Inflammatory Mechanisms of HCC Development
Abstract
:1. Introduction
2. Microenvironmental Factors in HCC
2.1. DNA Alterations in HCC
2.2. Chronic Inflammation
2.3. Inflammation and Tissue Remodeling
2.4. Inflammation and the Immune Response in HCC
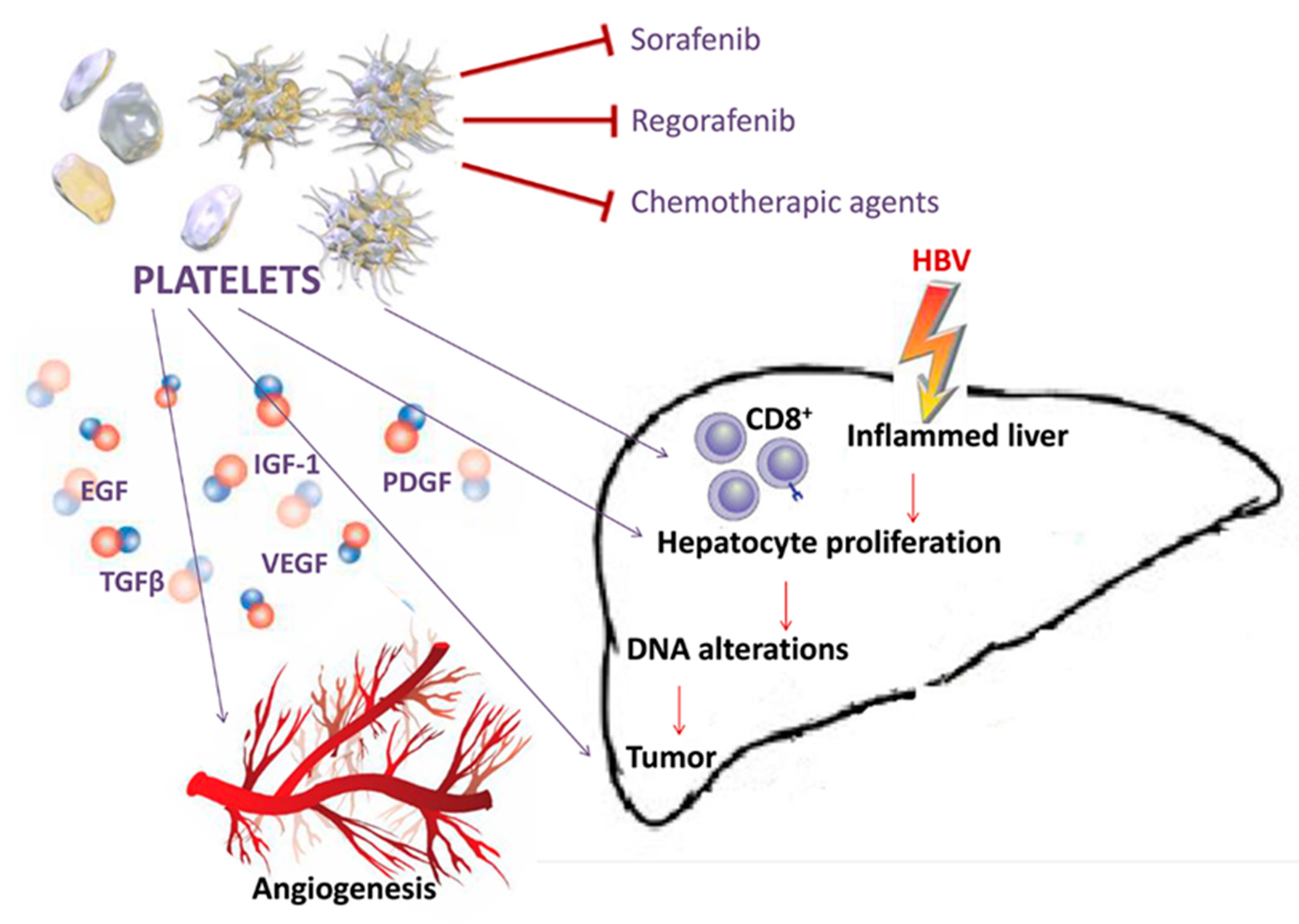
2.5. Other Microenvironmental Factors Involved in Hepatocarcinogenesis
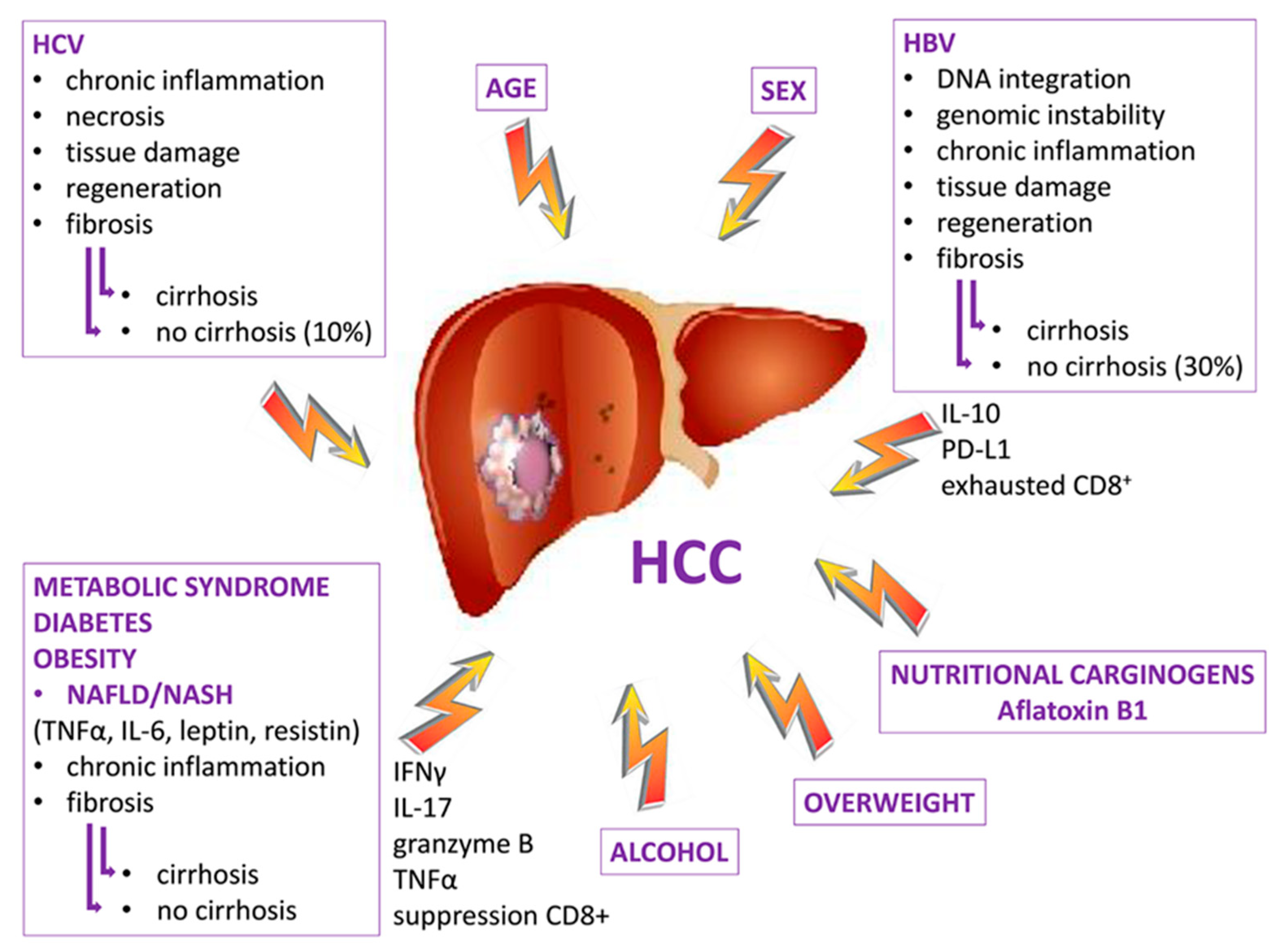
2.6. HCC Etiology and Chronic Inflammation
3. Preclinical Studies in HCC Immunotherapy
4. Clinical Studies in HCC Immunotherapy
5. Conclusions
Funding
Conflicts of Interest
References
- Woo, H.G.; Park, E.S.; Thorgeirsson, S.S.; Kim, Y.J. Exploring genomic profiles of hepatocellular carcinoma. Mol. Carcinog. 2011, 50, 235–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Multhoff, G.; Molls, M.; Radons, J. Chronic inflammation in cancer development. Front. Immunol. 2012, 2, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greten, T.F.; Wang, X.W.; Korangy, F. Current concepts of immune based treatments for patients with HCC: From basic science to novel treatment approaches. Gut 2015, 64, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Budzinska, M.A.; Maczurek, A.E.; Cheng, R.; Di Bartolomeo, A.; Warner, F.J.; McCaughan, G.W.; McLennan, S.V.; Shackel, N.A. Novel aspects of the liver microenvironment in hepatocellular carcinoma pathogenesis and development. Int. J. Mol. Sci. 2014, 15, 9422–9458. [Google Scholar] [CrossRef] [Green Version]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.C.; Llovet, J.M. The genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015, 149, 1226–1239. [Google Scholar] [CrossRef] [Green Version]
- de La Coste, A.; Romagnolo, B.; Billuart, P.; Renard, C.A.; Buendia, M.A.; Soubrane, O.; Fabre, M.; Chelly, J.; Beldjord, C.; Kahn, A.; et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 1998, 95, 8847–8851. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, A.; Totoki, Y.; Abe, T.; Boroevich, K.A.; Hosoda, F.; Nguyen, H.H.; Aoki, M.; Hosono, N.; Kubo, M.; Miya, F.; et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012, 44, 760–764. [Google Scholar] [CrossRef]
- Huang, J.; Deng, Q.; Wang, Q.; Li, K.Y.; Dai, J.H.; Li, N.; Zhu, Z.D.; Zhou, B.; Liu, X.Y.; Liu, R.F.; et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat. Genet. 2012, 44, 1117–1121. [Google Scholar] [CrossRef]
- Woo, H.G.; Wang, X.W.; Budhu, A.; Kim, Y.H.; Kwon, S.M.; Tang, Z.Y.; Sun, Z.; Harris, C.C.; Thorgeirsson, S.S. Association of TP53 mutations with stem cell-like gene expression and survival of patients with hepatocellular carcinoma. Gastroenterology 2011, 140, 1063–1070. [Google Scholar] [CrossRef] [Green Version]
- Bressac, B.; Kew, M.; Wands, J.; Ozturk, M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 1991, 350, 429–431. [Google Scholar] [CrossRef] [PubMed]
- Sung, W.K.; Zheng, H.; Li, S.; Chen, R.; Liu, X.; Li, Y.; Lee, N.P.; Lee, W.H.; Ariyaratne, P.N.; Tennakoon, C.; et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012, 44, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Brechot, C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: Old and new paradigms. Gastroenterology 2004, 127, S56–S61. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, J.; Yang, Z.; Kang, J.; Jiang, S.; Zhang, T.; Chen, T.; Li, M.; Lv, Q.; Chen, X.; et al. The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma. J. Hepatol. 2014, 60, 975–984. [Google Scholar] [CrossRef] [Green Version]
- Paterlini-Brechot, P.; Saigo, K.; Murakami, Y.; Chami, M.; Gozuacik, D.; Mugnier, C.; Lagorce, D.; Bréchot, C. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 2003, 22, 3911–3916. [Google Scholar] [CrossRef]
- Hino, O.; Tabata, S.; Hotta, Y. Evidence for increased in vitro recombination with insertion of human hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 1991, 88, 9248–9252. [Google Scholar] [CrossRef] [Green Version]
- Nault, J.C. Pathogenesis of hepatocellular carcinoma according to aetiology. Best Pract. Res. Clin. Gastroenterol. 2014, 28, 937–947. [Google Scholar] [CrossRef]
- Bartosch, B.; Thimme, R.; Blum, H.E.; Zoulim, F. Hepatitis C virus-induced hepatocarcinogenesis. J. Hepatol. 2009, 51, 810–820. [Google Scholar] [CrossRef] [Green Version]
- Joo, M.; Hahn, Y.S.; Kwon, M.; Sadikot, R.T.; Blackwell, T.S.; Christman, J.W. Hepatitis C virus core protein suppresses NF-kappaB activation and cyclooxygenase-2 expression by direct interaction with IkappaB kinase beta. J. Virol. 2005, 79, 7648–7657. [Google Scholar] [CrossRef] [Green Version]
- Seitz, H.K.; Simanowski, U.A.; Garzon, F.T.; Rideout, J.M.; Peters, T.J.; Koch, A.; Berger, M.R.; Einecke, H.; Maiwald, M. Possible role of acetaldehyde in ethanol-related rectal cocarcinogenesis in the rat. Gastroenterology 1990, 98, 406–413. [Google Scholar] [CrossRef]
- Bradford, B.U.; Kono, H.; Isayama, F.; Kosyk, O.; Wheeler, M.D.; Akiyama, T.E.; Bleye, L.; Krausz, K.W.; Gonzalez, F.J.; Koop, D.R.; et al. Cytochrome P450 CYP2E1, but not nicotinamide adenine dinucleotide phosphate oxidase, is required for ethanol-induced oxidative DNA damage in rodent liver. Hepatology 2005, 41, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Theruvathu, J.A.; Jaruga, P.; Nath, R.G.; Dizdaroglu, M.; Brooks, P.J. Polyamines stimulate the formation of mutagenic 1, N2-propanodeoxyguanosine adducts from acetaldehyde. Nucleic Acids Res. 2005, 33, 3513–3520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nahon, P.; Sutton, A.; Rufat, P.; Ziol, M.; Akouche, H.; Laguillier, C.; Charnaux, N.; Ganne-Carrié, N.; Grando-Lemaire, V.; N’Kontchou, G.; et al. Myeloperoxidase andm superoxide dismutase 2 polymorphisms comodulate the risk of hepatocellular carcinoma and death in alcoholic cirrhosis. Hepatology 2009, 50, 1484–1493. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M.; Gabriel, S.; Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010, 11, 685–696. [Google Scholar] [CrossRef]
