mTOR and STAT3 Pathway Hyper-Activation is Associated with Elevated Interleukin-6 Levels in Patients with Shwachman-Diamond Syndrome: Further Evidence of Lymphoid Lineage Impairment
Abstract
1. Introduction
2. Results
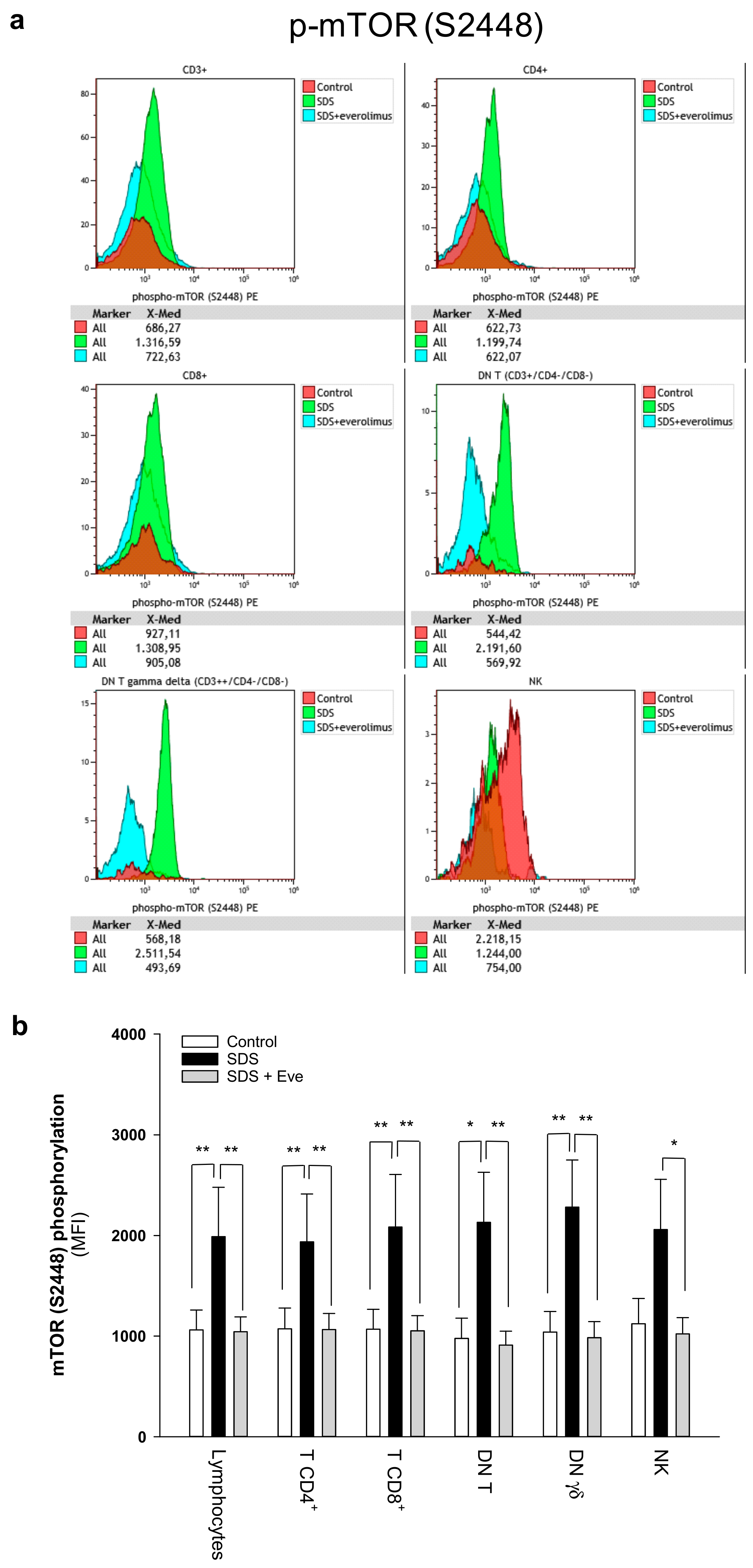
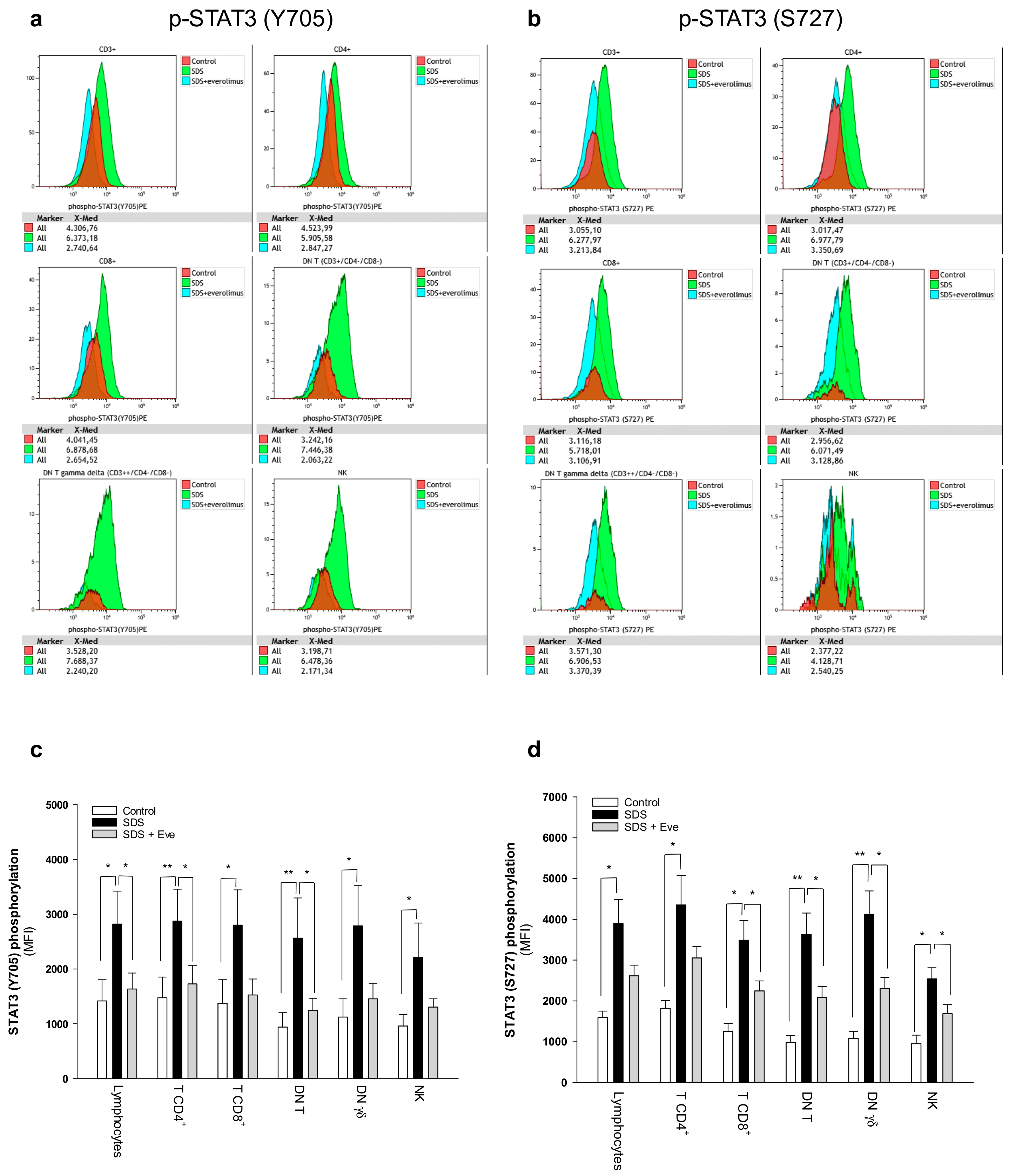
2.1. mTOR-STAT3 Pathway is Hyper-Activated Also in SDS Lymphocyte Subsets and Everolimus Can Reduce This Process In Vitro
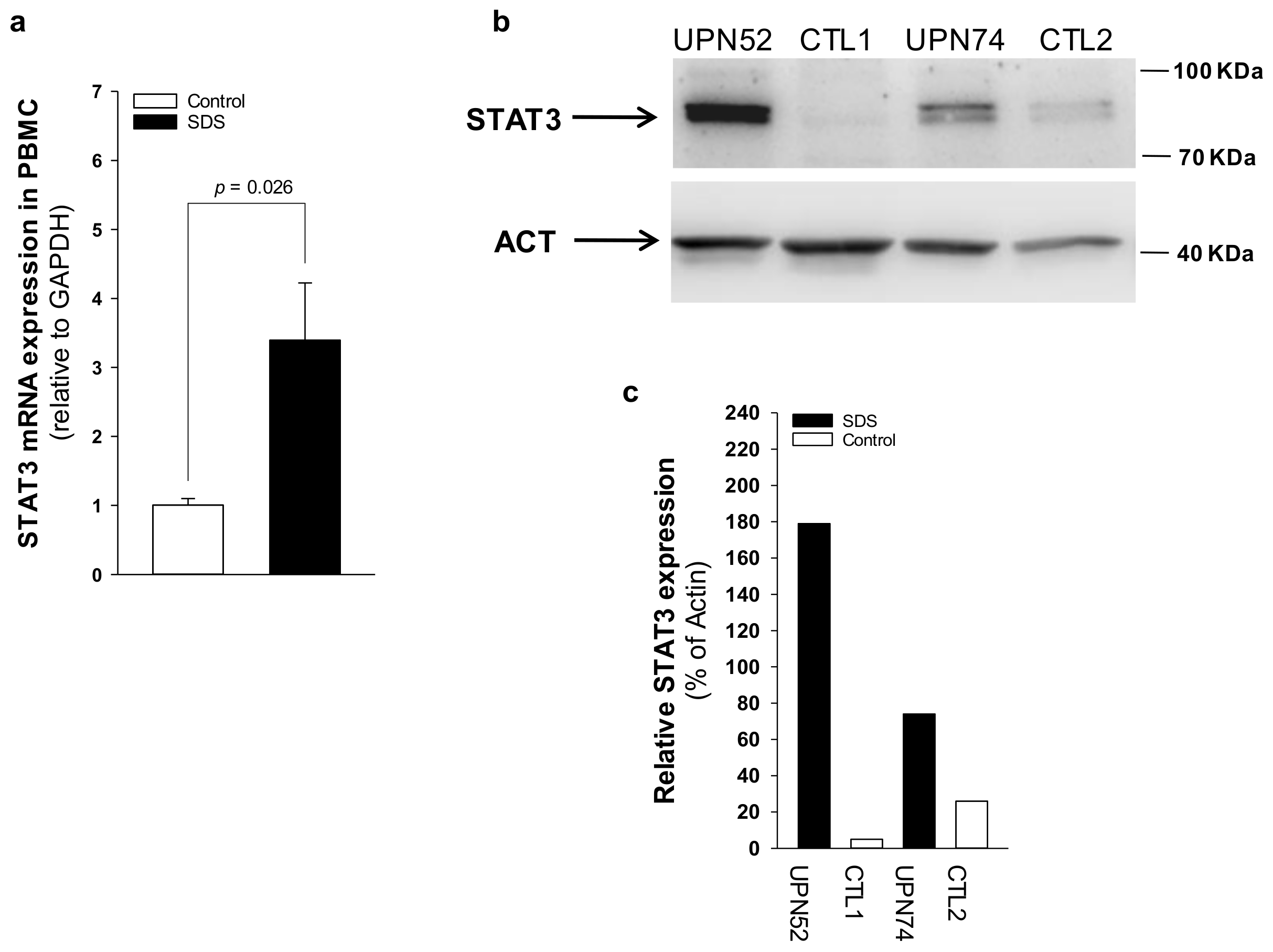
2.2. IL-6 Expression Is Upregulated in SDS
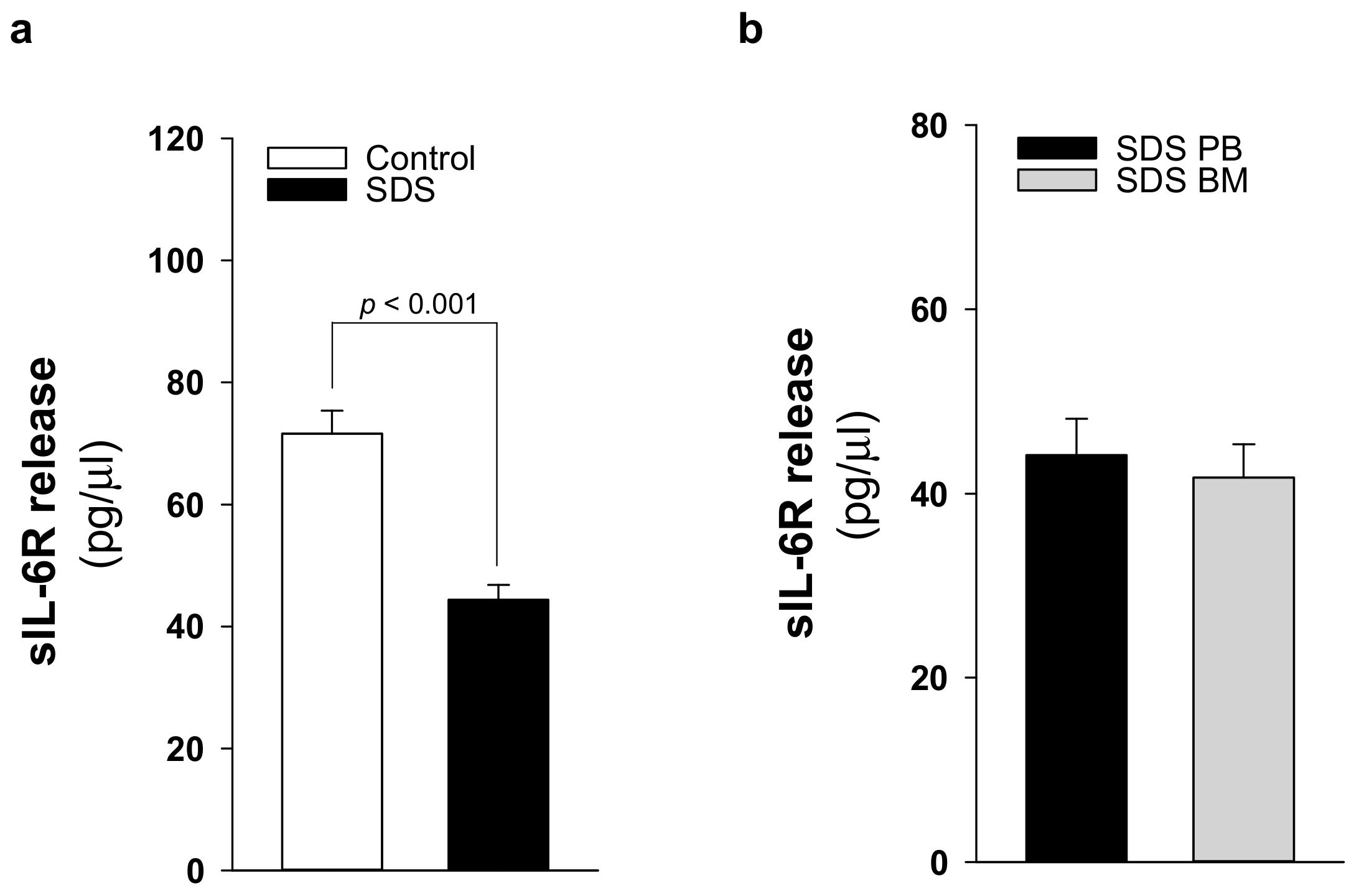
2.3. Patients with SDS ShowReduced Levels of Soluble IL-6 Receptor
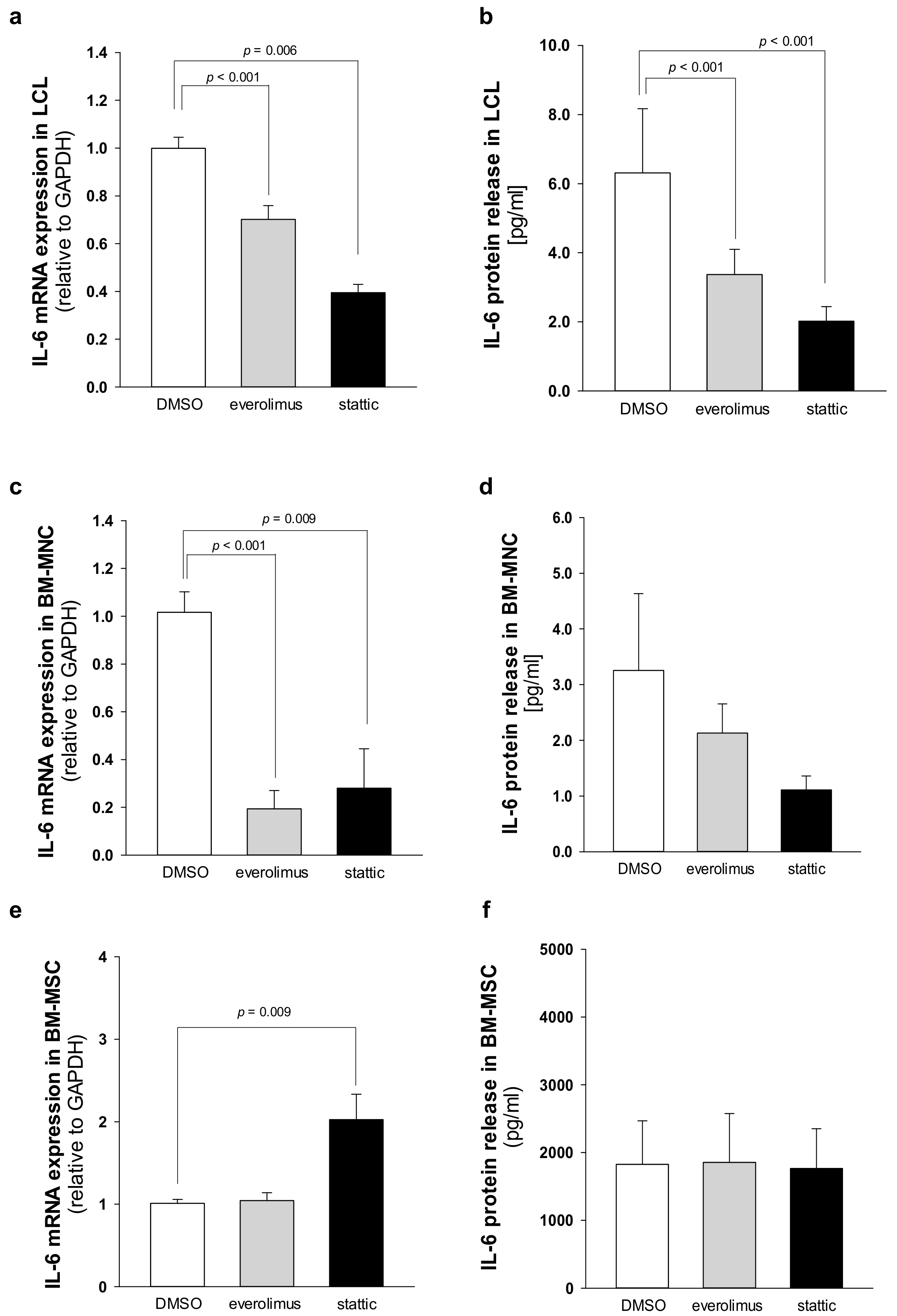
2.4. Elevated IL-6 Gene Expressionin Hematopoietic CellsIs Primarily Driven by mTOR-STAT3 Pathway in SDS
3. Discussion
4. Materials and Methods
4.1. Human Subjects
4.2. Plasma Isolation
4.3. Cell Cultures
4.4. Flow Cytometry
4.5. qRT-PCR
4.6. IL-6 and sIL-6R Detection
4.7. Western Blot
4.8. Gene Silencing
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dror, Y.; Donadieu, J.; Koglmeier, J.; Dodge, J.; Toiviainen-Salo, S.; Makitie, O.; Kerr, E.; Zeidler, C.; Shimamura, A.; Shah, N.; et al. Draft consensus guidelines for diagnosis and treatment of shwachman-diamond syndrome. Ann. N Y Acad. Sci. 2011, 1242, 40–55. [Google Scholar] [CrossRef]
- Kargas, V.; Castro-Hartmann, P.; Escudero-Urquijo, N.; Dent, K.; Hilcenko, C.; Sailer, C.; Zisser, G.; Marques-Carvalho, M.J.; Pellegrino, S.; Wawiórka, L.; et al. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. Elife 2019, 8, e44904. [Google Scholar] [CrossRef] [PubMed]
- Weis, F.; Giudice, E.; Churcher, M.; Jin, L.; Hilcenko, C.; Wong, C.C.; Traynor, D.; Kay, R.R.; Warren, A.J. Mechanism of eif6 release from the nascent 60s ribosomal subunit. Nat. Struct. Mol. Biol. 2015, 22, 914–919. [Google Scholar] [CrossRef]
- Finch, A.J.; Hilcenko, C.; Basse, N.; Drynan, L.F.; Goyenechea, B.; Menne, T.F.; González Fernández, A.; Simpson, P.; D’Santos, C.S.; Arends, M.J.; et al. Uncoupling of gtp hydrolysis from eif6 release on the ribosome causes shwachman-diamond syndrome. Genes Dev. 2011, 25, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Corey, S.J.; Minden, M.D.; Barber, D.L.; Kantarjian, H.; Wang, J.C.; Schimmer, A.D. Myelodysplastic syndromes: The complexity of stem-cell diseases. Nat. Rev. Cancer 2007, 7, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Donadieu, J.; Leblanc, T.; Bader Meunier, B.; Barkaoui, M.; Fenneteau, O.; Bertrand, Y.; Maier-Redelsperger, M.; Micheau, M.; Stephan, J.L.; Phillipe, N.; et al. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the french severe chronic neutropenia study group. Haematologica 2005, 90, 45–53. [Google Scholar] [PubMed]
