly. Mol. Cell 2017, 68, 698–714. [Google Scholar] [CrossRef]
- Lei, M.; Podell, E.R.; Cech, T.R. Structure of human pot1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004, 11, 1223–1229. [Google Scholar] [CrossRef]
- Loayza, D.; Parsons, H.; Donigian, J.; Hoke, K.; de Lange, T. DNA binding features of human pot1: A nonamer 5′-tagggttag-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 2004, 279, 13241–13248. [Google Scholar] [CrossRef]
- Denchi, E.L.; de Lange, T. Protection of telomeres through independent control of atm and atr by trf2 and pot1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef]
- Chen, C.; Gu, P.; Wu, J.; Chen, X.; Niu, S.; Sun, H.; Wu, L.; Li, N.; Peng, J.; Shi, S.; et al. Structural insights into pot1-tpp1 interaction and pot1 c-terminal mutations in human cancer. Nat. Commun. 2017, 8, 14929. [Google Scholar] [CrossRef]
- Hockemeyer, D.; Palm, W.; Else, T.; Daniels, J.P.; Takai, K.K.; Ye, J.Z.; Keegan, C.E.; de Lange, T.; Hammer, G.D. Telomere protection by mammalian pot1 requires interaction with tpp1. Nat. Struct. Mol. Biol. 2007, 14, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Pike, A.M.; Strong, M.A.; Ouyang, J.P.T.; Greider, C.W. Tin2 functions with tpp1/pot1 to stimulate telomerase processivity. Mol. Cell. Biol. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Martinez, P.; Thanasoula, M.; Carlos, A.R.; Gomez-Lopez, G.; Tejera, A.M.; Schoeftner, S.; Dominguez, O.; Pisano, D.G.; Tarsounas, M.; Blasco, M.A. Mammalian rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 2010, 12, 768–780. [Google Scholar] [CrossRef]
- Rai, R.; Chen, Y.; Lei, M.; Chang, S. Trf2-rap1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat. Commun. 2016, 7, 10881. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.R.; Li, H.; Nicholls, C.; Liu, J.P. Telomere protein complexes and interactions with telomerase in telomere maintenance. Front. Biosci. 2011, 16, 187–207. [Google Scholar] [CrossRef]
- Arnoult, N.; Karlseder, J. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 2015, 22, 859–866. [Google Scholar] [CrossRef]
- Rice, C.; Skordalakes, E. Structure and function of the telomeric cst complex. Comput. Struct. Biotechnol. J. 2016, 14, 161–167. [Google Scholar] [CrossRef]
- Feng, X.; Hsu, S.J.; Kasbek, C.; Chaiken, M.; Price, C.M. Ctc1-mediated c-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res. 2017, 45, 4281–4293. [Google Scholar] [CrossRef]
- Chen, L.Y.; Redon, S.; Lingner, J. The human cst complex is a terminator of telomerase activity. Nature 2012, 488, 540–544. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, B.; Li, T.; Liu, R.; Xiao, Y.; Geng, X.; Li, G.; Liu, Q.; Price, C.M.; Liu, Y.; et al. Mammalian cst averts replication failure by preventing g-quadruplex accumulation. Nucleic Acids Res. 2019, 47, 5243–5259. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Wang, Y.; Diao, J.; Price, C.M. Dynamic DNA binding, junction recognition and g4 melting activity underlie the telomeric and genome-wide roles of human cst. Nucleic Acids Res. 2017, 45, 12311–12324. [Google Scholar] [CrossRef] [PubMed]
- Mirman, Z.; Lottersberger, F.; Takai, H.; Kibe, T.; Gong, Y.; Takai, K.; Bianchi, A.; Zimmermann, M.; Durocher, D.; de Lange, T. 53bp1-rif1-shieldin counteracts dsb resection through cst- and polalpha-dependent fill-in. Nature 2018, 560, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Chastain, M.; Zhou, Q.; Shiva, O.; Fadri-Moskwik, M.; Whitmore, L.; Jia, P.; Dai, X.; Huang, C.; Ye, P.; Chai, W. Human cst facilitates genome-wide rad51 recruitment to gc-rich repetitive sequences in response to replication stress. Cell Rep. 2016, 16, 1300–1314. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Jia, S.; Takasugi, T.; Smith, E.; Nandakumar, J.; Hendrickson, E.; Chang, S. Ctc1-stn1 coordinates g- and c-strand synthesis to regulate telomere length. Aging Cell 2018, 17, e12783. [Google Scholar] [CrossRef]
- Zimmermann, M.; Kibe, T.; Kabir, S.; de Lange, T. Trf1 negotiates ttaggg repeat-associated replication problems by recruiting the blm helicase and the tpp1/pot1 repressor of atr signaling. Genes Dev. 2014, 28, 2477–2491. [Google Scholar] [CrossRef]
- Opresko, P.L.; Otterlei, M.; Graakjaer, J.; Bruheim, P.; Dawut, L.; Kolvraa, S.; May, A.; Seidman, M.M.; Bohr, V.A. The werner syndrome helicase and exonuclease cooperate to resolve telomeric d loops in a manner regulated by trf1 and trf2. Mol. Cell 2004, 14, 763–774. [Google Scholar] [CrossRef]
- Higa, M.; Fujita, M.; Yoshida, K. DNA replication origins and fork progression at mammalian telomeres. Genes 2017, 8, 112. [Google Scholar] [CrossRef]
- Diotti, R.; Loayza, D. Shelterin complex and associated factors at human telomeres. Nucleus 2011, 2, 119–135. [Google Scholar] [CrossRef]
- Vannier, J.B.; Sandhu, S.; Petalcorin, M.I.; Wu, X.; Nabi, Z.; Ding, H.; Boulton, S.J. Rtel1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 2013, 342, 239–242. [Google Scholar] [CrossRef]
- Mefford, H.C.; Trask, B.J. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 2002, 3, 91–102. [Google Scholar] [CrossRef]
- Linardopoulou, E.V.; Williams, E.M.; Fan, Y.; Friedman, C.; Young, J.M.; Trask, B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 2005, 437, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Young, E.; Pastor, S.; Rajagopalan, R.; McCaffrey, J.; Sibert, J.; Mak, A.C.Y.; Kwok, P.Y.; Riethman, H.; Xiao, M. High-throughput single-molecule mapping links subtelomeric variants and long-range haplotypes with specific telomeres. Nucleic Acids Res. 2017, 45, e73. [Google Scholar] [CrossRef] [PubMed]
- Cubiles, M.D.; Barroso, S.; Vaquero-Sedas, M.I.; Enguix, A.; Aguilera, A.; Vega-Palas, M.A. Epigenetic features of human telomeres. Nucleic Acids Res. 2018, 46, 2347–2355. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, P.E.; Tobi, E.W.; Balog, J.; Schouten, S.G.; Kremer, D.; El Bouazzaoui, F.; Henneman, P.; Putter, H.; Eline Slagboom, P.; Heijmans, B.T.; et al. Chromatin remodeling of human subtelomeres and terra promoters upon cellular senescence: Commonalities and differences between chromosomes. Epigenetics 2013, 8, 512–521. [Google Scholar] [CrossRef][Green Version]
- Luke, B.; Lingner, J. Terra: Telomeric repeat-containing rna. EMBO J. 2009, 28, 2503–2510. [Google Scholar] [CrossRef]
- Rippe, K.; Luke, B. Terra and the state of the telomere. Nat. Struct. Mol. Biol. 2015, 22, 853–858. [Google Scholar] [CrossRef]