- Llovet, J.M.; Villanueva, A.; Lachenmayer, A.; Finn, R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 2015, 12, 408–424. [Google Scholar] [CrossRef] [PubMed]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.B.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef]
- Sawey, E.T.; Chanrion, M.; Cai, C.; Wu, G.; Zhang, J.; Zender, L.; Zhao, A.; Busuttil, R.W.; Yee, H.; Stein, L.; et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell 2011, 19, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Cleary, S.P.; Jeck, W.R.; Zhao, X.; Chen, K.; Selitsky, S.R.; Savich, G.L.; Tan, T.X.; Wu, M.C.; Getz, G.; Lawrence, M.S.; et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013, 58, 1693–1702. [Google Scholar] [CrossRef]
- Chen, W.; Jiang, J.; Wang, P.P.; Gong, L.; Chen, J.; Du, W.; Bi, K.; Diao, H. Identifying hepatocellular carcinoma driver genes by integrative pathway crosstalk and protein interaction network. DNA Cell Biol. 2019, 38, 1112–1124. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Zhang, H.; Sun, B.; Karin, M. The immunobiology of hepatocellular carcinoma in humans and mice: Basic concepts and therapeutic implications. J. Hepatol. 2020, 72, 167–182. [Google Scholar] [CrossRef]
- Liao, H.; Chen, W.; Dai, Y.; Richardson, J.J.; Guo, J.; Yuan, K.; Zeng, Y.; Xie, K. Expression of programmed cell death-ligands in hepatocellular carcinoma: Correlation with immune microenvironment and survival outcomes. Front. Oncol. 2019, 9, 883. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver; European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2012, 56, 908–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sangro, B.; Gomez-Martin, C.; de la Mata, M.; Inarrairaegui, M.; Garralda, E.; Barrera, P.; Riezu-Boj, J.I.; Larrea, E.; Alfaro, C.; Sarobe, P.; et al. A clinical trial of CTLA-4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J. Hepatol. 2013, 59, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef] [Green Version]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Iannacone, M.; Sitia, G.; Isogawa, M.; Marchese, P.; Castro, M.G.; Lowenstein, P.R.; Chisari, F.V.; Ruggeri, Z.M.; Guidotti, L.G. Platelets mediate cytotoxic T lymphocyte-induced liver damage. Nat. Med. 2005, 11, 1167–1169. [Google Scholar] [CrossRef] [Green Version]
- Carr, B.I.; Lin, C.Y.; Lu, S.N. Platelet-related phenotypic patterns in hepatocellular carcinoma patients. Semin. Oncol. 2014, 41, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Carr, B.I.; Guerra, V. Thrombocytosis and hepatocellular carcinoma. Dig. Dis. Sci. 2013, 58, 1790–1796. [Google Scholar] [CrossRef] [Green Version]
- Leslie, M. Cell biology. Beyond clotting: The powers of platelets. Science 2010, 328, 562–564. [Google Scholar] [CrossRef] [PubMed]
- Borsig, L. The role of platelet activation in tumor metastasis. Expert Rev. Anticancer Ther. 2008, 8, 1247–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, R.; Nakano, Y.; Ohkohchi, N. Platelet administration via the portal vein promotes liver regeneration in rats after 70% hepatectomy. Ann. Surg. 2011, 253, 759–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Zhou, X.D.; Liu, Y.K.; Ren, N.; Chen, J.; Zhao, Y. Platelets promote the adhesion of human hepatoma cells with a highly metastatic potential to extracellular matrix protein: Involvement of platelet P-selectin and GP IIb-IIIa. J. Cancer Res. Clin. Oncol. 2002, 128, 283–287. [Google Scholar]
- Lalor, P.F.; Herbert, J.; Bicknell, R.; Adams, D.H. Hepatic sinusoidal endothelium avidly binds platelets in an integrin-dependent manner, leading to platelet and endothelial activation and leukocyte recruitment. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G469–G478. [Google Scholar] [CrossRef] [Green Version]
- Shetty, S.; Lalor, P.F.; Adams, D.H. Liver sinusoidal endothelial cells—Gatekeepers of hepatic immunity. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 555–567. [Google Scholar] [CrossRef]
- Pancoska, P.; Carr, B.I. Macro-and micro-environmental factors in clinical hepatocellular cancer. Semin. Oncol. 2014, 41, 185–194. [Google Scholar] [CrossRef] [Green Version]
- D’Alessandro, R.; Messa, C.; Refolo, M.G.; Carr, B.I. Modulation of sensitivity and resistance to multikinase inhibitors by microenvironmental platelet factors in HCC. Expert Opin. Pharm. 2015, 16, 2773–2780. [Google Scholar] [CrossRef]
- D’Alessandro, R.; Refolo, M.G.; Lippolis, C.; Carella, N.; Messa, C.; Cavallini, A.; Carr, B.I. Modulation of regorafenib effects on HCC cell lines by epidermal growth factor. Cancer Chemother. Pharm. 2015, 75, 1237–1245. [Google Scholar] [CrossRef] [Green Version]
- Lippolis, C.; Refolo, M.G.; D’Alessandro, R.; Carella, N.; Messa, C.; Cavallini, A.; Carr, B.I. Resistance to multikinase inhibitor actions mediated by insulin like growth factor-1. J. Exp. Clin. Cancer Res. 2015, 34, 90. [Google Scholar] [CrossRef] [Green Version]