- Lindsley, R.C.; Saber, W.; Mar, B.G.; Redd, R.; Wang, T.; Haagenson, M.D.; Grauman, P.V.; Hu, Z.H.; Spellman, S.R.; Lee, S.J.; et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N. Engl. J. Med. 2017, 376, 536–547. [Google Scholar] [CrossRef]
- Curran, E.K.; Godfrey, J.; Kline, J. Mechanisms of immune tolerance in leukemia and lymphoma. Trends Immunol. 2017, 38, 513–525. [Google Scholar] [CrossRef]
- D’Acquisto, F.; Crompton, T. Cd3+cd4-cd8- (double negative) t cells: Saviours or villains of the immune response? Biochem. Pharmacol. 2011, 82, 333–340. [Google Scholar] [CrossRef]
- Chen, B.; Lee, J.B.; Kang, H.; Minden, M.D.; Zhang, L. Targeting chemotherapy-resistant leukemia by combining dnt cellular therapy with conventional chemotherapy. J. Exp. Clin. Cancer Res. 2018, 37, 88. [Google Scholar] [CrossRef]
- Lee, J.; Minden, M.D.; Chen, W.C.; Streck, E.; Chen, B.; Kang, H.; Arruda, A.; Ly, D.; Der, S.D.; Kang, S.; et al. Allogeneic human double negative t cells as a novel immunotherapy for acute myeloid leukemia and its underlying mechanisms. Clin. Cancer Res. 2018, 24, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, X.; Liu, X.; Kline, D.E.; Teague, R.M.; Gajewski, T.F.; Kline, J. Cd40 ligation reverses t cell tolerance in acute myeloid leukemia. J. Clin. Investig. 2013, 123, 1999–2010. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, J.J.; Holland, S.M.; Staudt, L.M. Jaks and stats in immunity, immunodeficiency, and cancer. N. Engl. J. Med. 2013, 368, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the il-6/jak/stat3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between cancer and immune cells: Role of stat3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, A.; Kortylewski, M.; Kujawski, M.; Zhang, C.; Reckamp, K.; Armstrong, B.; Wang, L.; Kowolik, C.; Deng, J.; Figlin, R.; et al. Targeting stat3 in the myeloid compartment drastically improves the in vivo antitumor functions of adoptively transferred t cells. Cancer Res. 2010, 70, 7455–7464. [Google Scholar] [CrossRef]