- Lopez de Silanes, I.; Grana, O.; De Bonis, M.L.; Dominguez, O.; Pisano, D.G.; Blasco, M.A. Identification of terra locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 2014, 5, 4723. [Google Scholar] [CrossRef]
- Montero, J.J.; Lopez de Silanes, I.; Grana, O.; Blasco, M.A. Telomeric rnas are essential to maintain telomeres. Nat. Commun. 2016, 7, 12534. [Google Scholar] [CrossRef]
- Montero, J.J.; López-Silanes, I.; Megías, D.; Fraga, M.F.; Castells-García, Á.; Blasco, M.A. Terra recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 2018, 9, 1548. [Google Scholar] [CrossRef]
- Mazzolini, R.; Gonzalez, N.; Garcia-Garijo, A.; Millanes-Romero, A.; Peiro, S.; Smith, S.; Garcia de Herreros, A.; Canudas, S. Snail1 transcription factor controls telomere transcription and integrity. Nucleic Acids Res. 2018, 46, 146–158. [Google Scholar] [CrossRef]
- Riethman, H. Human subtelomeric copy number variations. Cytogenet. Genome Res. 2008, 123, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Calderon Mdel, C.; Rey, M.D.; Cabrera, A.; Prieto, P. The subtelomeric region is important for chromosome recognition and pairing during meiosis. Sci. Rep. 2014, 4, 6488. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, S.; Nishihara, Y.; Kugou, K.; Ohta, K.; Kanoh, J. Subtelomeres constitute a safeguard for gene expression and chromosome homeostasis. Nucleic Acids Res. 2017, 45, 10333–10349. [Google Scholar] [CrossRef] [PubMed]
- van Emden, T.S.; Forn, M.; Forne, I.; Sarkadi, Z.; Capella, M.; Martin Caballero, L.; Fischer-Burkart, S.; Bronner, C.; Simonetta, M.; Toczyski, D.; et al. Shelterin and subtelomeric DNA sequences control nucleosome maintenance and genome stability. EMBO Rep. 2019, 20. [Google Scholar] [CrossRef]
- Daigaku, Y.; Keszthelyi, A.; Muller, C.A.; Miyabe, I.; Brooks, T.; Retkute, R.; Hubank, M.; Nieduszynski, C.A.; Carr, A.M. A global profile of replicative polymerase usage. Nat. Struct. Mol. Biol. 2015, 22, 192–198. [Google Scholar] [CrossRef]
- Pursell, Z.F.; Isoz, I.; Lundstrom, E.B.; Johansson, E.; Kunkel, T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science (N. Y.) 2007, 317, 127–130. [Google Scholar] [CrossRef]
- Duderstadt, K.E.; Geertsema, H.J.; Stratmann, S.A.; Punter, C.M.; Kulczyk, A.W.; Richardson, C.C.; van Oijen, A.M. Simultaneous real-time imaging of leading and lagging strand synthesis reveals the coordination dynamics of single replisomes. Mol. Cell 2016, 64, 1035–1047. [Google Scholar] [CrossRef]
- Turner, K.J.; Vasu, V.; Griffin, D.K. Telomere biology and human phenotype. Cells 2019, 8, 73. [Google Scholar] [CrossRef]
- Ohki, R.; Tsurimoto, T.; Ishikawa, F. In vitro reconstitution of the end replication problem. Mol. Cell. Biol. 2001, 21, 5753–5766. [Google Scholar] [CrossRef]
- Huffman, K.E.; Levene, S.D.; Tesmer, V.M.; Shay, J.W.; Wright, W.E. Telomere shortening is proportional to the size of the g-rich telomeric 3′-overhang. J. Biol. Chem. 2000, 275, 19719–19722. [Google Scholar] [CrossRef]
- Hayflick, L. The illusion of cell immortality. Br. J. Cancer 2000, 83, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Wright, W.E. Role of telomeres and telomerase in cancer. Semin. Cancer Biol. 2011, 21, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Martens, U.M.; Chavez, E.A.; Poon, S.S.; Schmoor, C.; Lansdorp, P.M. Accumulation of short telomeres in human fibroblasts prior to replicative senescence. Exp. Cell Res. 2000, 256, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K. How long does telomerase extend telomeres? Regulation of telomerase release and telomere length homeostasis. Curr. Genet. 2018, 64, 1177–1181. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A. Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 2005, 6, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, T.B.; Sá, A.; Lopes, J.M.; Sobrinho-Simões, M.; Soares, P.; Vinagre, J. Telomere maintenance mechanisms in cancer. Genes 2018, 9, 241. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Cech, T.R. Human telomerase: Biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015, 29, 1095–1105. [Google Scholar] [CrossRef]
- Wu, R.A.; Tam, J.; Collins, K. DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J. 2017, 36, 1908–1927. [Google Scholar] [CrossRef]
- Collins, K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006, 7, 484–494. [Google Scholar] [CrossRef]
- Podlevsky, J.D.; Chen, J.J. It all comes together at the ends: Telomerase structure, function, and biogenesis. Mutat. Res. 2012, 730, 3–11. [Google Scholar] [CrossRef]
- Gu, B.; Bessler, M.; Mason, P.J. Dyskerin, telomerase and the DNA damage response. Cell Cycle 2009, 8, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Arndt, G.M.; MacKenzie, K.L. New prospects for targeting telomerase beyond the telomere. Nat. Rev. Cancer 2016, 16, 508–524. [Google Scholar] [CrossRef] [PubMed]
- Egan, E.D.; Collins, K. An enhanced h/aca rnp assembly mechanism for human telomerase rna. Mol. Cell. Biol. 2012, 32, 2428–2439. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.L.; Farina, H.G.; Gomez, D.E. Telomerase regulation: A key to inhibition? (review). Int. J. Oncol. 2013, 43, 1351–1356. [Google Scholar] [CrossRef]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A human telomerase holoenzyme protein required for cajal body localization and telomere synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef]
- Hockemeyer, D.; Collins, K. Control of telomerase action at human telomeres. Nat. Struct. Mol. Biol. 2015, 22, 848–852. [Google Scholar] [CrossRef]
- Venteicher, A.S.; Meng, Z.; Mason, P.J.; Veenstra, T.D.; Artandi, S.E. Identification of atpases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 2008, 132, 945–957. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Calado, R.T.; Ly, H.; Kajigaya, S.; Baerlocher, G.M.; Chanock, S.J.; Lansdorp, P.M.; Young, N.S. Mutations in tert, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 2005, 352, 1413–1424. [Google Scholar] [CrossRef]
- Gomez, D.E.; Armando, R.G.; Farina, H.G.; Menna, P.L.; Cerrudo, C.S.; Ghiringhelli, P.D.; Alonso, D.F. Telomere structure and telomerase in health and disease (review). Int. J. Oncol. 2012, 41, 1561–1569. [Google Scholar] [CrossRef]
- Armstrong, C.A.; Tomita, K. Fundamental mechanisms of telomerase action in yeasts and mammals: Understanding telomeres and telomerase in cancer cells. Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Zaug, A.J.; Kufer, R.; Cech, T.R. Dynamics of human telomerase recruitment depend on template- telomere base pairing. Mol. Biol. Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W. Regulating telomere length from the inside out: The replication fork model. Genes Dev. 2016, 30, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Marcand, S.; Brevet, V.; Gilson, E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999, 18, 3509–3519. [Google Scholar] [CrossRef] [PubMed]