- Nault, J.C.; Ningarhari, M.; Rebouissou, S.; Zucman-Rossi, J. The role of telomeres and telomerase in cirrhosis and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Youssef, N.; Paradis, V.; Ferlicot, S.; Bedossa, P. In situ detection of telomerase enzymatic activity in human hepatocellular carcinogenesis. J. Pathol. 2001, 194, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.C.; Calderaro, J.; Di Tommaso, L.; Balabaud, C.; Zafrani, E.S.; Bioulac-Sage, P.; Roncalli, M.; Zucman-Rossi, J. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 2014, 60, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Dhar, D.; Nakagawa, H.; Font-Burgada, J.; Ogata, H.; Jiang, Y.; Shalapour, S.; Seki, E.; Yost, S.E.; Jepsen, K.; et al. Identification of Liver Cancer Progenitors Whose Malignant Progression Depends on Autocrine IL-6 Signaling. Cell 2013, 155, 384–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, T.W.; Yevsa, T.; Woller, N.; Hoenicke, L.; Wuestefeld, T.; Dauch, D.; Hohmeyer, A.; Gereke, M.; Rudalska, R.; Potapova, A.; et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 2011, 479, 547–551. [Google Scholar] [CrossRef]
- Kim, A.K.; Dziura, J.; Strazzabosco, M. Nonsteroidal anti-inflammatory drug use, chronic liver disease, and hepatocellular carcinoma: The egg of Columbus or another illusion? Hepatology 2013, 58, 819–821. [Google Scholar] [CrossRef]
- Cervello, M.; Montalto, G. Cyclooxygenases in hepatocellular carcinoma. World J. Gastroenterol. 2006, 12, 5113–5121. [Google Scholar] [CrossRef]
- Tao, Y.; Li, Y.; Liu, X.; Deng, Q.; Yu, Y.; Yang, Z. Nonsteroidal anti-inflammatory drugs, especially aspirin, are linked to lower risk and better survival of hepatocellular carcinoma: A meta-analysis. Cancer Manag. Res. 2018, 10, 2695–2709. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.L.; Wang, M.D.; Zhou, X.; Qin, C.J.; Fu, G.B.; Tang, L.; Wu, H.; Huang, S.; Zhao, L.H.; Zeng, M.; et al. Blocking preferential glucose uptake sensitizes liver tumor-initiating cells to glucose restriction and sorafenib treatment. Cancer Lett. 2017, 388, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Zhu, X.D.; Ao, J.Y.; Ye, B.G.; Zhang, Y.Y.; Chai, Z.T.; Wang, C.H.; Shi, W.K.; Cao, M.Q.; Li, X.L.; et al. Colony-stimulating factor-1-induced AIF1 expression in tumorassociated macrophages enhances the progression of hepatocellular carcinoma. Oncoimmunology 2017, 6, e1333213. [Google Scholar] [CrossRef] [Green Version]
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut microbiota promotes obesity-associated liver cancer through PGE2-mediated suppression of antitumor immunity. Cancer Discov. 2017, 7, 522–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.B.; Zhuang, X.; Ju, S.; Xiang, X.; Mu, J.; Liu, Y.; Jiang, H.; Zhang, L.; Mobley, J.; McClain, C.; et al. Exosome-like nanoparticles from intestinal mucosal cells carry prostaglandin E2 and suppress activation of liver NKT cells. J. Immunol. 2013, 190, 3579–3589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Inokuchi, S.; Roh, Y.S.; Song, J.; Loomba, R.; Park, E.J.; Seki, E. Transforming growth factor-beta signaling in hepatocytes promotes hepatic fibrosis and carcinogenesis in mice with hepatocyte-specific deletion of TAK1. Gastroenterology 2013, 144, 1042–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, W.; Liu, X.; Ma, H.; Zhang, H.; Song, X.; Gao, L.; Liang, X.; Ma, C. Tim-3 fosters HCC development by enhancing TGF-beta-mediated alternative activation of macrophages. Gut 2015, 64, 1593–1604. [Google Scholar] [CrossRef]
- Puche, J.E.; Saiman, Y.; Friedman, S.L. Hepatic stellate cells and liver fibrosis. Compr. Physiol. 2013, 3, 1473–1492. [Google Scholar]
- Park, S.-A.; Kim, M.-J.; Park, S.-Y.; Kim, J.-S.; Lim, W.; Nam, J.-S.; Sheen, Y.Y. TIMP-1 mediatesTGF-dependent crosstalk between hepatic stellate and cancer cells via FAK signaling. Sci. Rep. 2015, 5, 16492. [Google Scholar] [CrossRef]
- Dituri, F.; Cossu, C.; Mancarella, S.; Giannelli, G. The Interactivity between TGF and BMP signaling in organogenesis, fibrosis, and cancer. Cells 2019, 8, 1130. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.X.; Ling, Y.; Wang, H.Y. Role of non-resolving inflammation in hepatocellular carcinoma development and progression. npj Precis. Oncol. 2018, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, E.; Achen, M.G.; Breier, G.; Risau, W. Hypoxia-induced transcriptional activation and increased mRNA stability of vascular endothelial growth factor in C6 glioma cells. J. Biol. Chem. 1995, 270, 19761–19766. [Google Scholar] [CrossRef] [Green Version]
- Forsythe, J.A.; Jiang, B.H.; Iyer, N.V.; Agani, F.; Leung, S.W.; Koos, R.D.; Semenza, G.L. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 1996, 16, 4604–4613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, R.K. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science 2005, 307, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.C.; Chen, J.; Ding, Y.T. Hypoxic and highly angiogenic non-tumor tissues surrounding hepatocellular carcinoma: The “niche” of endothelial progenitor cells. Int. J. Mol. Sci. 2010, 11, 2901–2909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.A.; Chang, L.L.; Zhu, H.; He, Q.J.; Yang, B. Hypoxic microenvironment and hepatocellular carcinoma treatment. Hepatoma Res. 2018, 4, 26. [Google Scholar] [CrossRef]
- Chouaib, S.; Noman, M.Z.; Kosmatopoulos, K.; Curran, M.A. Hypoxic stress: Obstacles and opportunities for innovative immunotherapy of cancer. Oncogene 2017, 36, 439–445. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.B.; Park, J.B.; Song, T.J.; Choi, S.Y. Molecular mechanism of HIF-1-independent VEGF expression in a hepatocellular carcinoma cell line. Int. J. Mol. Med. 2011, 28, 449–454. [Google Scholar]
