Gene Expression Alterations and Molecular Analysis of CHEK1 in Solid Tumors
Abstract
1. Introduction
2. Results
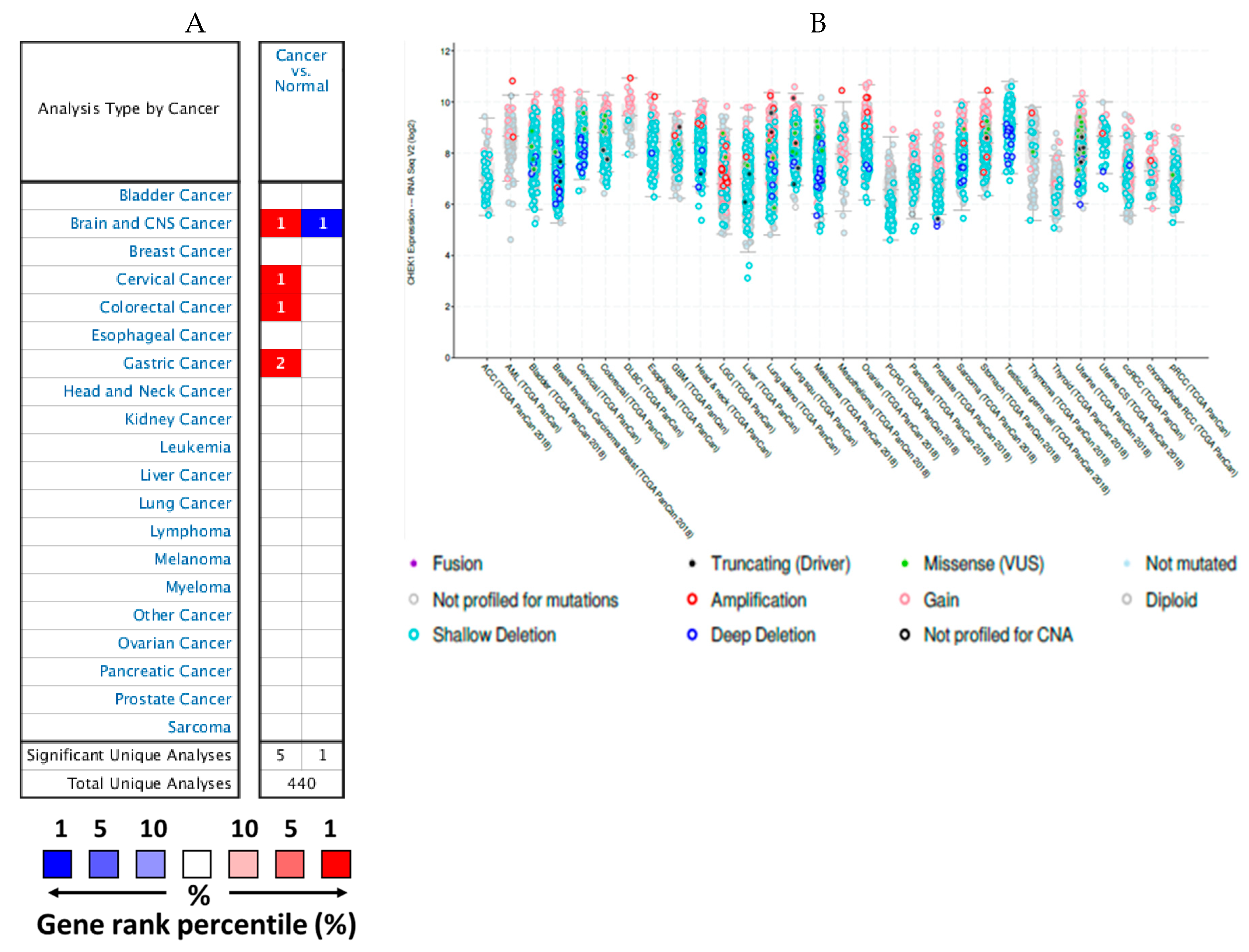
2.1. mRNA Expression Levels of CHEK1 in Cancers (Oncomine)
2.2. Expression Levels of CHEK1 in Selected Solid Tumors
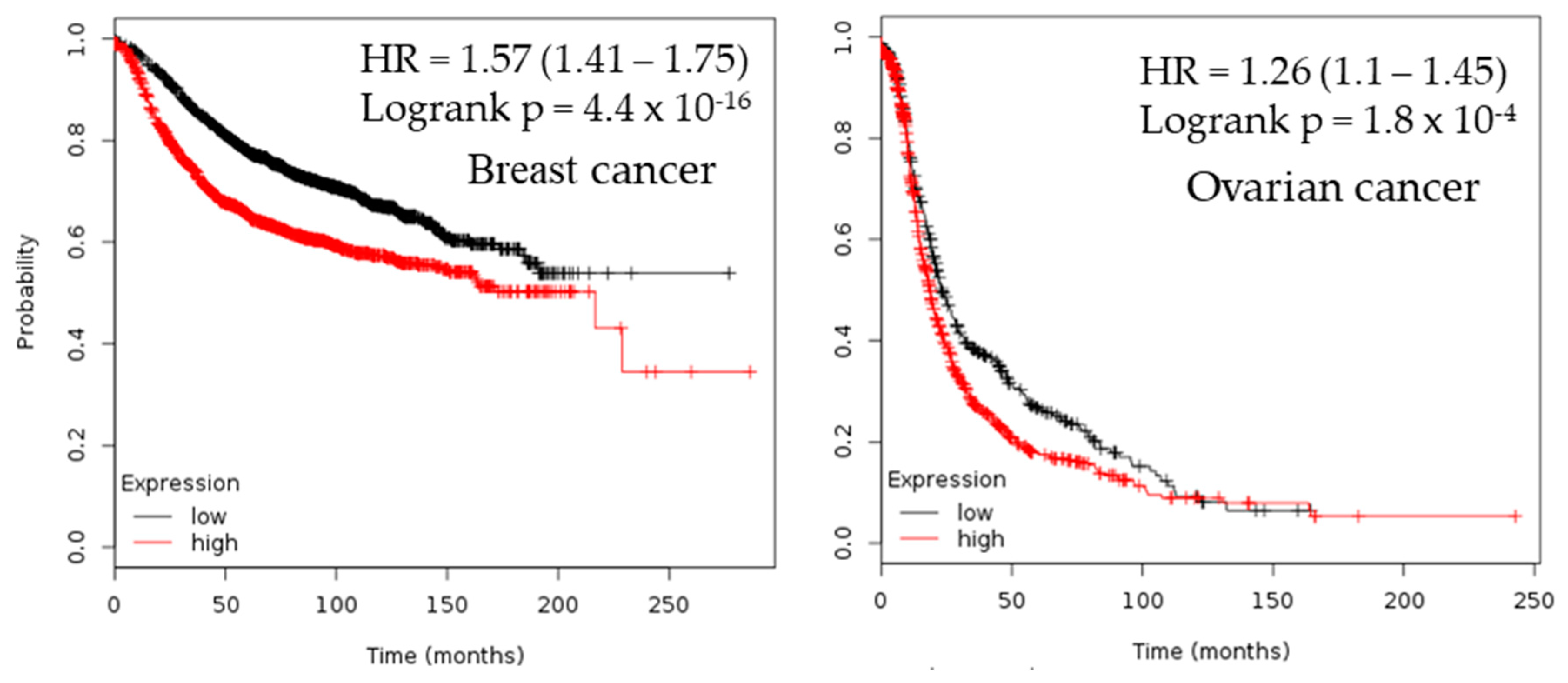
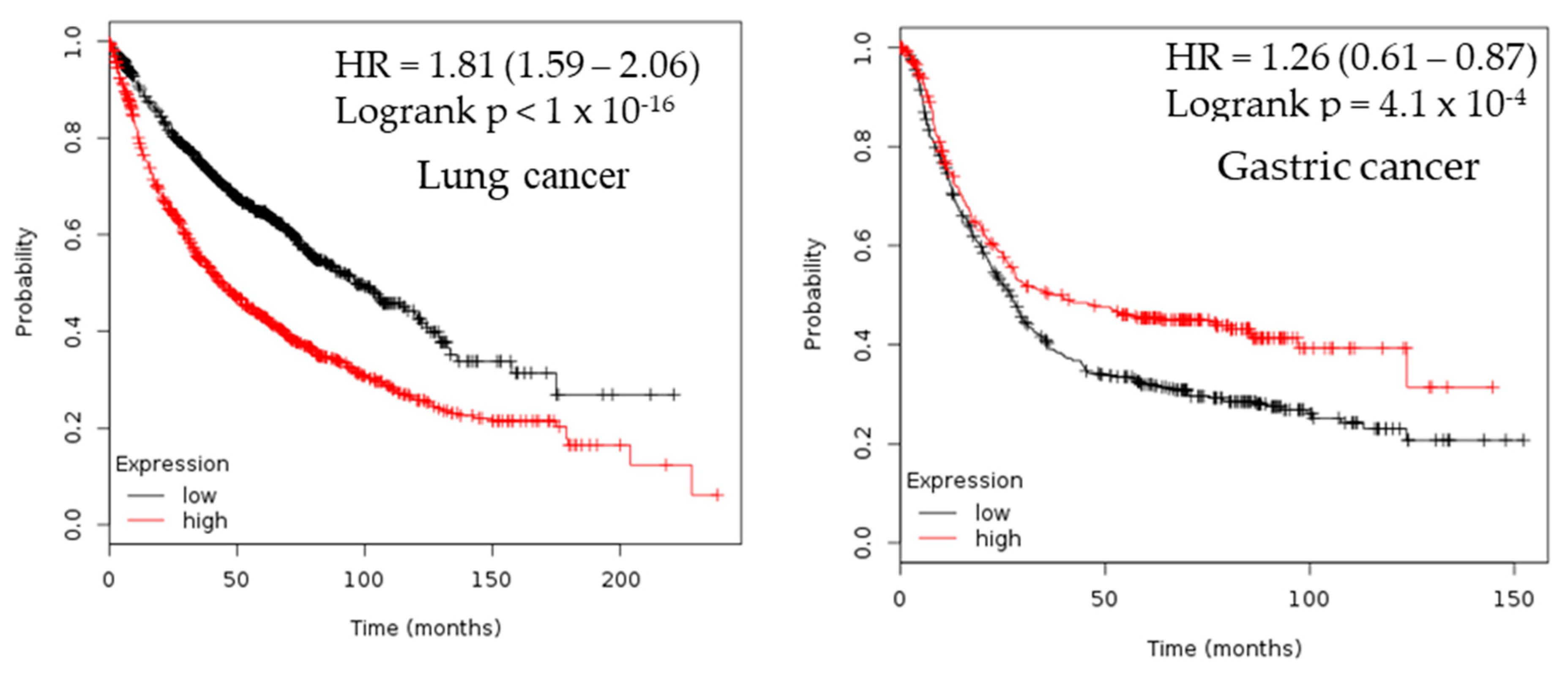
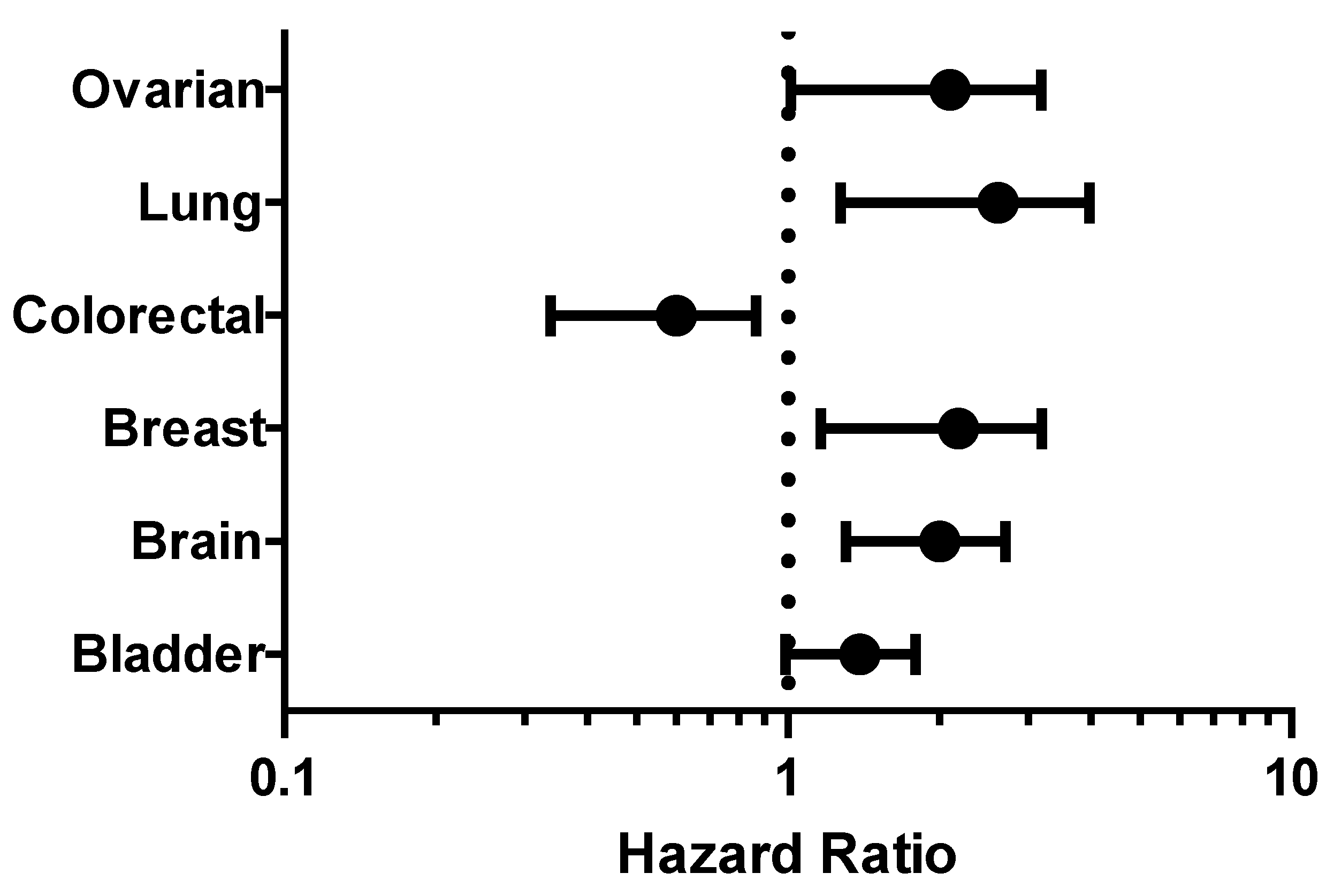
2.3. Survival Analysis
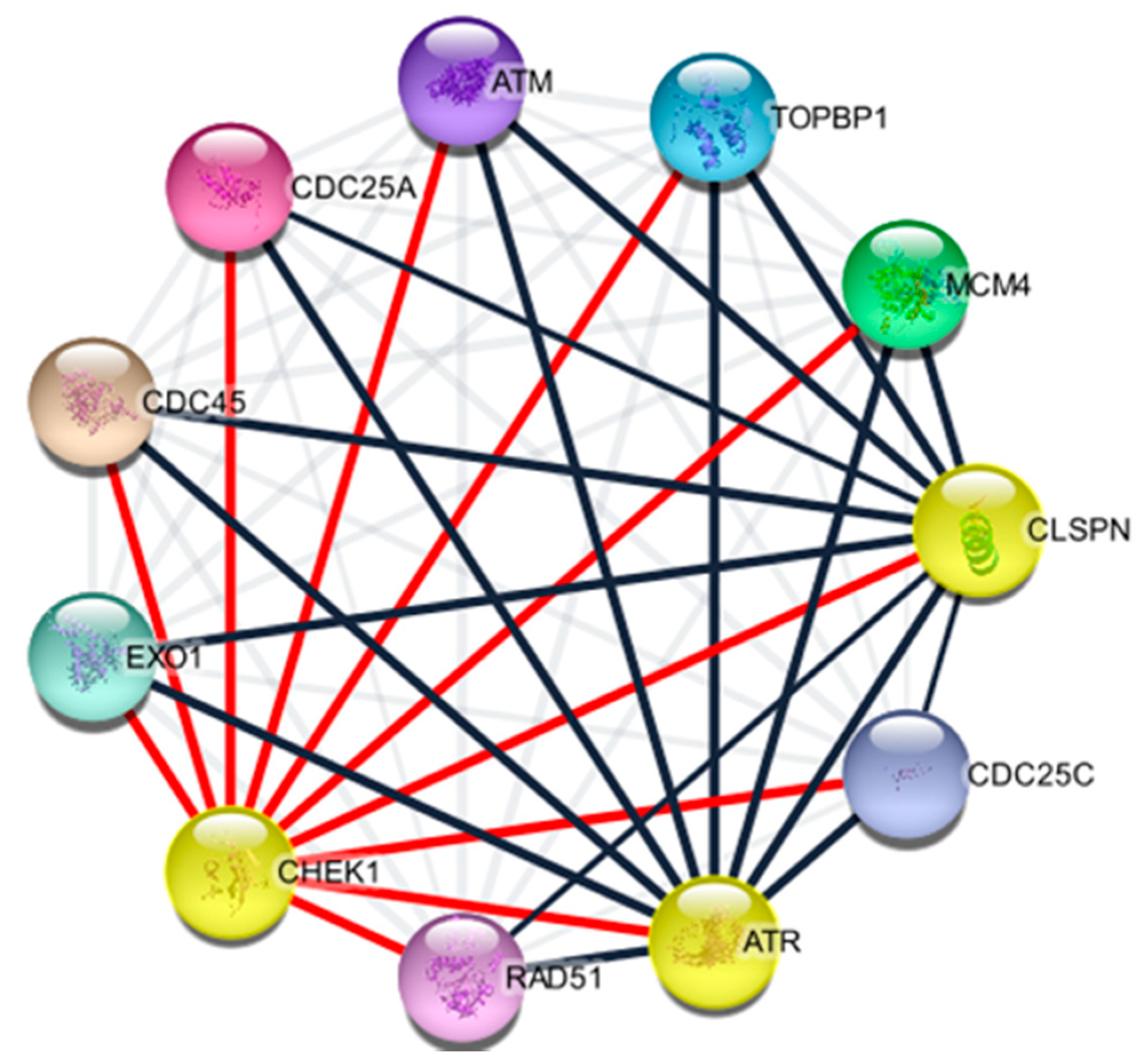
2.4. Protein Components of CHEK1
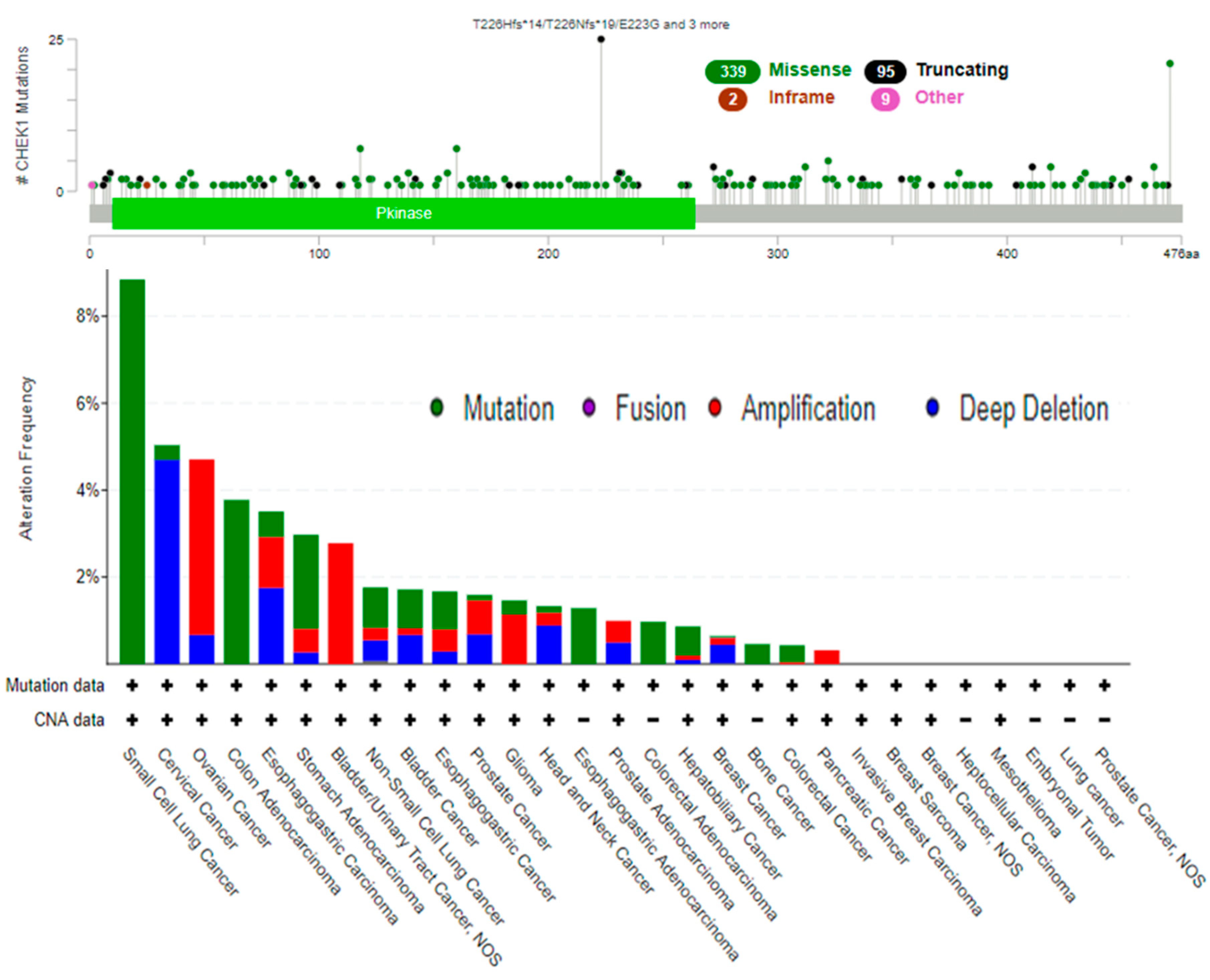
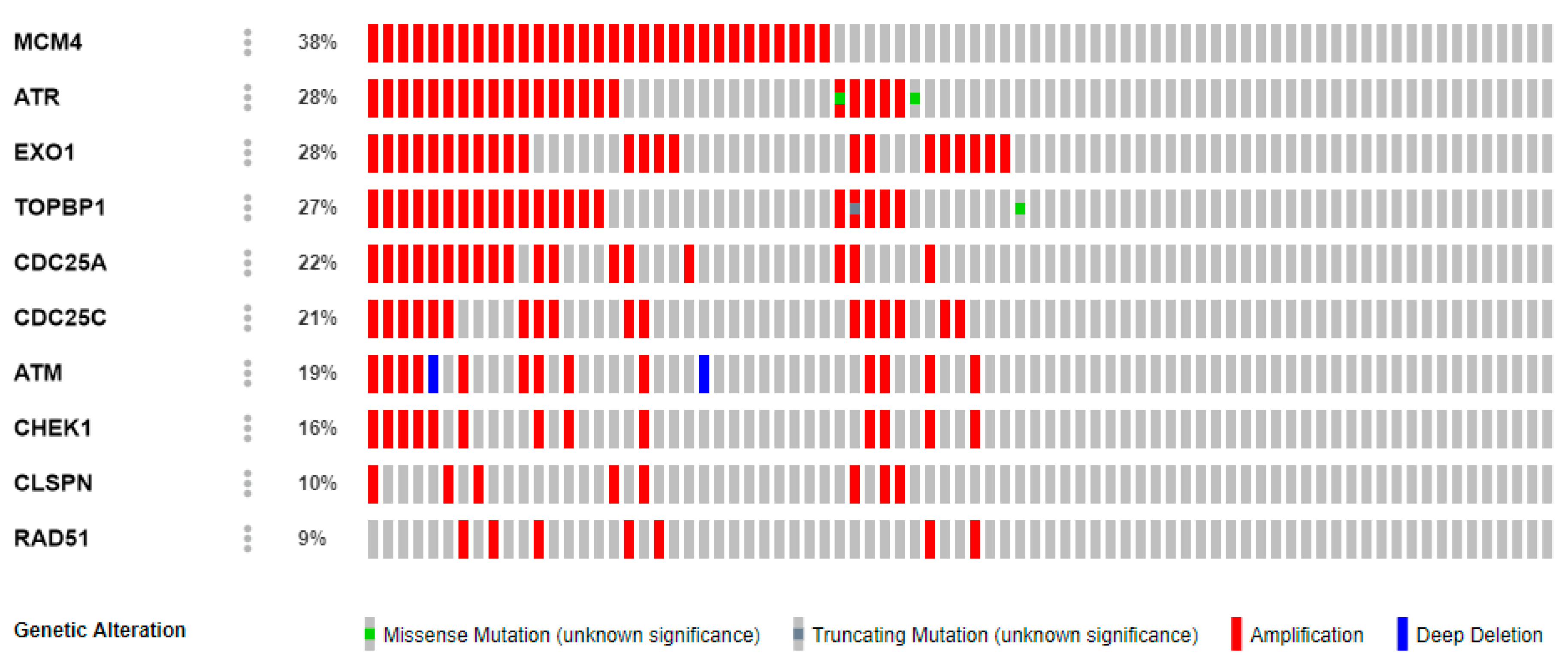
2.5. Genetic Alterations of CHEK1
2.6. Co-Expression Profile of CHEK1 in Colorectal Cancer
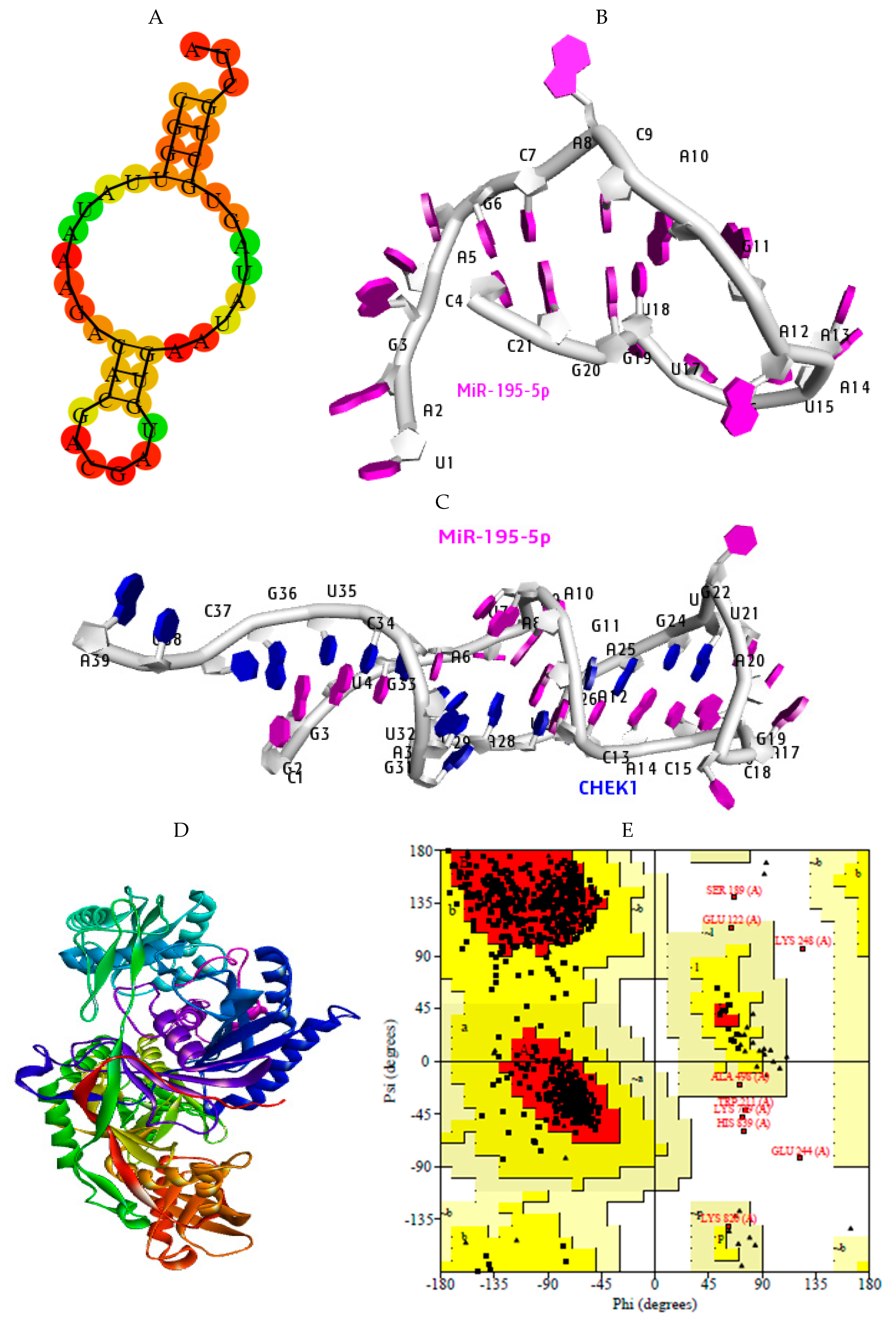
2.7. Structural Preparation of miR-195-5p-CHEK1 and Human Argonaute Protein Preparation
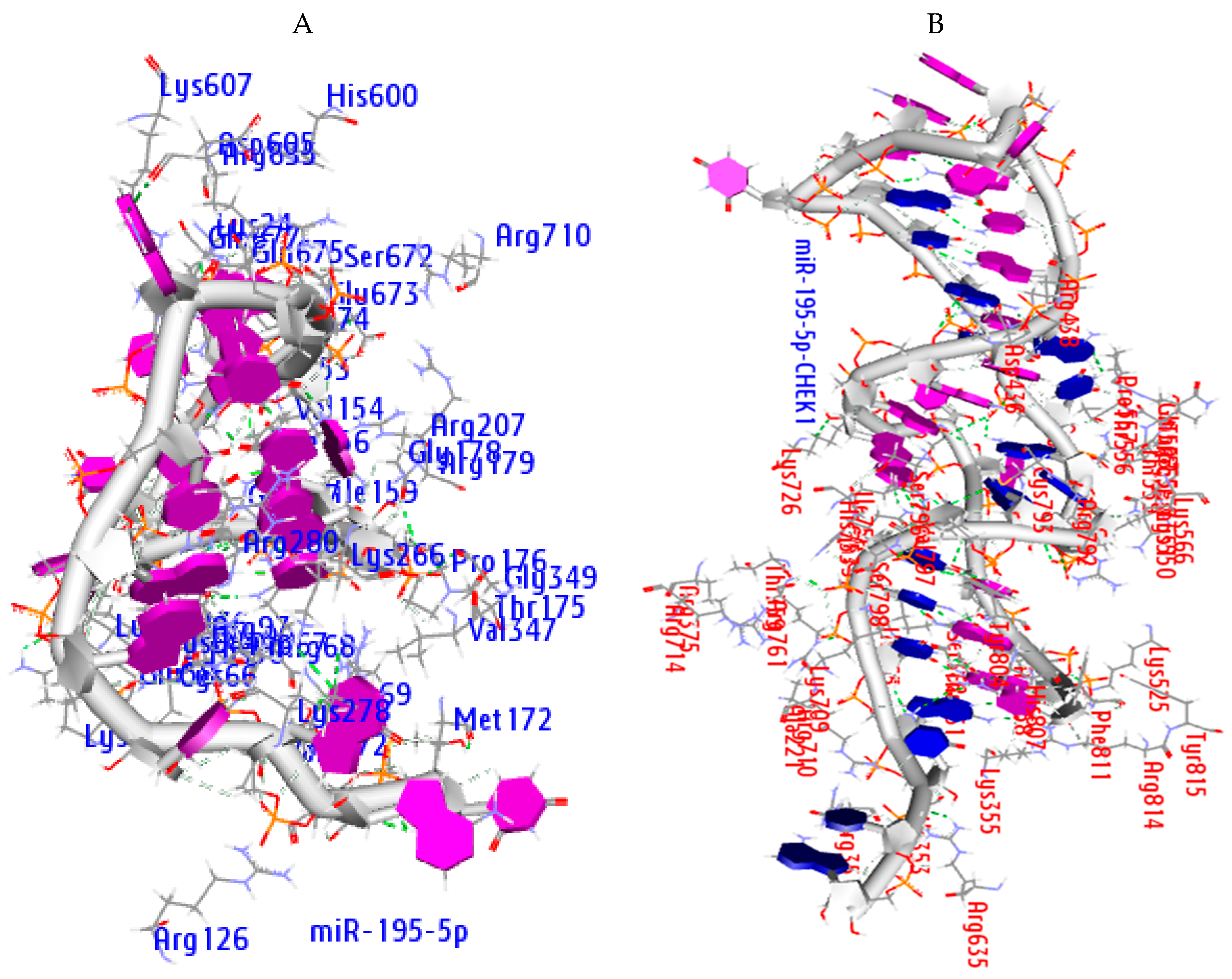