- Marcand, S.; Brevet, V.; Mann, C.; Gilson, E. Cell cycle restriction of telomere elongation. Curr. Biol. 2000, 10, 487–490. [Google Scholar] [CrossRef]
- Teixeira, M.T.; Arneric, M.; Sperisen, P.; Lingner, J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 2004, 117, 323–335. [Google Scholar] [CrossRef]
- Cristofari, G.; Lingner, J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006, 25, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Barthel, F.P.; Wei, W.; Tang, M.; Martinez-Ledesma, E.; Hu, X.; Amin, S.B.; Akdemir, K.C.; Seth, S.; Song, X.; Wang, Q.; et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017, 49, 349–357. [Google Scholar] [CrossRef]
- Cao, Y.; Bryan, T.M.; Reddel, R.R. Increased copy number of the tert and terc telomerase subunit genes in cancer cells. Cancer Sci. 2008, 99, 1092–1099. [Google Scholar] [CrossRef]
- Zhang, A.; Zheng, C.; Hou, M.; Lindvall, C.; Li, K.J.; Erlandsson, F.; Bjorkholm, M.; Gruber, A.; Blennow, E.; Xu, D. Deletion of the telomerase reverse transcriptase gene and haploinsufficiency of telomere maintenance in cri du chat syndrome. Am. J. Hum. Genet. 2003, 72, 940–948. [Google Scholar] [CrossRef]
- Fan, X.; Wang, Y.; Kratz, J.; Brat, D.J.; Robitaille, Y.; Moghrabi, A.; Perlman, E.J.; Dang, C.V.; Burger, P.C.; Eberhart, C.G. Htert gene amplification and increased mrna expression in central nervous system embryonal tumors. Am. J. Pathol. 2003, 162, 1763–1769. [Google Scholar] [CrossRef]
- Saretzki, G.; Petersen, S.; Petersen, I.; Kolble, K.; von Zglinicki, T. Htert gene dosage correlates with telomerase activity in human lung cancer cell lines. Cancer Lett. 2002, 176, 81–91. [Google Scholar] [CrossRef]
- Leão, R.; Apolónio, J.D.; Lee, D.; Figueiredo, A.; Tabori, U.; Castelo-Branco, P. Mechanisms of human telomerase reverse transcriptase (htert) regulation: Clinical impacts in cancer. J. Biomed. Sci. 2018, 25, 22. [Google Scholar] [CrossRef] [PubMed]
- Gay-Bellile, M.; Veronese, L.; Combes, P.; Eymard-Pierre, E.; Kwiatkowski, F.; Dauplat, M.M.; Cayre, A.; Privat, M.; Abrial, C.; Bignon, Y.J.; et al. Tert promoter status and gene copy number gains: Effect on tert expression and association with prognosis in breast cancer. Oncotarget 2017, 8, 77540–77551. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Kjellin, H.; Sofiadis, A.; Fotouhi, O.; Juhlin, C.C.; Backdahl, M.; Zedenius, J.; Xu, D.; Lehtio, J.; Larsson, C. Genetic and epigenetic background and protein expression profiles in relation to telomerase activation in medullary thyroid carcinoma. Oncotarget 2016, 7, 21332–21346. [Google Scholar] [CrossRef]
- Zhu, C.Q.; Cutz, J.C.; Liu, N.; Lau, D.; Shepherd, F.A.; Squire, J.A.; Tsao, M.S. Amplification of telomerase (htert) gene is a poor prognostic marker in non-small-cell lung cancer. Br. J. Cancer 2006, 94, 1452–1459. [Google Scholar] [CrossRef]
- Valentijn, L.J.; Koster, J.; Zwijnenburg, D.A.; Hasselt, N.E.; van Sluis, P.; Volckmann, R.; van Noesel, M.M.; George, R.E.; Tytgat, G.A.; Molenaar, J.J.; et al. Tert rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat. Genet. 2015, 47, 1411–1414. [Google Scholar] [CrossRef]
- Peifer, M.; Hertwig, F.; Roels, F.; Dreidax, D.; Gartlgruber, M.; Menon, R.; Kramer, A.; Roncaioli, J.L.; Sand, F.; Heuckmann, J.M.; et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 2015, 526, 700–704. [Google Scholar] [CrossRef]
- Heidenreich, B.; Kumar, R. Altered tert promoter and other genomic regulatory elements: Occurrence and impact. Int. J. Cancer 2017, 141, 867–876. [Google Scholar] [CrossRef]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. Tert promoter mutations in familial and sporadic melanoma. Science (N. Y.) 2013, 339, 959–961. [Google Scholar] [CrossRef]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent tert promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef]
- Andres-Lencina, J.J.; Rachakonda, S.; Garcia-Casado, Z.; Srinivas, N.; Skorokhod, A.; Requena, C.; Soriano, V.; Kumar, R.; Nagore, E. Tert promoter mutation subtypes and survival in stage i and ii melanoma patients. Int. J. Cancer 2019, 144, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Nagore, E.; Rachakonda, S.; Kumar, R. Tert promoter mutations in melanoma survival. Oncotarget 2019, 10, 1546–1548. [Google Scholar] [CrossRef] [PubMed]
- Nagore, E.; Heidenreich, B.; Rachakonda, S.; Garcia-Casado, Z.; Requena, C.; Soriano, V.; Frank, C.; Traves, V.; Quecedo, E.; Sanjuan-Gimenez, J.; et al. Tert promoter mutations in melanoma survival. Int. J. Cancer 2016, 139, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.J.; Rube, H.T.; Kreig, A.; Mancini, A.; Fouse, S.D.; Nagarajan, R.P.; Choi, S.; Hong, C.; He, D.; Pekmezci, M.; et al. Cancer. The transcription factor gabp selectively binds and activates the mutant tert promoter in cancer. Science 2015, 348, 1036–1039. [Google Scholar] [CrossRef] [PubMed]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of tert promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef]
- Huang, D.S.; Wang, Z.; He, X.J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Meng, Q.; Ye, Z.Y.; Wang, W.; Jiang, X.T.; et al. Recurrent tert promoter mutations identified in a large-scale study of multiple tumour types are associated with increased tert expression and telomerase activation. Eur. J. Cancer (Oxf. Engl. ) 2015, 51, 969–976. [Google Scholar] [CrossRef]
- Heidenreich, B.; Nagore, E.; Rachakonda, P.S.; Garcia-Casado, Z.; Requena, C.; Traves, V.; Becker, J.; Soufir, N.; Hemminki, K.; Kumar, R. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nat. Commun. 2014, 5, 3401. [Google Scholar] [CrossRef]
- Heidenreich, B.; Rachakonda, P.S.; Hosen, I.; Volz, F.; Hemminki, K.; Weyerbrock, A.; Kumar, R. Tert promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget 2015, 6, 10617–10633. [Google Scholar] [CrossRef]
- Hosen, I.; Rachakonda, P.S.; Heidenreich, B.; de Verdier, P.J.; Ryk, C.; Steineck, G.; Hemminki, K.; Kumar, R. Mutations in tert promoter and fgfr3 and telomere length in bladder cancer. Int. J. Cancer 2015, 137, 1621–1629. [Google Scholar] [CrossRef]
- Chiba, K.; Johnson, J.Z.; Vogan, J.M.; Wagner, T.; Boyle, J.M.; Hockemeyer, D. Cancer-associated tert promoter mutations abrogate telomerase silencing. Elife 2015, 4. [Google Scholar] [CrossRef]