- Xiong, X.X.; Qiu, X.Y.; Hu, D.X.; Chen, X.Q. Advances in hypoxia-mediated mechanisms in hepatocellular carcinoma. Mol. Pharm. 2017, 92, 246–255. [Google Scholar] [CrossRef] [Green Version]
- Guise, C.P.; Mowday, A.M.; Ashoorzadeh, A.; Yuan, R.; Lin, W.H.; Wu, D.H.; Smaill, J.B.; Patterson, A.V.; Ding, K. Bioreductive prodrugs as cancer therapeutics: Targeting tumor hypoxia. Chin. J. Cancer 2014, 33, 80–86. [Google Scholar] [CrossRef]
- Moen, I.; Stuhr, L.E. Hyperbaric oxygen therapy and cancer—A review. Target Oncol. 2012, 7, 233–242. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Zhang, X.; Cheng, W.; Chen, J.; Ma, C.; Yang, B.; Hua, Z.C. Tumor-specific delivery of histidine-rich glycoprotein suppresses tumor growth and metastasis by anti-angiogenesis and vessel normalization. Curr. Gene Ther. 2014, 14, 75–85. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.X. Inflammatory immune responses in tumor microenvironment and metastasis of hepatocellular carcinoma. Cancer Microenviron. 2012, 5, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zhang, J.X.; Wang, H.; Wang, G.L.; Hu, Q.G.; Zheng, Q.C. Hepatocellular carcinoma and macrophage interaction induced tumor immunosuppression via Treg requires TLR4 signaling. World J. Gastroenterol. 2012, 18, 2938–2947. [Google Scholar] [CrossRef]
- Chen, K.J.; Lin, S.Z.; Zhou, L.; Xie, H.Y.; Zhou, W.H.; Taki-Eldin, A.; Zheng, S.S. Selective recruitment of regulatory T cell through CCR6-CCL20 in hepatocellular carcinoma fosters tumor progression and predicts poor prognosis. PLoS ONE 2011, 6, e24671. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Xu, D.; Liu, Z.; Shi, M.; Zhao, P.; Fu, B.; Zhang, Z.; Yang, H.; Zhang, H.; Zhou, C.; et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 2007, 132, 2328–2339. [Google Scholar] [CrossRef]
- Behboudi, S.; Boswell, S.; Williams, R. Cell-mediated immune responses to alpha-fetoprotein and other antigens in hepatocellular carcinoma. Liver Int. 2010, 30, 521–526. [Google Scholar] [CrossRef]
- Deng, Y.; Cheng, J.; Fu, B.; Liu, W.; Chen, G.; Zhang, Q.; Yang, Y. Hepatic carcinoma-associated fibroblasts enhance immune suppression by facilitating the generation of myeloid-derived suppressor cells. Oncogene 2017, 36, 1090–1101. [Google Scholar] [CrossRef] [PubMed]
- Nan, J.; Xing, Y.F.; Hu, B.; Tang, J.X.; Dong, H.M.; He, Y.M.; Ruan, D.Y.; Ye, Q.J.; Cai, J.R.; Ma, X.K.; et al. Endoplasmic reticulum stress induced LOX-1(+) CD15(+) polymorphonuclear myeloid-derived suppressor cells in hepatocellular carcinoma. Immunology 2018, 154, 144–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, Y.; Shimosegawa, T. Significant roles of regulatory T cells and myeloid derived suppressor cells in hepatitis B virus persistent infection and hepatitis B virus-related HCCs. Int. J. Mol. Sci. 2015, 16, 3307–3322. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhou, J.; Liu, X.; Feng, Y.; Yang, W.; Wu, F.; Cheung, O.K.; Sun, H.; Zeng, X.; Tang, W. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, Z.; Wang, L.; Tian, G.; Tian, J.; Yang, Z.; Cao, G.; Zhou, H.; Zhao, L.; Wu, Z.; et al. Critical role of myeloid-derived suppressor cells in tumor induced liver immune suppression through inhibition of NKT cell function. Front. Immunol. 2017, 8, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, M.K.; Sinha, P.; Clements, V.K.; Rodriguez, P.; Ostrand-Rosenberg, S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 2010, 70, 68–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dardalhon, V.; Anderson, A.C.; Karman, J.; Apetoh, L.; Chandwaskar, R.; Lee, D.H.; Cornejo, M.; Nishi, N.; Yamauchi, A.; Quintana, F.J.; et al. Tim-3/galectin-9 pathway: Regulation of Th1 immunity through promotion of CD11b+Ly-6G+ myeloid cells. J. Immunol. 2010, 185, 1383–1392. [Google Scholar] [CrossRef] [Green Version]
- Hoechst, B.; Voigtlaender, T.; Ormandy, L.; Gamrekelashvili, J.; Zhao, F.; Wedemeye, H.; Lehner, F.; Manns, M.P.; Greten, T.F.; Korangy, F. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology 2009, 50, 799–807. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Movahedi, K.; Laoui, D.; Gysemans, C.; Baeten, M.; Stangé, G.; Van den Bossche, J.; Mack, M.; Pipeleers, D.; In’t Veld, P.; De Baetselier, P.; et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010, 70, 5728–5739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolas-Avila, J.A.; Adrover, J.M.; Hidalgo, A. Neutrophils in homeostasis, immunity, and Cancer. Immunity 2017, 46, 15–28. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.T.; Yuan, J.H.; Ma, J.Z.; Yang, W.J.; Liu, X.N.; Yin, Y.P.; Liu, Y.; Pan, W.; Sun, S.H. CTGF secreted by mesenchymal-like hepatocellular carcinoma cells plays a role in the polarization of macrophages in hepatocellular carcinoma progression. BioMed Pharm. 2017, 95, 111–119. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, J.; Xu, D.; Gao, X.M.; Zhang, Z.; Hsu, J.L.; Li, C.W.; Lim, S.O.; Sheng, Y.Y.; Zhang, Y.; et al. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut 2019, 68, 1653–1666. [Google Scholar] [CrossRef]
- Zhou, J.; Ding, T.; Pan, W.; Zhu, L.Y.; Li, L.; Zheng, L. Increased intratumoral regulatory T cells are related to intratumoral macrophages and poor prognosis in hepatocellular carcinoma patients. Int. J. Cancer 2009, 125, 1640–1648. [Google Scholar] [CrossRef] [PubMed]