- Kujawski, M.; Zhang, C.; Herrmann, A.; Reckamp, K.; Scuto, A.; Jensen, M.; Deng, J.; Forman, S.; Figlin, R.; Yu, H. Targeting stat3 in adoptively transferred t cells promotes their in vivo expansion and antitumor effects. Cancer Res. 2010, 70, 9599–9610. [Google Scholar] [CrossRef]
- Kumari, N.; Dwarakanath, B.S.; Das, A.; Bhatt, A.N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016, 37, 11553–11572. [Google Scholar] [CrossRef]
- Chalaris, A.; Garbers, C.; Rabe, B.; Rose-John, S.; Scheller, J. The soluble interleukin 6 receptor: Generation and role in inflammation and cancer. Eur. J. Cell Biol. 2011, 90, 484–494. [Google Scholar] [CrossRef]
- Audet, J.; Miller, C.L.; Rose-John, S.; Piret, J.M.; Eaves, C.J. Distinct role of gp130 activation in promoting self-renewal divisions by mitogenically stimulated murine hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 2001, 98, 1757–1762. [Google Scholar] [CrossRef]
- Walker, F.; Zhang, H.H.; Matthews, V.; Weinstock, J.; Nice, E.C.; Ernst, M.; Rose-John, S.; Burgess, A.W. Il6/sil6r complex contributes to emergency granulopoietic responses in g-csf- and gm-csf-deficient mice. Blood 2008, 111, 3978–3985. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ye, A.; Bi, L.; Wu, J.; Yu, K.; Zhang, S. Th17 cells and interleukin-17 increase with poor prognosis in patients with acute myeloid leukemia. Cancer Sci. 2014, 105, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Correa, B.; Bergua, J.M.; Campos, C.; Gayoso, I.; Arcos, M.J.; Bañas, H.; Morgado, S.; Casado, J.G.; Solana, R.; Tarazona, R. Cytokine profiles in acute myeloid leukemia patients at diagnosis: Survival is inversely correlated with il-6 and directly correlated with il-10 levels. Cytokine 2013, 61, 885–891. [Google Scholar] [CrossRef] [PubMed]
- Yokogami, K.; Wakisaka, S.; Avruch, J.; Reeves, S.A. Serine phosphorylation and maximal activation of stat3 during cntf signaling is mediated by the rapamycin target mtor. Curr. Biol. 2000, 10, 47–50. [Google Scholar] [CrossRef]
- Dodd, K.M.; Yang, J.; Shen, M.H.; Sampson, J.R.; Tee, A.R. Mtorc1 drives hif-1α and vegf-a signalling via multiple mechanisms involving 4e-bp1, s6k1 and stat3. Oncogene 2015, 34, 2239–2250. [Google Scholar] [CrossRef]
- Bezzerri, V.; Vella, A.; Calcaterra, E.; Finotti, A.; Gasparello, J.; Gambari, R.; Assael, B.M.; Cipolli, M.; Sorio, C. New insights into the shwachman-diamond syndrome-related haematological disorder: Hyper-activation of mtor and stat3 in leukocytes. Sci. Rep. 2016, 6, 33165. [Google Scholar] [CrossRef]