2.8. Molecular Docking Analysis
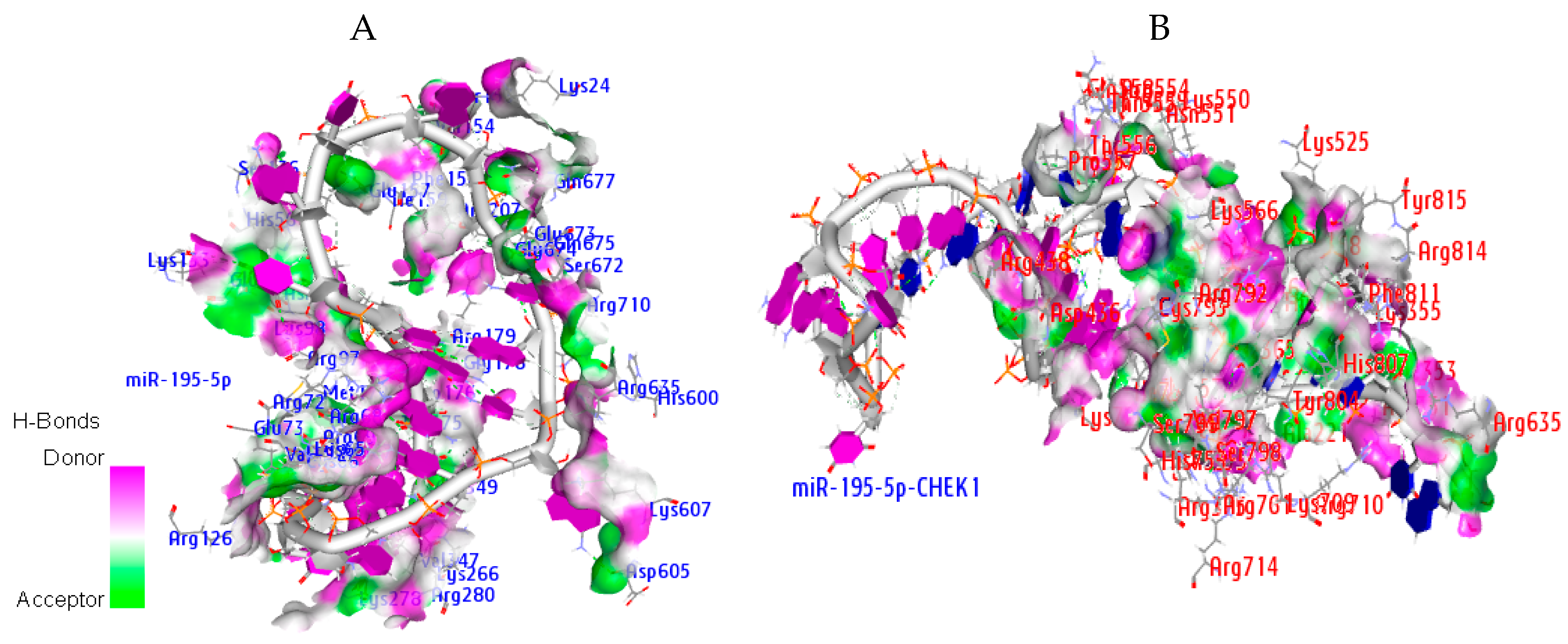
2.9. Hydrogen Bond Interaction
3. Discussion
4. Materials and Methods
4.1. CHEK1 Expression Analysis (Oncomine Database)
4.2. Identification of CHEK1 Associated Proteins
4.3. Prognostic Analysis of CHEK1
4.4. Survival Analysis of CHEK1 in Some Selected Solid Tumors
4.5. Complex Genomic Exploration of CHEK1
4.6. Molecular Binding Analysis
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Salas, L.A.; Johnson, K.C.; Koestler, D.C.; O’Sullivan, D.E.; Christensen, B.C. Integrative epigenetic and genetic pan-cancer somatic alteration portraits. Epigenetics 2017, 12, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Shen, C.; Tan, M.; Li, M.; Song, X.; Wang, C. Systematic analysis of gene expression alterations and clinical outcomes of adenylate cyclase-associated protein in cancer. Oncotarget 2017, 8, 27216. [Google Scholar] [PubMed]