- Andzinski, L.; Kasnitz, N.; Stahnke, S.; Wu, C.F.; Gereke, M.; von Köckritz-Blickwede, M.; Schilling, B.; Brandau, S.; Weiss, S.; Jablonska, J. Type I IFNs induce anti-tumor polarization of tumor associated neutrophils in mice and human. Int. J. Cancer 2016, 138, 1982–1993. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, J.; Shaul, M.E.; Mishalian, I.; Hovav, A.H.; Levy, L.; Zolotriov, L.; Granot, Z.; Fridlender, Z.G. Tumor-associated neutrophils induce apoptosis of nonactivated CD8 T-cells in a TNFalpha and NO-dependent mechanism, promoting a tumor-supportive environment. Oncoimmunology 2017, 6, e1356965. [Google Scholar] [CrossRef] [PubMed]
- Chew, V.; Chen, J.; Lee, D.; Loh, E.; Lee, J.; Lim, K.H.; Weber, A.; Slankamenac, K.; Poon, R.T.; Yang, H.; et al. Chemokine-driven lymphocyte infiltration: An early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut 2012, 61, 427–438. [Google Scholar] [CrossRef] [Green Version]
- Flecken, T.; Schmidt, N.; Hild, S.; Gostick, E.; Drognitz, O.; Zeiser, R.; Schemmer, P.; Bruns, H.; Eiermann, T.; Price, D.A.; et al. Immunodominance and functional alterations of tumor-associated antigen specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology 2014, 59, 1415–1426. [Google Scholar] [CrossRef] [Green Version]
- Kurebayashi, Y.; Ojima, H.; Tsujikawa, H.; Kubota, N.; Maehara, J.; Abe, Y.; Kitago, M.; Shinoda, M.; Kitagawa, Y.; Sakamoto, M. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018, 68, 1025–1041. [Google Scholar] [CrossRef]
- Zhou, S.L.; Zhou, Z.J.; Hu, Z.Q.; Huan, X.W.; Wang, Z.; Chen, E.B.; Fan, J.; Cao, Y.; Dai, Z.; Zhou, J. Tumor-associated neutrophils recruit macrophages and T regulatory cells to promote progression of hepatocellular carcinoma and resistance to Sorafenib. Gastroenterology 2016, 150, 1646–1658. [Google Scholar] [CrossRef] [Green Version]
- Quan, H.; Zhou, F.; Nie, D.; Chen, Q.; Cai, X.; Shan, X.; Zhou, Z.; Chen, K.; Huang, A.; Li, S.; et al. Hepatitis C virus core protein epigenetically silences SFRP1 and enhances HCC aggressiveness by inducing epithelial-mesenchymal transition. Oncogene 2013. [Google Scholar] [CrossRef] [Green Version]
- Turati, F.; Trichopoulos, D.; Polesel, J.; Bravi, F.; Rossi, M.; Talamini, R.; Franceschi, S.; Montella, M.; Trichopoulou, A.; La Vecchia, C.; et al. Mediterranean diet and hepatocellular carcinoma. J. Hepatol. 2014, 60, 606–611. [Google Scholar] [CrossRef]
- Fox, J.G.; Feng, Y.; Theve, E.J.; Raczynski, A.R.; Fiala, J.L.; Doernte, A.L.; Williams, M.; McFaline, J.L.; Essigmann, J.M.; Schauer, D.B.; et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut 2010, 59, 88–97. [Google Scholar] [CrossRef]
- Lv, L.H.; Wan, Y.L.; Lin, Y.; Zhang, W.; Yang, M.; Li, G.L.; Lin, H.M.; Shang, C.Z.; Chen, Y.J.; Min, J. Anticancer drugs cause release of exosomes with heat shock proteins from human hepatocellular carcinoma cells that elicit effective natural killer cell antitumor responses in vitro. J. Biol. Chem. 2012, 287, 15874–15885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridget, P.; Fong, K.L.; Kelley, R.K. Immunotherapy in hepatocellular carcinoma: The complex interface between inflammation, fibrosis, and the immune response. J. Immunother. Cancer 2019, 7, 267. [Google Scholar]
- Homann, C.; Varming, K.; Hogase, K.; Mollnes, T.E.; Graudal, N.; Thomsen, A.C.; Garred, P. Acquired C3 deficiency in patients with alcoholic cirrhosis predisposes to infection and increased mortality. Gut 1997, 40, 544–549. [Google Scholar] [CrossRef]
- Homann, C.; Garred, P.; Hasselqvist, P.; Graudal, N.; Thiel, S.; Thomsen, A.C. Mannan-binding protein and complement dependent opsonization in alcoholic cirrhosis. Liver 1995, 15, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Chew, V.; Lai, L.; Pan, L.; Lim, C.J.; Li, J.; Ong, R.; Chua, C.; Leong, J.Y.; Lim, K.H.; Toh, H.C.; et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using highdimensional proteomic and transcriptomic analyses. Proc. Natl. Acad. Sci. USA 2017, 114, E5900–E5909. [Google Scholar] [CrossRef] [Green Version]
- Snelgrove, S.L.; Abeynaike, L.D.; Thevalingam, S.; Deane, A.; Hickey, M.J. Regulatory T Cell Transmigration and Intravascular Migration Undergo Mechanistically Distinct Regulation at Different Phases of the Inflammatory Response. J Immunol. 2019, 203, 2850–2861. [Google Scholar] [CrossRef]
- Wu, K.; Kryczek, I.; Chen, L.; Zou, W.; Welling, T.H. Kupffer cell suppression of CD8+ T cells in human hepatocellular carcinoma is mediated by B7-H1/programmed death-1 interactions. Cancer Res. 2009, 69, 8067–8075. [Google Scholar] [CrossRef] [Green Version]
- Do, A.L.; Wong, C.R.; Nguyen, L.H.; Nguyen, V.G.; Trinh, H.; Nguyen, M.H. Hepatocellular carcinoma incidence in noncirrhotic patients with chronic hepatitis B and patients with cirrhosis of all etiologies. J. Clin. Gastroenterol. 2014, 48, 644–649. [Google Scholar] [CrossRef]
- Kao, J.H.; Chen, P.J.; Lai, M.Y.; Chen, D.S. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 2003, 124, 327–334. [Google Scholar] [CrossRef]
- Reig, M.; Marino, Z.; Perello, C.; Inarrairaegui, M.; Ribeiro, A.; Lens, S.; Díaz, A.; Vilana, R.; Darnell, A.; Varela, M.; et al. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J. Hepatol. 2016, 65, 719–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirei, K.; Kanda, T.; Nakamura, H.; Matsuoka, S.; Takayama, T.; Sugitani, M.; Moriyama, M. Persistent hepatic inflammation plays a role in hepatocellular carcinoma after sustained virological response in patients with HCV infection. Int. J. Med. Sci. 2018, 15, 466–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitaoka, S.; Shiota, G.; Kawasaki, H. Serum levels of interleukin-10, interleukin-12 and soluble interleukin-2 receptor in chronic liver disease type C. Hepatogastroenterology 2003, 50, 1569–1574. [Google Scholar]