- Chapuis, N.; Tamburini, J.; Green, A.S.; Willems, L.; Bardet, V.; Park, S.; Lacombe, C.; Mayeux, P.; Bouscary, D. Perspectives on inhibiting mtor as a future treatment strategy for hematological malignancies. Leukemia 2010, 24, 1686–1699. [Google Scholar] [CrossRef]
- Hoshii, T.; Matsuda, S.; Hirao, A. Pleiotropic roles of mtor complexes in haemato-lymphopoiesis and leukemogenesis. J. Biochem. 2014, 156, 73–83. [Google Scholar] [CrossRef]
- Porta, C.; Paglino, C.; Mosca, A. Targeting pi3k/akt/mtor signaling in cancer. Front. Oncol. 2014, 4, 64. [Google Scholar] [CrossRef]
- Witzig, T.E.; Reeder, C.; Han, J.J.; LaPlant, B.; Stenson, M.; Tun, H.W.; Macon, W.; Ansell, S.M.; Habermann, T.M.; Inwards, D.J.; et al. The mtorc1 inhibitor everolimus has antitumor activity in vitro and produces tumor responses in patients with relapsed t-cell lymphoma. Blood 2015, 126, 328–335. [Google Scholar] [CrossRef]
- Cai, Y.; Xue, F.; Qin, H.; Chen, X.; Liu, N.; Fleming, C.; Hu, X.; Zhang, H.G.; Chen, F.; Zheng, J.; et al. Differential roles of the mtor-stat3 signaling in dermal γδ t cell effector function in skin inflammation. Cell Rep. 2019, 27, 3034–3048. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S. Il-6 trans-signaling via the soluble il-6 receptor: Importance for the pro-inflammatory activities of il-6. Int. J. Biol. Sci. 2012, 8, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Erices, A.; Conget, P.; Rojas, C.; Minguell, J.J. Gp130 activation by soluble interleukin-6 receptor/interleukin-6 enhances osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. Exp. Cell Res. 2002, 280, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Pricola, K.L.; Kuhn, N.Z.; Haleem-Smith, H.; Song, Y.; Tuan, R.S. Interleukin-6 maintains bone marrow-derived mesenchymal stem cell stemness by an erk1/2-dependent mechanism. J. Cell Biochem. 2009, 108, 577–588. [Google Scholar] [CrossRef]
- Calabrese, L.H.; Rose-John, S. Il-6 biology: Implications for clinical targeting in rheumatic disease. Nat. Rev. Rheumatol. 2014, 10, 720–727. [Google Scholar] [CrossRef]
- Altman, J.K.; Sassano, A.; Platanias, L.C. Targeting mtor for the treatment of aml. New agents and new directions. Oncotarget 2011, 2, 510–517. [Google Scholar] [CrossRef]
- Dinner, S.; Platanias, L.C. Targeting the mtor pathway in leukemia. J. Cell Biochem. 2016, 117, 1745–1752. [Google Scholar] [CrossRef]