- Ball, M.W.; Gorin, M.A.; Drake, C.G.; Hammers, H.J.; Allaf, M.E. The landscape of whole-genome alterations and pathologic features in genitourinary malignancies: An analysis of the cancer genome atlas. Eur. Urol. Focus 2017, 3, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Tho, L.M.; Xu, N.; Gillespie, D.A. The ATM–Chk2 and ATR–Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010, 108, 73–112. [Google Scholar] [PubMed]
- Chen, Z.; Xiao, Z.; Gu, W.Z.; Xue, J.; Bui, M.H.; Kovar, P.; Li, G.; Wang, G.; Tao, Z.F.; Tong, Y. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int. J. Cancer 2006, 119, 2784–2794. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 31–33. [Google Scholar] [CrossRef]
- García, V.; Lara-Chica, M.; Cantarero, I.; Sterner, O.; Calzado, M.A.; Muñoz, E. Galiellalactone induces cell cycle arrest and apoptosis through the ATM/ATR pathway in prostate cancer cells. Oncotarget 2016, 7, 4490. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Teng, L.; Legerski, R.J. DNA damage checkpoint recovery and cancer development. Exp. Cell Res. 2015, 334, 350–358. [Google Scholar] [CrossRef]
- Consortium, A.P.G. AACR Project GENIE: Powering precision medicine through an international consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef]
- Katsel, P.; Tan, W.; Fam, P.; Purohit, D.P.; Haroutunian, V. Cycle checkpoint abnormalities during dementia: A plausible association with the loss of protection against oxidative stress in Alzheimer’s disease. PLoS ONE 2013, 8, e68361. [Google Scholar] [CrossRef]