- Huang, Y.S.; Hwang, S.J.; Chan, C.Y.; Wu, J.C.; Chao, Y.; Chang, F.Y.; Lee, S.D. Serum levels of cytokines in hepatitis C-related liver disease: A longitudinal study. Zhonghua Yi Xue Za Zhi (Taipei) 1999, 62, 327–333. [Google Scholar] [PubMed]
- Song, L.H.; Binh, V.Q.; Duy, D.N.; Kun, J.F.; Bock, T.C.; Kremsner, P.G.; Luty, A.J. Serum cytokine profiles associated with clinical presentation in Vietnamese infected with hepatitis B virus. J. Clin. Virol. 2003, 28, 93–103. [Google Scholar] [CrossRef]
- Lim, C.J.; Lee, Y.H.; Pan, L.; Lai, L.; Chua, C.; Wasser, M.; Lim, T.K.H.; Yeong, J.; Toh, H.C.; Lee, S.Y.; et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus related hepatocellular carcinoma. Gut 2019, 68, 916–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, I.C.; Chang, J.; Kim, K.; Park, S.M. Aspirin use and risk of hepatocellular carcinoma in a national cohort study of Korean adults. Sci. Rep. 2018, 8, 4968. [Google Scholar] [CrossRef]
- Hernandez-Vargas, H.; Lambert, M.P.; Le Calvez-Kelm, F.; Gouysse, G.; McKayChopin, S.; Tavtigian, S.V.; Scoazec, J.Y.; Herceg, Z. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS ONE 2010, 5, e9749. [Google Scholar] [CrossRef]
- Sia, D.; Jiao, Y.; Martinez-Quetglas, I.; Kuchuk, O.; Villacorta-Martin, C.; Castro de Moura, M.; Putra, J.; Camprecios, G.; Bassaganyas, L.; Akers, N.; et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 2017, 153, 812–826. [Google Scholar] [CrossRef] [Green Version]
- Boutari, C.; Perakakis, N.; Mantzoros, C.S. Association of adipokines with development and progression of nonalcoholic fatty liver disease. Endocrinol. Metab. (Seoul) 2018, 33, 33–43. [Google Scholar] [CrossRef]
- Haas, J.T.; Vonghia, L.; Mogilenko, D.A.; Verrijken, A.; Molendi-Coste, O.; Fleury, S.; Deprince, A.; Nikitin, A.; Woitrain, E.; Ducrocq-Geoffroy, L.; et al. Transcriptional network analysis implicates altered hepatic immune function in NASH development and resolution. Nat. Metab. 2019, 1, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Shalapour, S.; Lin, X.J.; Bastian, I.N.; Brain, J.; Burt, A.D.; Aksenov, A.A.; Vrbanac, A.F.; Li, W.; Perkins, A.; Matsutani, T.; et al. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 2017, 551, 340–345. [Google Scholar] [CrossRef]
- Chen, Y.; Ramjiawan, R.R.; Reiberger, T.; Ng, M.R.; Hato, T.; Huang, Y.; Ochiai, H.; Kitahara, S.; Unan, E.C.; Reddy, T.P.; et al. CXCR4 inhibition in tumor microenvironment facilitates anti-programmed death receptor-1 immunotherapy in sorafenib-treated hepatocellular carcinoma in mice. Hepatology 2015, 61, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Brown, Z.J.; Yu, S.J.; Heinrich, B.; Ma, C.; Fu, Q.; Sandhu, M.; Agdashian, D.; Zhang, Q.; Korangy, F.; Greten, T.F. Indoleamine 2,3-dioxygenase provides adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma. Cancer Immunol. Immunother. 2018, 67, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Kato, Y.; Ozawa, Y.; Kodama, K.; Ito, J.; Ichikawa, K.; Yamada, K.; Hori, Y.; Tabata, K.; Takase, K.; et al. Immunomodulatory activity of Lenvatinib contributes to antitumor activity in the Hepa1-6 hepatocellular carcinoma model. Cancer Sci. 2018, 109, 3993–4002. [Google Scholar] [CrossRef] [PubMed]
- Llopiz, D.; Ruiz, M.; Villanueva, L.; Iglesias, T.; Silva, L.; Egea, J.; Lasarte, J.J.; Pivette, P.; Trochon-Joseph, V.; Vasseur, B.; et al. Enhanced anti-tumor efficacy of checkpoint inhibitors in combination with the histone deacetylase inhibitor Belinostat in a murine hepatocellular carcinoma model. Cancer Immunol. Immunother. 2019, 68, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Xin, B.; Wu, P.; Lin, C.H.; Peng, C.; Wang, G.; Lee, J.; Lu, L.F.; Feng, G.S. An efficient combination immunotherapy for primary liver cancer by harmonized activation of innate and adaptive immunity in mice. Hepatology 2019, 69, 2518–2532. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.J.; Ma, C.; Heinrich, B.; Brown, Z.; Sandhu, M.; Zhang, Q.; Fu, Q.; Agdashian, D.; Rosato, U.; Korangy, F.; et al. Targeting the crosstalk between cytokine-induced killer cells and myeloid-derived suppressor cells in hepatocellular carcinoma. J. Hepatol. 2019, 70, 449–457. [Google Scholar] [CrossRef]
- Liu, Z.; Lu, Z.; Jing, R.; Zuo, B.; Gao, X.; Han, G.; Qi, H.; Wu, L.; Liu, Y.; Yin, H. Alarmin augments the antitumor immunity of lentiviral vaccine in ectopic, orthotopic and autochthonous hepatocellular carcinoma mice. Theranostic 2019, 9, 4006–4018. [Google Scholar] [CrossRef]
- Liu, H.; Xu, Y.; Xiang, J.; Long, L.; Green, S.; Yang, Z.; Zimdahl, B.; Lu, J.; Cheng, N.; Horan, L.H.; et al. Targeting alpha-fetoprotein (AFP)-MHC complex with CAR T-cell therapy for liver cancer. Clin. Cancer Res. 2017, 23, 478–488. [Google Scholar] [CrossRef] [Green Version]
- Batra, S.A.; Rathi, P.; Guo, L.; Courtney, A.N.; Fleurence, J.; Balzeau, J.; Shaik, R.S.; Nguyen, T.P.; Wu, M.F.; Bulsara, S.; et al. Glypican-3-specific CAR T cells co-expressing IL15 and IL21 have superior expansion and antitumor activity against hepatocellular carcinoma. Cancer Immunol. Res. 2020, 8, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Abou-Alfa, G.K.; Chan, S.L.; Furuse, J.; Galle, P.R.; Kelley, R.K.; Qin, S.; Armstrong, J.; Darilay, A.; Vlahovic, G.; Negro, A.; et al. A randomized, multicenter phase 3 study of durvalumab (D) and tremelimumab (T) as first-line treatment in patients with unresectable hepatocellular carcinoma (HCC): HIMALAYA study. J. Clin. Oncol. 2018, 36. [Google Scholar] [CrossRef]