- Bezzerri, V.; Vella, A.; Gennaro, G.D.; Ortolani, R.; Nicolis, E.; Cesaro, S.; Fabrizzi, B.; Bronte, V.; Corey, S.J.; Cipolli, M. Peripheral blood immunophenotyping in a large cohort of patients with shwachman-diamond syndrome. Pediatr. Blood Cancer 2019, 66, e27597. [Google Scholar] [CrossRef]
- Durant, L.; Watford, W.T.; Ramos, H.L.; Laurence, A.; Vahedi, G.; Wei, L.; Takahashi, H.; Sun, H.W.; Kanno, Y.; Powrie, F.; et al. Diverse targets of the transcription factor stat3 contribute to t cell pathogenicity and homeostasis. Immunity 2010, 32, 605–615. [Google Scholar] [CrossRef]
- Hossain, D.M.; Dos Santos, C.; Zhang, Q.; Kozlowska, A.; Liu, H.; Gao, C.; Moreira, D.; Swiderski, P.; Jozwiak, A.; Kline, J.; et al. Leukemia cell-targeted stat3 silencing and tlr9 triggering generate systemic antitumor immunity. Blood 2014, 123, 15–25. [Google Scholar] [CrossRef]
- Dror, Y.; Ginzberg, H.; Dalal, I.; Cherepanov, V.; Downey, G.; Durie, P.; Roifman, C.M.; Freedman, M.H. Immune function in patients with shwachman-diamond syndrome. Br. J. Haematol. 2001, 114, 712–717. [Google Scholar] [CrossRef]
- Raaijmakers, M.H.; Mukherjee, S.; Guo, S.; Zhang, S.; Kobayashi, T.; Schoonmaker, J.A.; Ebert, B.L.; Al-Shahrour, F.; Hasserjian, R.P.; Scadden, E.O.; et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 2010, 464, 852–857. [Google Scholar] [CrossRef]
- Santamaría, C.; Muntión, S.; Rosón, B.; Blanco, B.; López-Villar, O.; Carrancio, S.; Sánchez-Guijo, F.M.; Díez-Campelo, M.; Alvarez-Fernández, S.; Sarasquete, M.E.; et al. Impaired expression of dicer, drosha, sbds and some micrornas in mesenchymal stromal cells from myelodysplastic syndrome patients. Haematologica 2012, 97, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Geyh, S.; Oz, S.; Cadeddu, R.P.; Fröbel, J.; Brückner, B.; Kündgen, A.; Fenk, R.; Bruns, I.; Zilkens, C.; Hermsen, D.; et al. Insufficient stromal support in mds results from molecular and functional deficits of mesenchymal stromal cells. Leukemia 2013, 27, 1841–1851. [Google Scholar] [CrossRef]
- Schroeder, T.; Geyh, S.; Germing, U.; Haas, R. Mesenchymal stromal cells in myeloid malignancies. Blood Res. 2016, 51, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Severin, F.; Frezzato, F.; Visentin, A.; Martini, V.; Trimarco, V.; Carraro, S.; Tibaldi, E.; Brunati, A.M.; Piazza, F.; Semenzato, G.; et al. In chronic lymphocytic leukemia the jak2/stat3 pathway is constitutively activated and its inhibition leads to cll cell death unaffected by the protective bone marrow microenvironment. Cancers 2019, 11, 1939. [Google Scholar] [CrossRef] [PubMed]