- Rachakonda, S.; Kong, H.; Srinivas, N.; Garcia-Casado, Z.; Requena, C.; Fallah, M.; Heidenreich, B.; Planelles, D.; Traves, V.; Schadendorf, D.; et al. Telomere length, telomerase reverse transcriptase promoter mutations, and melanoma risk. Genes Chromosomes Cancer 2018, 57, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Rachakonda, P.S.; Hosen, I.; de Verdier, P.J.; Fallah, M.; Heidenreich, B.; Ryk, C.; Wiklund, N.P.; Steineck, G.; Schadendorf, D.; Hemminki, K.; et al. Tert promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl. Acad. Sci. USA 2013, 110, 17426–17431. [Google Scholar] [CrossRef] [PubMed]
- Chiba, K.; Lorbeer, F.K.; Shain, A.H.; McSwiggen, D.T.; Schruf, E.; Oh, A.; Ryu, J.; Darzacq, X.; Bastian, B.C.; Hockemeyer, D. Mutations in the promoter of the telomerase gene tert contribute to tumorigenesis by a two-step mechanism. Science 2017, 357, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Diplas, B.H.; He, X.; Brosnan-Cashman, J.A.; Liu, H.; Chen, L.H.; Wang, Z.; Moure, C.J.; Killela, P.J.; Loriaux, D.B.; Lipp, E.S.; et al. The genomic landscape of tert promoter wildtype-idh wildtype glioblastoma. Nat. Commun. 2018, 9, 2087. [Google Scholar] [CrossRef]
- Stern, J.L.; Paucek, R.D.; Huang, F.W.; Ghandi, M.; Nwumeh, R.; Costello, J.C.; Cech, T.R. Allele-specific DNA methylation and its interplay with repressive histone marks at promoter-mutant tert genes. Cell Rep. 2017, 21, 3700–3707. [Google Scholar] [CrossRef]
- Lee, D.D.; Leao, R.; Komosa, M.; Gallo, M.; Zhang, C.H.; Lipman, T.; Remke, M.; Heidari, A.; Nunes, N.M.; Apolonio, J.D.; et al. DNA hypermethylation within tert promoter upregulates tert expression in cancer. J. Clin. Investig. 2019, 129, 223–229. [Google Scholar] [CrossRef]
- Kyo, S.; Takakura, M.; Fujiwara, T.; Inoue, M. Understanding and exploiting htert promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008, 99, 1528–1538. [Google Scholar] [CrossRef]
- Castelo-Branco, P.; Choufani, S.; Mack, S.; Gallagher, D.; Zhang, C.; Lipman, T.; Zhukova, N.; Walker, E.J.; Martin, D.; Merino, D.; et al. Methylation of the tert promoter and risk stratification of childhood brain tumours: An integrative genomic and molecular study. Lancet. Oncol. 2013, 14, 534–542. [Google Scholar] [CrossRef]
- Castelo-Branco, P.; Leao, R.; Lipman, T.; Campbell, B.; Lee, D.; Price, A.; Zhang, C.; Heidari, A.; Stephens, D.; Boerno, S.; et al. A cancer specific hypermethylation signature of the tert promoter predicts biochemical relapse in prostate cancer: A retrospective cohort study. Oncotarget 2016, 7, 57726–57736. [Google Scholar] [CrossRef]
- Fan, Y.; Lee, S.; Wu, G.; Easton, J.; Yergeau, D.; Dummer, R.; Vogel, P.; Kirkwood, J.M.; Barnhill, R.L.; Pappo, A.; et al. Telomerase expression by aberrant methylation of the tert promoter in melanoma arising in giant congenital nevi. J. Invest. Derm. 2016, 136, 339–342. [Google Scholar] [CrossRef]
- Lindsey, J.C.; Schwalbe, E.C.; Potluri, S.; Bailey, S.; Williamson, D.; Clifford, S.C. Tert promoter mutation and aberrant hypermethylation are associated with elevated expression in medulloblastoma and characterise the majority of non-infant shh subgroup tumours. Acta Neuropathol. 2014, 127, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Stern, J.L.; Theodorescu, D.; Vogelstein, B.; Papadopoulos, N.; Cech, T.R. Mutation of the tert promoter, switch to active chromatin, and monoallelic tert expression in multiple cancers. Genes Dev. 2015, 29, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, S.; Stohr, B.A. The role of telomere biology in cancer. Annu. Rev. Pathol. 2013, 8, 49–78. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Gupta, J.; Bacchetti, S.; Reddel, R.R. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995, 14, 4240–4248. [Google Scholar] [CrossRef]
- Draskovic, I.; Arnoult, N.; Steiner, V.; Bacchetti, S.; Lomonte, P.; Londono-Vallejo, A. Probing pml body function in alt cells reveals spatiotemporal requirements for telomere recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 15726–15731. [Google Scholar] [CrossRef]
- Fogli, A.; Demattei, M.V.; Corset, L.; Vaurs-Barriere, C.; Chautard, E.; Biau, J.; Kemeny, J.L.; Godfraind, C.; Pereira, B.; Khalil, T.; et al. Detection of the alternative lengthening of telomeres pathway in malignant gliomas for improved molecular diagnosis. J. Neuro-Oncol. 2017, 135, 381–390. [Google Scholar] [CrossRef]
- Lau, L.M.; Dagg, R.A.; Henson, J.D.; Au, A.Y.; Royds, J.A.; Reddel, R.R. Detection of alternative lengthening of telomeres by telomere quantitative pcr. Nucleic Acids Res. 2013, 41, e34. [Google Scholar] [CrossRef]
- Yost, K.E.; Clatterbuck Soper, S.F.; Walker, R.L.; Pineda, M.A.; Zhu, Y.J.; Ester, C.D.; Showman, S.; Roschke, A.V.; Waterfall, J.J.; Meltzer, P.S. Rapid and reversible suppression of alt by daxx in osteosarcoma cells. Sci. Rep. 2019, 9, 4544. [Google Scholar] [CrossRef]
- Dyer, M.A.; Qadeer, Z.A.; Valle-Garcia, D.; Bernstein, E. Atrx and daxx: Mechanisms and mutations. Cold Spring Harb. Perspect. Med. 2017, 7. [Google Scholar] [CrossRef]
- Mason-Osann, E.; Dai, A.; Floro, J.; Lock, Y.J.; Reiss, M.; Gali, H.; Matschulat, A.; Labadorf, A.; Flynn, R.L. Identification of a novel gene fusion in alt positive osteosarcoma. Oncotarget 2018, 9, 32868–32880. [Google Scholar] [CrossRef]
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suva, M.L.; Benes, C.H.; et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to atr inhibitors. Science 2015, 347, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.L.; Westra, W.H.; et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011, 179, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Hjelmborg, J.B.; Dalgard, C.; Moller, S.; Steenstrup, T.; Kimura, M.; Christensen, K.; Kyvik, K.O.; Aviv, A. The heritability of leucocyte telomere length dynamics. J. Med. Genet. 2015, 52, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Delgado, D.A.; Zhang, C.; Gleason, K.; Demanelis, K.; Chen, L.S.; Gao, J.; Roy, S.; Shinkle, J.; Sabarinathan, M.; Argos, M.; et al. The contribution of parent-to-offspring transmission of telomeres to the heritability of telomere length in humans. Hum. Genet. 2019, 138, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Mayhew, A.J.; Meyre, D. Assessing the heritability of complex traits in humans: Methodological challenges and opportunities. Curr. Genom. 2017, 18, 332–340. [Google Scholar] [CrossRef]