| Animal Model | Results | Reference |
|---|---|---|
| MUP-uPA transgenic mice | HCC progenitor cells showed autocrine IL-6 signaling that stimulates in vivo growth and malignant progression | [54] |
| C.B-17 wild-type mice with an intact immunity, C.B-17 SCID mice with an impaired adaptive immune response, C.B-17 SCID/beige mice with defects in NK-cell and macrophage function | Impaired immune-mediated clearance of pre-malignant senescent hepatocytes secreting chemo- and cytokines resulted in the development of murine HCCs | [55] |
| Rat model of choline-deficient, L-amino acid-defined diet (CDAA)-Male Sprague-Dawley rats Nude mice | Aspirin or nimesulide administration decreased the number of preneoplastic and neoplastic nodules Celecoxib treatment was highly effective in inhibiting the multiplicity and size of liver preneoplastic lesions COX (cyclooxygenase)-2 inhibitors (celecoxib and meloxicam) enhanced tumor cell apoptosis and reduced proliferation | [57] |
| IL-6–/–/TLR-4–/– C57BL/6J mice | Hepatic stem/progenitor marker CD133 was responsible for driving and maintaining HCC. CD133 expression can be induced by IL-6 and hypoxic conditions in a STAT3-dependent manner | [59] |
| RAW264.7-shNC/shAIF1 cells and Hepa1–6 cells injected in C57BL/6 mice | Mouse cytokine antibody array analysis showed that macrophages overexpressing AIF1 (Allograft Inflammatory Factor 1) secreted high levels of CXCL16, which is reported to facilitate the migration and invasion of HCC | [60] |
| C57/BL6 mice | The gut microbiota–driven COX-2 pathway produced PG (Prostaglandin)-E2 which plays a pivotal role in suppressing antitumor immunity and promoting HCC onset | [61] |
| C57BL/6 mice | Obesity induced alterations of gut microbiota increasing the levels of DCA (deoxycholic acid); the enterohepatic circulation of DCA provoked senescence-associated secretory phenotype in hepatic stellate cells, which in turn secreted inflammatory and tumour-promoting factors in the liver facilitating HCC development | [62] |
| C57BL/6 mice | Pro-inflammatory conditions enhanced the migration of PGE2 carried by nanoparticles from the intestine to the liver, where they induced the inactivation of natural killer T cells (cancer cells escape from immune control) | [63] |
| Tak1ΔHep mice | TGF-β promoted HCC development by inducing hepatocyte apoptosis and compensatory proliferation in early phases of tumorigenesis, and inducing expression of anti-apoptotic, pro-oncogenic and angiogenic factors during tumor progression. | [64] |
| BALB/c mice | Tim-3 (immune regulator, involved in many inflammation-related diseases) expression in tumor-associated macrophages promoted HCC growth | [65] |
| Name | Phase | Line of Treatment | Strategy | Primary Endpoint |
|---|---|---|---|---|
| NCT03841201 | 2 | I | Nivolumab (anti-PD-1/PD-L1) + Lenvatinib (VEGFRs inhibitor) | ORR (Objective response rate) Safety and tolerability |
| NCT03630640 | 2 | neo + adj | Nivolumab | RFS (Recurrence Free Survival) |
| NCT02576509 | 3 | I | Nivolumab vs. Sorafenib (Raf inhibotor) | OS (Overall Survival) |
| NCT03203304 | 1 | II | Nivolumab Ipilimumab (anti CTLA-4) | AEs (Adverse Events) |
| NCT01658878 | 1/2 | I | Nivolumab Sorafenib Nivolumab + Ipilimumab Nivolumab + Cabozantinib (RTK inhibitor) Nivolumab + Ipilimumab + Cabozantinib | AEsORR |
| NCT03510871 | 2 | neo | Nivolumab + Ipilimumab | Tumor shrinkage |
| NCT03841110 | 1 | II | FT500 (NK cell product) +/− Nivolumab, Pembrolizumab (anti-PD-1/PD-L1), Atezolizumab (anti PD-L1), Cyclophosphamide, Fludarabine | DLT (Dose Limiting Toxicities) |
| NCT03682276 | 1/2 | I | Ipilimumab + Nivolumab | Delay to surgery AEs |
| NCT03228667 | 2 | II | ALT-803 (IL-15 superagonist) + Pembrolizumab ALT-803 + Nivolumab ALT-803 + Atezolizumab ALT-803 + Avelumab (anti PD-L1) | ORR |
| NCT04134559 | 2 | II | Pembrolizumab | irBOR (immune-related Best Overall Response) |
| NCT02595866 | 1 | II | Pembrolizumab | AEs ECIs (Events of Clinical Interest) |
| NCT03337841 | 2 | neo + adj | Pembrolizumab | One-year RFS |
| NCT04099277 | 1 | II | LY343515 1+/− Pembrolizumab | DLT |
| NCT03222076 | 2 | neo | Nivolumab +/− Ipilimumab | AEs |
| NCT03383458 | 3 | adj | Nivolumab | RFS |
| NCT03655002 | 1 | II | Nivolumab, Cyclophosphamide, IRX-2 (cytokine-based biologic agent) | Safety |
| NCT03812562 | 1 | I | Yttrium Y 90 glass microspheres, Nivolumab | RR (Recurrence Rate) |
| NCT03867084 | 3 | adj | Pembrolizumab | RFS OS |
| NCT03755739 | 2/3 | I | Pembrolizumab | OS |
| NCT02702401 | 3 | II | Pembrolizumab | PFS OS |
| NCT03062358 | 3 | II | Pembrolizumab | OS |
| NCT02702414 | 2 | II | Pembrolizumab | ORR |
| NCT03006926 | 1 | II | Pembrolizumab + Levantinib | AEs DLT |
| NCT03713593 | 3 | I | Levantinib +/− Pembrolizumab | OS |
| NCT02940496 | 1/2 | II | Pembrolizumab Pembrolizumab + elbasvir/grazoprevir + ribavirin (antiviral drugs) | DLT |
| NCT03511222 | 2 | I | Vorolanib (antiangiogenic agent) + Pembrolizumab | RP2D (Recommended phase II dose) |
| NCT03299946 | 1 | neo | Cabozantinib + Nivolumab | AEs proceed to surgery |
| NCT03412773 | 3 | I | Tislelizumab (anti PD-1/PD-L1) | OS ORR |
| Name | Phase | Line of Treatment | Strategy | Primary Endpoint |
|---|---|---|---|---|
| NCT03638141 | 2 | II | Durvalumab (anti PD-L1)+ Tremelimumab (anti CTLA-4) | ORR (Objective Response Rate) |