- Goto, H.; Izawa, I.; Li, P.; Inagaki, M. Novel regulation of checkpoint kinase 1: Is checkpoint kinase 1 a good candidate for anti-cancer therapy? Cancer Sci. 2012, 103, 1195–1200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hunter, T. Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 2014, 134, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.; Pabla, N.; Dong, Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell. Mol. Life Sci. 2013, 70, 4009–4021. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, F.; Codegoni, A.M.; Furlan, D.; Tibiletti, M.G.; Capella, C.; Broggini, M. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes Chromosomes Cancer 1999, 26, 176–180. [Google Scholar] [CrossRef]
- Menoyo, A.; Alazzouzi, H.; Espín, E.; Armengol, M.; Yamamoto, H.; Schwartz, S. Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001, 61, 7727–7730. [Google Scholar] [PubMed]
- Vassileva, V.; Millar, A.; Briollais, L.; Chapman, W.; Bapat, B. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer Res. 2002, 62, 4095–4099. [Google Scholar]
- Ng, C.-P.; Lee, H.C.; Ho, C.W.; Arooz, T.; Siu, W.Y.; Lau, A.; Poon, R.Y. Differential mode of regulation of the checkpoint kinases CHK1 and CHK2 by their regulatory domains. J. Biol. Chem. 2004, 279, 8808–8819. [Google Scholar] [CrossRef]
- Cho, S.H.; Toouli, C.D.; Fujii, G.H.; Crain, C.; Parry, D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle 2005, 4, 131–139. [Google Scholar] [CrossRef]
- Verlinden, L.; Bempt, I.V.; Eelen, G.; Drijkoningen, M.; Verlinden, I.; Marchal, K.; De Wolf-Peeters, C.; Christiaens, M.-R.; Michiels, L.; Bouillon, R. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor−/progesterone receptor−/HER-2− breast carcinomas. Cancer Res. 2007, 67, 6574–6581. [Google Scholar] [CrossRef]