- Oberg, H.H.; Wesch, D.; Grüssel, S.; Rose-John, S.; Kabelitz, D. Differential expression of cd126 and cd130 mediates different stat-3 phosphorylation in cd4+cd25- and cd25high regulatory t cells. Int. Immunol. 2006, 18, 555–563. [Google Scholar] [CrossRef]
- Scheller, J.; Rose-John, S. Interleukin-6 and its receptor: From bench to bedside. Med. Microbiol. Immunol. 2006, 195, 173–183. [Google Scholar] [CrossRef]
- Kondo, M.; Yamaoka, K.; Sakata, K.; Sonomoto, K.; Lin, L.; Nakano, K.; Tanaka, Y. Contribution of the interleukin-6/stat-3 signaling pathway to chondrogenic differentiation of human mesenchymal stem cells. Arthritis Rheumatol. 2015, 67, 1250–1260. [Google Scholar] [CrossRef]
- Bezzerri, V.; Cipolli, M. Shwachman-diamond syndrome: Molecular mechanisms and current perspectives. Mol. Diagn. Ther. 2018, 23, 281–290. [Google Scholar] [CrossRef]
- Lambert, C.; Genin, C. Cd3 bright lymphocyte population reveal gammadelta t cells. Cytom. B Clin. Cytom. 2004, 61, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Nguyen-Jackson, H.; Panopoulos, A.D.; Li, H.S.; Murray, P.J.; Watowich, S.S. Stat3 controls myeloid progenitor growth during emergency granulopoiesis. Blood 2010, 116, 2462–2471. [Google Scholar] [CrossRef] [PubMed]



| UPN | Gender | Age | Genotype | PMN (Cell/mm3) | Phenotype | Cytogenetics |
|---|---|---|---|---|---|---|
| 1 | M | 27 | 258+2T>C/183–184TA>CT | 1460 | PI, FTT, recurrent infections, HbF > 2%, bone malformation, thrombocytopenia | 46, XY, i(7)(q10) |
| 6 | M | 27 | 258+2T>C/101A>T | 3515 | PI, FTT, recurrent infections, bone malformation, thrombocytopenia, cognitive impairment | 46, XY, del(20)q |
| 13 | M | 19 | 258+2T>C/183–184TA>CT+258+2T>C | 1130 | PI, FTT, recurrent infections, bone malformation, thrombocytopenia, anemia, cognitive impairment | 46, XY, del(20)q |
| 26 | M | 16 | 258+2T>C/183–184TA>CT | 58 | PI, FTT, bone malformation, thrombocytopenia | 46, XY |
| 33 | F | 8 | 258+2T>C/183–184TA>CT | 1100 | PI, FTT, bone malformation, thrombocytopenia | 46, XX |
| 35 | F | 15 | 258+2T>C/183–184TA>CT | 970 | PI, FTT, thrombocytopenia, anemia | 46, XX |