| NCT03298451 | 3 | I | Durvalumab +/− Tremelimumab | OS (Overall Survival) |
| NCT03847428 | 3 | I | Durvalumab + Bevacizumab (anti-VEGFA) | RFS (Recurrence-Free Survival) |
| NCT03434379 | 3 | I | Atezolizumab (anti PD-L1) + Bevacizumab | PFS (Progression-Free Survival) OS |
| NCT02715531 | 1 | I | Atezolizumab + Bevacizumab | AEs (Adverse Events) OR (Objective Response) PFS |
| NCT03755791 | 3 | I | Cabozantinib (RTK inhibitor) + Atezolizumab | PFS OS |
| NCT03937830 | 2 | II | Durvalumab + Bevacizumab + Doxorubicin (TACE) | PFS |
| NCT02519348 | 2 | II | Durvalumab +/− Tremelimumab Tremelimumab +/− Durvalumab Durvalumab +/− Bevacizumab | AEs DLT (Dose Limiting Toxicity) |
| Name | Phase | Line of Treatment | Strategy | Primary Endpoint |
|---|---|---|---|---|
| NCT03864211 | 1/3 | II | Toriplimab (anti-PD-1) | AEs (Adverse Events) ORR (Overall Response Rate) |
| NCT03914352 | SHR-1210 (anti-PD-1) | OS (Overall Survival) DFS (Disease-Free Survival) | ||
| NCT03605706 | 3 | I | SHR-1210 + FOLFOX4 FOLFOX4 Sorafenib (Raf inhibitor) | OS |
| NCT04191889 | 2 | FOLFOX + Apatinib (VEGFR-2 inhibitor) + Camrelizumab (SHR-1210) | ORR | |
| NCT04152356 | I | PD-1 + Sorafenib | DFS | |
| NCT04220944 | 1 | I | Sintilimab (anti-PD-1) | PFS (Progression Free Survival) |
| NCT04174781 | 2 | Sintilimab | PFS | |
| NCT04167293 | 2/3 | I | Sintilimab | 24-week PFS |
| NCT04229355 | 3 | I | Lenvatinib (VEGFRs inhibotor)vs. PD-1 inhibitor | PFS |
| NCT03949231 | 3 | Toripalimab | OS | |
| NCT03966209 | 1 | II | JS001 (PD-1 inhibitor) | Adverse Events Rate Graft Rejection Rate |
| NCT03655613 | 1/2 | II | APL-501 (PD-1 inhibitor) + APL-101 (c-Met inhibitor) | DLT (Dose Limiting Toxicity) |
| NCT04172506 | 1/2 | II | AK105 (anti PD-1) | ORR |
| NCT03680508 | 2 | I | TSR-022 (anti-TIM-3) + TSR-042 (anti-PD-1) | OR (Objective Response) |
| NCT03939975 | 2 | II | Pembrolizumab (anti-PD-1/PD-L1) Nivolumab (anti-PD-1/PD-L1) JS001 | AEs Response |
| NCT02988440 | 1 | PDR001 (anti-PD-1)+ Sorafenib | AEs | |
| NCT02795429 | 2 | II | PDR001 +/− INC280 (c-Met/HGFR inhibitor) | DLT ORR |
| NCT03474640 | 1 | II | Toripalimab | AEs |
| Name | Phase | Line of Treatment | Strategy | Primary Endpoint |
|---|---|---|---|---|
| NCT02723942 | 1/2 | CAR-T cell immunotherapy targeting GPC3 | Radiological assessment of therapeutic effect | |
| NCT02905188 | 1 | I/II | GPC3-CAR (GLYCAR T cells) + Cytoxan and Fludarabine (lymphodepleting chemotherapy) | DLT (Dose Limiting Toxicity) |
| NCT03130712 | 1/2 | II | GPC3-CART cells | AEs (Adverse Events) |
| NCT03198546 | 1 | I | GPC3 and/or TGFβ CAR-T cells | DLT |
| NCT03013712 | 1/2 | EpCAM-CAR T cells | AEs | |
| NCT03575806 | 2 | II | Autologous Tcm (central memory T cells) | DFS (Disease-free Survival) Clinical Efficacy Safety |
| NCT02839954 | 1/2 | I | anti-MUC1 CAR-pNK cells (Chimeric Antigen Receptor NK cells with specificity for MUC1) | AEs |
| NCT04106167 | Allogeneic Natural Killer (NK) cells | OS (Overall Survival) | ||
| NCT01147380 | 1 | Liver NK cell inoculation | Side Effect | |
| NCT03093688 | 1/2 | II | iNKT (invariant Natural Killer T) cells + PD-1 + CD8+ T cells | AEs ORR (Overall Response Rate) |
| NCT03319459 | 1 | II | FATE-NK100 (donor-derived NK) +/− Cetuximab (EGFR inhibitor)+/− Trastuzumab (anti-HER2) | DLT |
| NCT02882659 | 1 | II | Dendritic Killer Cell (DKC) | AEs DLT Safety |
| NCT02886897 | 1/2 | II | Autologous D-CIK (Dendritic and Cytokine-Induced Killer) cells + anti-PD-1 antibody | PFS (Progression Free Survival) |
| NCT02632006 | 1/2 | I | Pluripotent Killer T Cells expressing antibodies for PD-1 | OS |
| NCT00562666 | 1 | Autologous Gamma-delta T Lymphocytes | AEs | |
| NCT03132792 | 1 | II | Autologous genetically modified AFPᶜ³³² T cells | DLT AEs |
| NCT03592706 | 2/3 | II | Autologous IKC (Immune Killer Cells) | Change of tumor size PFS |
| NCT03998033 | 1/2 | II | ET140202 Receptor (+) T cells | AEs RP2D (Recommended Phase 2 Dose) |
| NCT02678013 | 3 | Cytotoxic T Lymphocytes (CTL) | RFS (Recurrence Free Survival) | |
| NCT03836352 | 2 | II | DPX-Survivac (T cell activating therapy) + Pembrolizumab (anti PD-1/PD-L1) +/− Cyclophosphamide | ORR AEs |
| Name | Phase | Line of Treatment | Strategy | Primary Endpoint |
|---|---|---|---|---|
| NCT03071094 | 1/2 | Pexastimogene Devacirepvec (Pexa Vec is a vaccinia virus based oncolytic immunotherapy designed to stimulate the immune system following infection and replication within tumor cells) + Nivolumab (anti PD-1/PD-L1) | Safety DLT (Dose Limiting Toxicity) Anti-tumor activity Efficacy | |
| NCT02562755 | 3 | Pexa-Vec + Sorafenib (Raf inhibitor) | OS (Overall Survival) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Refolo, M.G.; Messa, C.; Guerra, V.; Carr, B.I.; D’Alessandro, R. Inflammatory Mechanisms of HCC Development. Cancers 2020, 12, 641. https://doi.org/10.3390/cancers12030641
Refolo MG, Messa C, Guerra V, Carr BI, D’Alessandro R. Inflammatory Mechanisms of HCC Development. Cancers. 2020; 12(3):641. https://doi.org/10.3390/cancers12030641
Chicago/Turabian StyleRefolo, Maria Grazia, Caterina Messa, Vito Guerra, Brian Irving Carr, and Rosalba D’Alessandro. 2020. "Inflammatory Mechanisms of HCC Development" Cancers 12, no. 3: 641. https://doi.org/10.3390/cancers12030641
APA StyleRefolo, M. G., Messa, C., Guerra, V., Carr, B. I., & D’Alessandro, R. (2020). Inflammatory Mechanisms of HCC Development. Cancers, 12(3), 641. https://doi.org/10.3390/cancers12030641