- Mirabello, L.; Yu, K.; Kraft, P.; De Vivo, I.; Hunter, D.J.; Prescott, J.; Wong, J.Y.; Chatterjee, N.; Hayes, R.B.; Savage, S.A. The association of telomere length and genetic variation in telomere biology genes. Hum. Mutat. 2010, 31, 1050–1058. [Google Scholar] [CrossRef]
- De Meyer, T.; Vandepitte, K.; Denil, S.; De Buyzere, M.L.; Rietzschel, E.R.; Bekaert, S. A non-genetic, epigenetic-like mechanism of telomere length inheritance? Eur. J. Hum. Genet. EJHG 2014, 22, 10–11. [Google Scholar] [CrossRef]
- Dugdale, H.L.; Richardson, D.S. Heritability of telomere variation: It is all about the environment! Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef]
- Broer, L.; Codd, V.; Nyholt, D.R.; Deelen, J.; Mangino, M.; Willemsen, G.; Albrecht, E.; Amin, N.; Beekman, M.; de Geus, E.J.; et al. Meta-analysis of telomere length in 19,713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur. J. Hum. Genet. EJHG 2013, 21, 1163–1168. [Google Scholar] [CrossRef]
- Aviv, A. The mitochondrial genome, paternal age and telomere length in humans. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef]
- Eisenberg, D.T. Inconsistent inheritance of telomere length (tl): Is offspring tl more strongly correlated with maternal or paternal tl? Eur. J. Hum. Genet. EJHG 2014, 22, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Njajou, O.T.; Cawthon, R.M.; Damcott, C.M.; Wu, S.H.; Ott, S.; Garant, M.J.; Blackburn, E.H.; Mitchell, B.D.; Shuldiner, A.R.; Hsueh, W.C. Telomere length is paternally inherited and is associated with parental lifespan. Proc. Natl. Acad. Sci. USA 2007, 104, 12135–12139. [Google Scholar] [CrossRef] [PubMed]
- Wulaningsih, W.; Hardy, R.; Wong, A.; Kuh, D. Parental age and offspring leukocyte telomere length and attrition in midlife: Evidence from the 1946 british birth cohort. Exp. Gerontol. 2018, 112, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Daniali, L.; Benetos, A.; Susser, E.; Kark, J.D.; Labat, C.; Kimura, M.; Desai, K.; Granick, M.; Aviv, A. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat. Commun. 2013, 4, 1597. [Google Scholar] [CrossRef]
- Benetos, A.; Kark, J.D.; Susser, E.; Kimura, M.; Sinnreich, R.; Chen, W.; Steenstrup, T.; Christensen, K.; Herbig, U.; von Bornemann Hjelmborg, J.; et al. Tracking and fixed ranking of leukocyte telomere length across the adult life course. Aging Cell 2013, 12, 615–621. [Google Scholar] [CrossRef]
- Pooley, K.A.; Tyrer, J.; Shah, M.; Driver, K.E.; Leyland, J.; Brown, J.; Audley, T.; McGuffog, L.; Ponder, B.A.; Pharoah, P.D.; et al. No association between tert-clptm1l single nucleotide polymorphism rs401681 and mean telomere length or cancer risk. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1862–1865. [Google Scholar] [CrossRef]
- Pooley, K.A.; Bojesen, S.E.; Weischer, M.; Nielsen, S.F.; Thompson, D.; Amin Al Olama, A.; Michailidou, K.; Tyrer, J.P.; Benlloch, S.; Brown, J.; et al. A genome-wide association scan (gwas) for mean telomere length within the cogs project: Identified loci show little association with hormone-related cancer risk. Hum. Mol. Genet. 2013, 22, 5056–5064. [Google Scholar] [CrossRef]
- Codd, V.; Mangino, M.; van der Harst, P.; Braund, P.S.; Kaiser, M.; Beveridge, A.J.; Rafelt, S.; Moore, J.; Nelson, C.; Soranzo, N.; et al. Common variants near terc are associated with mean telomere length. Nat. Genet. 2010, 42, 197–199. [Google Scholar] [CrossRef]
- Codd, V.; Nelson, C.P.; Albrecht, E.; Mangino, M.; Deelen, J.; Buxton, J.L.; Hottenga, J.J.; Fischer, K.; Esko, T.; Surakka, I.; et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 2013, 45, 422–427. [Google Scholar] [CrossRef]
- Levy, D.; Neuhausen, S.L.; Hunt, S.C.; Kimura, M.; Hwang, S.J.; Chen, W.; Bis, J.C.; Fitzpatrick, A.L.; Smith, E.; Johnson, A.D.; et al. Genome-wide association identifies obfc1 as a locus involved in human leukocyte telomere biology. Proc. Natl. Acad. Sci. USA 2010, 107, 9293–9298. [Google Scholar] [CrossRef]
- Mangino, M.; Hwang, S.J.; Spector, T.D.; Hunt, S.C.; Kimura, M.; Fitzpatrick, A.L.; Christiansen, L.; Petersen, I.; Elbers, C.C.; Harris, T.; et al. Genome-wide meta-analysis points to ctc1 and znf676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 2012, 21, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Chen, M.; Shete, S.; Amos, C.I.; Kamat, A.; Ye, Y.; Lin, J.; Dinney, C.P.; Wu, X. A genome-wide association study identifies a locus on chromosome 14q21 as a predictor of leukocyte telomere length and as a marker of susceptibility for bladder cancer. Cancer Prev. Res. 2011, 4, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Cheng, R.; Honig, L.S.; Feitosa, M.; Kammerer, C.M.; Kang, M.S.; Schupf, N.; Lin, S.J.; Sanders, J.L.; Bae, H.; et al. Genome wide association and linkage analyses identified three loci-4q25, 17q23.2, and 10q11.21-associated with variation in leukocyte telomere length: The long life family study. Front. Genet. 2013, 4, 310. [Google Scholar] [CrossRef] [PubMed]
- Mangino, M.; Brouilette, S.; Braund, P.; Tirmizi, N.; Vasa-Nicotera, M.; Thompson, J.R.; Samani, N.J. A regulatory snp of the bicd1 gene contributes to telomere length variation in humans. Hum. Mol. Genet. 2008, 17, 2518–2523. [Google Scholar] [CrossRef]
- Prescott, J.; Kraft, P.; Chasman, D.I.; Savage, S.A.; Mirabello, L.; Berndt, S.I.; Weissfeld, J.L.; Han, J.; Hayes, R.B.; Chanock, S.J.; et al. Genome-wide association study of relative telomere length. PLoS ONE 2011, 6, e19635. [Google Scholar] [CrossRef]
- Vasa-Nicotera, M.; Brouilette, S.; Mangino, M.; Thompson, J.R.; Braund, P.; Clemitson, J.R.; Mason, A.; Bodycote, C.L.; Raleigh, S.M.; Louis, E.; et al. Mapping of a major locus that determines telomere length in humans. Am. J. Hum. Genet. 2005, 76, 147–151. [Google Scholar] [CrossRef]
- Andrew, T.; Aviv, A.; Falchi, M.; Surdulescu, G.L.; Gardner, J.P.; Lu, X.; Kimura, M.; Kato, B.S.; Valdes, A.M.; Spector, T.D. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am. J. Hum. Genet. 2006, 78, 480–486. [Google Scholar] [CrossRef]
- Mangino, M.; Richards, J.B.; Soranzo, N.; Zhai, G.; Aviv, A.; Valdes, A.M.; Samani, N.J.; Deloukas, P.; Spector, T.D. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J. Med. Genet. 2009, 46, 451–454. [Google Scholar] [CrossRef]
- Saxena, R.; Bjonnes, A.; Prescott, J.; Dib, P.; Natt, P.; Lane, J.; Lerner, M.; Cooper, J.A.; Ye, Y.; Li, K.W.; et al. Genome-wide association study identifies variants in casein kinase ii (csnk2a2) to be associated with leukocyte telomere length in a punjabi sikh diabetic cohort. Circ. Cardiovasc. Genet. 2014, 7, 287–295. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, L.; Li, Z.; Zhou, D.; Liu, W.; Shen, Q.; Wu, Y.; Zhang, D.; Hu, X.; Wang, T.; et al. A genome-wide association study identifies a locus on tert for mean telomere length in han chinese. PLoS ONE 2014, 9, e85043. [Google Scholar] [CrossRef]