| 37 | F | 10 | 258+2T>C/183–184TA>CT | 1280 | PI, FTT, recurrent infections | 46, XX, i(7)(q10) |
| 43 | M | 22 | 258+2T>C/258+2T>C+533–549+403del | 970 | PI, FTT, bone malformation, thrombocytopenia, cognitive impairment | 46, XY |
| 47 | M | 12 | 258+2T>C/183–184TA>CT | 550 | PI, FTT, recurrent infections, bone malformation, thrombocytopenia | 46, XY |
| 52 | M | 10 | 258+2T>C/183–184TA>CT+258+2T>C | 1070 | PI, FTT, bone malformation | 46, XY |
| 56 | F | 15 | 258+2T>C/183–184TA>CT | 1840 | PI, FTT, recurrent infections, HbF > 2%, bone malformation, thrombocytopenia | 46, XX |
| 57 | F | 40 | 258+2T>C/G63C | 500 | PI, FTT, HbF > 2%, bone malformation, thrombocytopenia, cognitive impairment | 46, XX |
| 58 | M | 12 | 258+2T>C/183–184TA>CT | 390 | PI, FTT, HbF > 2%, bone malformation, thrombocytopenia, anemia | 46, XY |
| 63 | M | 15 | 258+2T>C/183–184TA>CT+258+2T>C | 536 | PI, FTT, recurrent infections, HbF > 2%, bone malformation, thrombocytopenia | 46, XY |
| 65 | M | 19 | 258+2T>C/258+2T>C | 1390 | PI, recurrent infections, bone malformation, thrombocytopenia, cognitive impairment | 46, XY, del(20)q |
| 66 | M | 23 | 258+2T>C/183–184TA>CT | 1340 | PI, bone malformation, thrombocytopenia, cognitive impairment | 46, XY |
| 67 | M | 8 | 258+2T>C/183–184TA>CT | 500 | PI, FTT, HbF > 2%, bone malformation | 46, XY |
| 68 | M | 22 | 258+2T>C/183–184TA>CT+258+2T>C | 600 | PI, FTT, recurrent infections, bone malformation, thrombocytopenia, cognitive impairment | 46, XY |
| 69 | F | 8 | 258+2T>C/183–184TA>CT | 770 | PI, FTT, HbF > 2%, anemia, cognitive impairment | 46, XX |
| 72 | M | 28 | 258+2T>C/183–184TA>CT | 380 | PI, FTT, recurrent infections, HbF > 2%, bone malformation, thrombocytopenia, anemia, cognitive impairment | 46, XY |
| 73 | F | 7 | 258+2T>C/183–184TA>CT | 520 | PI, FTT, HbF > 2%, thrombocytopenia | 46, XX |
| 74 | M | 9 | 258+2T>C/183–184TA>CT | 1430 | PI, FTT, HbF > 2%, cognitive impairment | 46, XY |
| 75 | F | 7 | 258+2T>C/183–184TA>CT | 1000 | PI, FTT, HbF > 2%, bone malformation, thrombocytopenia, cognitive impairment | 46, XX |
| 80 | M | 7 | 258+2T>C/183–184TA>CT | 680 | PI, FTT, recurrent infections, bone malformation, anemia, cognitive impairment | 46, XY |
| 82 | M | 16 | 258+2T>C/183–184TA>CT | 300 | PI, FTT, recurrent infections, bone malformation, thrombocytopenia, anemia, cognitive impairment | 46, XY |
| 87 | M | 18 | 258+2T>C/183–184TA>CT | 880 | PI, FTT, recurrent infections, bone malformation, cognitive impairment | 46, XY |