- Zhang, C.; Kibriya, M.G.; Jasmine, F.; Roy, S.; Gao, J.; Sabarinathan, M.; Shinkle, J.; Delgado, D.; Ahmed, A.; Islam, T.; et al. A study of telomere length, arsenic exposure, and arsenic toxicity in a bangladeshi cohort. Environ. Res. 2018, 164, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.M.; Whitehead, T.P.; de Smith, A.J.; Smirnov, I.V.; Park, M.; Endicott, A.A.; Francis, S.S.; Codd, V.; Group, E.C.T.; Samani, N.J.; et al. Common genetic variants associated with telomere length confer risk for neuroblastoma and other childhood cancers. Carcinogenesis 2016, 37, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Bojesen, S.E.; Pooley, K.A.; Johnatty, S.E.; Beesley, J.; Michailidou, K.; Tyrer, J.P.; Edwards, S.L.; Pickett, H.A.; Shen, H.C.; Smart, C.E.; et al. Multiple independent variants at the tert locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013, 45, 371–384, 384e371-372. [Google Scholar] [CrossRef] [PubMed]
- Mangino, M.; Christiansen, L.; Stone, R.; Hunt, S.C.; Horvath, K.; Eisenberg, D.T.; Kimura, M.; Petersen, I.; Kark, J.D.; Herbig, U.; et al. Dcaf4, a novel gene associated with leucocyte telomere length. J. Med. Genet. 2015, 52, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Hsu, S.J.; Bhattacharjee, A.; Wang, Y.; Diao, J.; Price, C.M. Ctc1-stn1 terminates telomerase while stn1-ten1 enables c-strand synthesis during telomere replication in colon cancer cells. Nat. Commun. 2018, 9, 2827. [Google Scholar] [CrossRef]
- Gu, P.; Min, J.N.; Wang, Y.; Huang, C.; Peng, T.; Chai, W.; Chang, S. Ctc1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012, 31, 2309–2321. [Google Scholar] [CrossRef]
- Gu, P.; Chang, S. Functional characterization of human ctc1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease coats plus. Aging Cell 2013, 12, 1100–1109. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, Z.; Stong, N.; Plasschaert, R.; Moczan, A.; Chen, H.S.; Hu, S.; Wikramasinghe, P.; Davuluri, R.V.; Bartolomei, M.S.; et al. A role for ctcf and cohesin in subtelomere chromatin organization, terra transcription, and telomere end protection. EMBO J. 2012, 31, 4165–4178. [Google Scholar] [CrossRef]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The emerging roles of terra in telomere maintenance and genome stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef]
- Kim, W.; Lee, S.; Son, Y.; Ko, C.; Ryu, W.S. Ddb1 stimulates viral transcription of hepatitis b virus via hbx-independent mechanisms. J. Virol. 2016, 90, 9644–9653. [Google Scholar] [CrossRef]
- Alonso, M.M.; Fueyo, J.; Yung, W.K.; Gomez-Manzano, C. E2f1 and telomerase: Alliance in the dark side. Cell Cycle 2006, 5, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, Z.; Wang, Y.; Stinchcombe, T.E.; Owzar, K.; Han, Y.; Hung, R.J.; Brhane, Y.; McLaughlin, J.; Brennan, P.; et al. Functional variants in dcaf4 associated with lung cancer risk in european populations. Carcinogenesis 2017, 38, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Rog, O.; Smolikov, S.; Krauskopf, A.; Kupiec, M. The yeast vps genes affect telomere length regulation. Curr. Genet. 2005, 47, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Ly, L.L.; Yoshida, H.; Yamaguchi, M. Nuclear transcription factor y and its roles in cellular processes related to human disease. Am. J. Cancer Res. 2013, 3, 339–346. [Google Scholar]
- Kote-Jarai, Z.; Saunders, E.J.; Leongamornlert, D.A.; Tymrakiewicz, M.; Dadaev, T.; Jugurnauth-Little, S.; Ross-Adams, H.; Al Olama, A.A.; Benlloch, S.; Halim, S.; et al. Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with tert expression. Hum. Mol. Genet. 2013, 22, 2520–2528. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, B.; Zhang, M.; Parikh, H.; Jia, J.; Chung, C.C.; Sampson, J.N.; Hoskins, J.W.; Hutchinson, A.; Burdette, L.; et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the tert-clptm1l region on chromosome 5p15.33. Hum. Mol. Genet. 2014, 23, 6616–6633. [Google Scholar] [CrossRef]
- Fang, J.; Jia, J.; Makowski, M.; Xu, M.; Wang, Z.; Zhang, T.; Hoskins, J.W.; Choi, J.; Han, Y.; Zhang, M.; et al. Functional characterization of a multi-cancer risk locus on chr5p15.33 reveals regulation of tert by znf148. Nat. Commun. 2017, 8, 15034. [Google Scholar] [CrossRef]
- Jones, A.M.; Beggs, A.D.; Carvajal-Carmona, L.; Farrington, S.; Tenesa, A.; Walker, M.; Howarth, K.; Ballereau, S.; Hodgson, S.V.; Zauber, A.; et al. Terc polymorphisms are associated both with susceptibility to colorectal cancer and with longer telomeres. Gut 2012, 61, 248–254. [Google Scholar] [CrossRef]
- Starkweather, A.R.; Alhaeeri, A.A.; Montpetit, A.; Brumelle, J.; Filler, K.; Montpetit, M.; Mohanraj, L.; Lyon, D.E.; Jackson-Cook, C.K. An integrative review of factors associated with telomere length and implications for biobehavioral research. Nurs. Res. 2014, 63, 36–50. [Google Scholar] [CrossRef]
- Patel, C.J.; Manrai, A.K.; Corona, E.; Kohane, I.S. Systematic correlation of environmental exposure and physiological and self-reported behaviour factors with leukocyte telomere length. Int. J. Epidemiol. 2017, 46, 44–56. [Google Scholar] [CrossRef]
- Cassidy, A.; De Vivo, I.; Liu, Y.; Han, J.; Prescott, J.; Hunter, D.J.; Rimm, E.B. Associations between diet, lifestyle factors, and telomere length in women. Am. J. Clin. Nutr. 2010, 91, 1273–1280. [Google Scholar] [CrossRef]
- Reichert, S.; Stier, A. Does oxidative stress shorten telomeres in vivo? A review. Biol. Lett. 2017, 13. [Google Scholar] [CrossRef]
- Fouquerel, E.; Lormand, J.; Bose, A.; Lee, H.T.; Kim, G.S.; Li, J.; Sobol, R.W.; Freudenthal, B.D.; Myong, S.; Opresko, P.L. Oxidative guanine base damage regulates human telomerase activity. Nat. Struct. Mol. Biol. 2016, 23, 1092–1100. [Google Scholar] [CrossRef]
- Barnes, R.P.; Fouquerel, E.; Opresko, P.L. The impact of oxidative DNA damage and stress on telomere homeostasis. Mech. Ageing Dev. 2019, 177, 37–45. [Google Scholar] [CrossRef]
- Victorelli, S.; Passos, J.F. Telomeres and cell senescence -size matters not. EBioMedicine 2017, 21, 14–20. [Google Scholar] [CrossRef]
- Mannan, T.; Ahmed, S.; Akhtar, E.; Ahsan, K.B.; Haq, A.; Kippler, M.; Vahter, M.; Raqib, R. Associations of arsenic exposure with telomere length and naive t cells in childhood-a birth cohort study. Toxicol. Sci. Off. J. Soc. Toxicol. 2018, 164, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Borghini, A.; Faita, F.; Mercuri, A.; Minichilli, F.; Bustaffa, E.; Bianchi, F.; Andreassi, M.G. Arsenic exposure, genetic susceptibility and leukocyte telomere length in an italian young adult population. Mutagenesis 2016, 31, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Kesaniemi, J.; Lavrinienko, A.; Tukalenko, E.; Boratynski, Z.; Kivisaari, K.; Mappes, T.; Milinevsky, G.; Moller, A.P.; Mousseau, T.A.; Watts, P.C. Exposure to environmental radionuclides associates with tissue-specific impacts on telomerase expression and telomere length. Sci. Rep. 2019, 9, 850. [Google Scholar] [CrossRef]
- He, L.; Chen, Z.; Dai, B.; Li, G.; Zhu, G. Low-level lead exposure and cardiovascular disease: The roles of telomere shortening and lipid disturbance. J. Toxicol. Sci. 2018, 43, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Astuti, Y.; Wardhana, A.; Watkins, J.; Wulaningsih, W.; Network, P.R. Cigarette smoking and telomere length: A systematic review of 84 studies and meta-analysis. Environ. Res. 2017, 158, 480–489. [Google Scholar] [CrossRef]
- Carroll, J.E.; Diez Roux, A.V.; Fitzpatrick, A.L.; Seeman, T. Low social support is associated with shorter leukocyte telomere length in late life: Multi-ethnic study of atherosclerosis. Psychosom. Med. 2013, 75, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Adler, N.; Pantell, M.S.; O’Donovan, A.; Blackburn, E.; Cawthon, R.; Koster, A.; Opresko, P.; Newman, A.; Harris, T.B.; Epel, E. Educational attainment and late life telomere length in the health, aging and body composition study. Brain Behav. Immun. 2013, 27, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Alexeeff, S.E.; Schaefer, C.A.; Kvale, M.N.; Shan, J.; Blackburn, E.H.; Risch, N.; Ranatunga, D.K.; Jorgenson, E.; Hoffmann, T.J.; Sakoda, L.C.; et al. Telomere length and socioeconomic status at neighborhood and individual levels among 80,000 adults in the genetic epidemiology research on adult health and aging cohort. Environ. Epidemiol. 2019, 3, e049. [Google Scholar] [CrossRef]
- Caini, S.; Raimondi, S.; Johansson, H.; De Giorgi, V.; Zanna, I.; Palli, D.; Gandini, S. Telomere length and the risk of cutaneous melanoma and non-melanoma skin cancer: A review of the literature and meta-analysis. J. Dermatol. Sci. 2015, 80, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Haycock, P.C.; Burgess, S.; Nounu, A.; Zheng, J.; Okoli, G.N.; Bowden, J.; Wade, K.H.; Timpson, N.J.; Evans, D.M.; Willeit, P.; et al. Association between telomere length and risk of cancer and non-neoplastic diseases: A mendelian randomization study. JAMA Oncol. 2017, 3, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Pooley, K.A.; Sandhu, M.S.; Tyrer, J.; Shah, M.; Driver, K.E.; Luben, R.N.; Bingham, S.A.; Ponder, B.A.; Pharoah, P.D.; Khaw, K.T.; et al. Telomere length in prospective and retrospective cancer case-control studies. Cancer Res. 2010, 70, 3170–3176. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, Y.; Qu, F.; He, S.; Huang, X.; Jiang, H.; Jin, T.; Wan, S.; Xing, J. Association between leukocyte telomere length and glioma risk: A case-control study. Neuro Oncol. 2014, 16, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Q.; Zhu, W.; Liu, T.; Xie, S.H.; Zhong, L.X.; Cai, Y.Y.; Li, X.N.; Liang, M.; Chen, W.; et al. The association of telomere length in peripheral blood cells with cancer risk: A systematic review and meta-analysis of prospective studies. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Rode, L.; Nordestgaard, B.G.; Bojesen, S.E. Long telomeres and cancer risk among 95 568 individuals from the general population. Int. J. Epidemiol. 2016, 45, 1634–1643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Doherty, J.A.; Burgess, S.; Hung, R.J.; Lindstrom, S.; Kraft, P.; Gong, J.; Amos, C.I.; Sellers, T.A.; Monteiro, A.N.; et al. Genetic determinants of telomere length and risk of common cancers: A mendelian randomization study. Hum. Mol. Genet. 2015, 24, 5356–5366. [Google Scholar] [CrossRef]
- McNally, E.J.; Luncsford, P.J.; Armanios, M. Long telomeres and cancer risk: The price of cellular immortality. J. Clin. Investig. 2019, 130, 3474–3481. [Google Scholar] [CrossRef] [PubMed]
- Aviv, A.; Anderson, J.J.; Shay, J.W. Mutations, cancer and the telomere length paradox. Trends Cancer 2017, 3, 253–258. [Google Scholar] [CrossRef]
- Hamada, T.; Yuan, C.; Bao, Y.; Zhang, M.; Khalaf, N.; Babic, A.; Morales-Oyarvide, V.; Cochrane, B.B.; Gaziano, J.M.; Giovannucci, E.L.; et al. Prediagnostic leukocyte telomere length and pancreatic cancer survival. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1868–1875. [Google Scholar] [CrossRef]
- Kachuri, L.; Helby, J.; Bojesen, S.E.; Christiani, D.C.; Su, L.; Wu, X.; Tardon, A.; Fernandez-Tardon, G.; Field, J.K.; Davies, M.P.; et al. Investigation of leukocyte telomere length and genetic variants in chromosome 5p15.33 as prognostic markers in lung cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Weischer, M.; Nordestgaard, B.G.; Cawthon, R.M.; Freiberg, J.J.; Tybjaerg-Hansen, A.; Bojesen, S.E. Short telomere length, cancer survival, and cancer risk in 47102 individuals. J. Natl. Cancer Inst. 2013, 105, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Duggan, C.; Risques, R.; Alfano, C.; Prunkard, D.; Imayama, I.; Holte, S.; Baumgartner, K.; Baumgartner, R.; Bernstein, L.; Ballard-Barbash, R.; et al. Change in peripheral blood leukocyte telomere length and mortality in breast cancer survivors. J. Natl. Cancer Inst. 2014, 106, 35. [Google Scholar] [CrossRef]
- Rachakonda, S.; Srinivas, N.; Mahmoudpour, S.H.; Garcia-Casado, Z.; Requena, C.; Traves, V.; Soriano, V.; Cardelli, M.; Pjanova, D.; Molven, A.; et al. Telomere length and survival in primary cutaneous melanoma patients. Sci. Rep. 2018, 8, 10947. [Google Scholar] [CrossRef]
- Holohan, B.; Wright, W.E.; Shay, J.W. Cell biology of disease: Telomeropathies: An emerging spectrum disorder. J. Cell Biol. 2014, 205, 289–299. [Google Scholar] [CrossRef]
- Armanios, M.; Blackburn, E.H. The telomere syndromes. Nat. Rev. Genet. 2012, 13, 693–704. [Google Scholar] [CrossRef]
- Sarek, G.; Marzec, P.; Margalef, P.; Boulton, S.J. Molecular basis of telomere dysfunction in human genetic diseases. Nat. Struct. Mol. Biol. 2015, 22, 867–874. [Google Scholar] [CrossRef]