| 91 | M | 4 | 258+2T>C/183–184TA>CT+258+2T>C | 1050 | PI, FTT, bone malformation | 46, XY |
| 94 | F | 19 | 258+2T>C/352A>G | 2420 | PI, HbF > 2%, thrombocytopenia | 46, XX |
| 104 | M | 10 | 258+2T>C/183–184TA>CT | 500 | PI, FTT, recurrent infections, HbF > 2%, bone malformation, thrombocytopenia | 46, XY |
| 106 | M | 36 | 258+2T>C/183–184TA>CT | 1210 | PI, FTT, bone malformation, anemia | 46, XY |
| 108 | M | 17 | 258+2T>C/183–184TA>CT | 970 | PS, FTT, bone malformation, anemia | 46, XY |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vella, A.; D’Aversa, E.; Api, M.; Breveglieri, G.; Allegri, M.; Giacomazzi, A.; Marinelli Busilacchi, E.; Fabrizzi, B.; Cestari, T.; Sorio, C.; et al. mTOR and STAT3 Pathway Hyper-Activation is Associated with Elevated Interleukin-6 Levels in Patients with Shwachman-Diamond Syndrome: Further Evidence of Lymphoid Lineage Impairment. Cancers 2020, 12, 597. https://doi.org/10.3390/cancers12030597
Vella A, D’Aversa E, Api M, Breveglieri G, Allegri M, Giacomazzi A, Marinelli Busilacchi E, Fabrizzi B, Cestari T, Sorio C, et al. mTOR and STAT3 Pathway Hyper-Activation is Associated with Elevated Interleukin-6 Levels in Patients with Shwachman-Diamond Syndrome: Further Evidence of Lymphoid Lineage Impairment. Cancers. 2020; 12(3):597. https://doi.org/10.3390/cancers12030597
Chicago/Turabian StyleVella, Antonio, Elisabetta D’Aversa, Martina Api, Giulia Breveglieri, Marisole Allegri, Alice Giacomazzi, Elena Marinelli Busilacchi, Benedetta Fabrizzi, Tiziana Cestari, Claudio Sorio, and et al. 2020. "mTOR and STAT3 Pathway Hyper-Activation is Associated with Elevated Interleukin-6 Levels in Patients with Shwachman-Diamond Syndrome: Further Evidence of Lymphoid Lineage Impairment" Cancers 12, no. 3: 597. https://doi.org/10.3390/cancers12030597
APA StyleVella, A., D’Aversa, E., Api, M., Breveglieri, G., Allegri, M., Giacomazzi, A., Marinelli Busilacchi, E., Fabrizzi, B., Cestari, T., Sorio, C., Bedini, G., D’Amico, G., Bronte, V., Poloni, A., Benedetti, A., Bovo, C., Corey, S. J., Borgatti, M., Cipolli, M., & Bezzerri, V. (2020). mTOR and STAT3 Pathway Hyper-Activation is Associated with Elevated Interleukin-6 Levels in Patients with Shwachman-Diamond Syndrome: Further Evidence of Lymphoid Lineage Impairment. Cancers, 12(3), 597. https://doi.org/10.3390/cancers12030597