- Wentzensen, I.M.; Mirabello, L.; Pfeiffer, R.M.; Savage, S.A. The association of telomere length and cancer: A meta-analysis. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Seow, W.J.; Cawthon, R.M.; Purdue, M.P.; Hu, W.; Gao, Y.T.; Huang, W.Y.; Weinstein, S.J.; Ji, B.T.; Virtamo, J.; Hosgood, H.D.; et al. Telomere length in white blood cell DNA and lung cancer: A pooled analysis of three prospective cohorts. Cancer Res. 2014, 74, 4090–4098. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Qureshi, A.A.; Prescott, J.; Guo, Q.; Ye, L.; Hunter, D.J.; De Vivo, I. A prospective study of telomere length and the risk of skin cancer. J. Invest. Derm. 2009, 129, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Prescott, J.; McGrath, M.; Lee, I.M.; Buring, J.E.; De Vivo, I. Telomere length and genetic analyses in population-based studies of endometrial cancer risk. Cancer 2010, 116, 4275–4282. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Du, M.; De Vivo, I.; Manson, J.E.; Liu, S.; McTiernan, A.; Curb, J.D.; Lessin, L.S.; Bonner, M.R.; Guo, Q.; et al. Shorter telomeres associate with a reduced risk of melanoma development. Cancer Res. 2011, 71, 6758–6763. [Google Scholar] [CrossRef] [PubMed]
- Iles, M.M.; Bishop, D.T.; Taylor, J.C.; Hayward, N.K.; Brossard, M.; Cust, A.E.; Dunning, A.M.; Lee, J.E.; Moses, E.K.; Akslen, L.A.; et al. The effect on melanoma risk of genes previously associated with telomere length. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Campa, D.; Matarazzi, M.; Greenhalf, W.; Bijlsma, M.; Saum, K.U.; Pasquali, C.; van Laarhoven, H.; Szentesi, A.; Federici, F.; Vodicka, P.; et al. Genetic determinants of telomere length and risk of pancreatic cancer: A pandora study. Int. J. Cancer 2019, 144, 1275–1283. [Google Scholar] [CrossRef]
- Srinivas, N.; Rachakonda, S.; Hielscher, T.; Calderazzo, S.; Rudnai, P.; Gurzau, E.; Koppova, K.; Fletcher, T.; Kumar, R. Telomere length, arsenic exposure and risk of basal cell carcinoma of skin. Carcinogenesis 2019, 40, 715–723. [Google Scholar] [CrossRef]
- Ivancich, M.; Schrank, Z.; Wojdyla, L.; Leviskas, B.; Kuckovic, A.; Sanjali, A.; Puri, N. Treating cancer by targeting telomeres and telomerase. Antioxidants 2017, 6, 15. [Google Scholar] [CrossRef]
- Gorenjak, V.; Akbar, S.; Stathopoulou, M.G.; Visvikis-Siest, S. The future of telomere length in personalized medicine. Front. Biosci. 2018, 23, 1628–1654. [Google Scholar]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent progress of targeted g-quadruplex-preferred ligands toward cancer therapy. Molecules (Basel Switz.) 2019, 24, 429. [Google Scholar] [CrossRef] [PubMed]
- Ruden, M.; Puri, N. Novel anticancer therapeutics targeting telomerase. Cancer Treat. Rev. 2013, 39, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Rankin, A.M.; Forman, L.; Sarkar, S.; Faller, D.V. Enhanced cytotoxicity from deoxyguanosine-enriched t-oligo in prostate cancer cells. Nucleic Acid Ther. 2013, 23, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Weng, D.; Cunin, M.C.; Song, B.; Price, B.D.; Eller, M.S.; Gilchrest, B.A.; Calderwood, S.K.; Gong, J. Radiosensitization of mammary carcinoma cells by telomere homolog oligonucleotide pretreatment. Breast Cancer Res. BCR 2010, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Faller, D.V. Telomere-homologous g-rich oligonucleotides sensitize human ovarian cancer cells to trail-induced growth inhibition and apoptosis. Nucleic Acid Ther. 2013, 23, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.; Levine, D.; Kishore, R.; Qin, G.; Thorne, T.; Lambers, E.; Sasi, S.P.; Yaar, M.; Gilchrest, B.A.; Goukassian, D.A. Inhibition of melanoma angiogenesis by telomere homolog oligonucleotides. J. Oncol. 2010, 2010, 928628. [Google Scholar] [CrossRef]
- Schrank, Z.; Khan, N.; Osude, C.; Singh, S.; Miller, R.J.; Merrick, C.; Mabel, A.; Kuckovic, A.; Puri, N. Oligonucleotides targeting telomeres and telomerase in cancer. Molecules (Basel Switz.) 2018, 23, 2267. [Google Scholar] [CrossRef]
- Wojdyla, L.; Stone, A.L.; Sethakorn, N.; Uppada, S.B.; Devito, J.T.; Bissonnette, M.; Puri, N. T-oligo as an anticancer agent in colorectal cancer. Biochem. Biophys. Res. Commun. 2014, 446, 596–601. [Google Scholar] [CrossRef]
- Mender, I.; Gryaznov, S.; Dikmen, Z.G.; Wright, W.E.; Shay, J.W. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2′-deoxyguanosine. Cancer Discov. 2015, 5, 82–95. [Google Scholar] [CrossRef]
- Zhang, G.; Wu, L.W.; Mender, I.; Barzily-Rokni, M.; Hammond, M.R.; Ope, O.; Cheng, C.; Vasilopoulos, T.; Randell, S.; Sadek, N.; et al. Induction of telomere dysfunction prolongs disease control of therapy-resistant melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 4771–4784. [Google Scholar] [CrossRef]
- Bejarano, L.; Schuhmacher, A.J.; Mendez, M.; Megias, D.; Blanco-Aparicio, C.; Martinez, S.; Pastor, J.; Squatrito, M.; Blasco, M.A. Inhibition of trf1 telomere protein impairs tumor initiation and progression in glioblastoma mouse models and patient-derived xenografts. Cancer Cell 2017, 32, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beccaria, M.; Martinez, P.; Mendez-Pertuz, M.; Martinez, S.; Blanco-Aparicio, C.; Canamero, M.; Mulero, F.; Ambrogio, C.; Flores, J.M.; Megias, D.; et al. Therapeutic inhibition of trf1 impairs the growth of p53-deficient k-rasg12v-induced lung cancer by induction of telomeric DNA damage. EMBO Mol. Med. 2015, 7, 930–949. [Google Scholar] [CrossRef] [PubMed]
- Di Maro, S.; Zizza, P.; Salvati, E.; De Luca, V.; Capasso, C.; Fotticchia, I.; Pagano, B.; Marinelli, L.; Gilson, E.; Novellino, E.; et al. Shading the trf2 recruiting function: A new horizon in drug development. J. Am. Chem. Soc. 2014, 136, 16708–16711. [Google Scholar] [CrossRef] [PubMed]
- Jager, K.; Walter, M. Therapeutic targeting of telomerase. Genes 2016, 7, 39. [Google Scholar] [CrossRef]
- Tahara, H.; Shin-Ya, K.; Seimiya, H.; Yamada, H.; Tsuruo, T.; Ide, T. G-quadruplex stabilization by telomestatin induces trf2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene 2006, 25, 1955–1966. [Google Scholar] [CrossRef]
- Zanetti, M. A second chance for telomerase reverse transcriptase in anticancer immunotherapy. Nat. Rev. Clin. Oncol. 2017, 14, 115–128. [Google Scholar] [CrossRef]
- Assani, G.; Xiong, Y.; Zhou, F.; Zhou, Y. Effect of therapies-mediated modulation of telomere and/or telomerase on cancer cells radiosensitivity. Oncotarget 2018, 9, 35008–35025. [Google Scholar] [CrossRef]
- Berardinelli, F.; Coluzzi, E.; Sgura, A.; Antoccia, A. Targeting telomerase and telomeres to enhance ionizing radiation effects in in vitro and in vivo cancer models. Mutat. Res. 2017, 773, 204–219. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).