- Madoz-Gúrpide, J.; Cañamero, M.; Sanchez, L.; Solano, J.; Alfonso, P.; Casal, J.I. A proteomics analysis of cell signaling alterations in colorectal cancer. Mol. Cell. Proteom. 2007, 6, 2150–2164. [Google Scholar] [CrossRef]
- Liang, Y.; Lin, S.-Y.; Brunicardi, F.C.; Goss, J.; Li, K. DNA damage response pathways in tumor suppression and cancer treatment. World J. Surg. 2009, 33, 661–666. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.-Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396. [Google Scholar] [CrossRef] [PubMed]
- Ie, Y.; Wei, R.-R.; Huang, G.-L.; Zhang, M.-Y.; Yuan, Y.-F.; Wang, H.-Y. Checkpoint kinase 1 is negatively regulated by miR-497 in hepatocellular carcinoma. Med. Oncol. 2014, 31, 844. [Google Scholar]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef]
- Sun, L.; Hui, A.-M.; Su, Q.; Vortmeyer, A.; Kotliarov, Y.; Pastorino, S.; Passaniti, A.; Menon, J.; Walling, J.; Bailey, R. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006, 9, 287–300. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef]
- Biewenga, P.; Buist, M.R.; Moerland, P.D.; van Themaat, E.V.L.; van Kampen, A.H.C.; ten Kate, F.J.W.; Baas, F. Gene expression in early stage cervical cancer. Gynecol. Oncol. 2008, 108, 520–526. [Google Scholar] [CrossRef]
- Sabates-Bellver, J.; Van der Flier, L.G.; de Palo, M.; Cattaneo, E.; Maake, C.; Rehrauer, H.; Laczko, E.; Kurowski, M.A.; Bujnicki, J.M.; Menigatti, M. Transcriptome profile of human colorectal adenomas. Mol. Cancer Res. 2007, 5, 1263–1275. [Google Scholar] [CrossRef]
- Cui, J.; Chen, Y.; Chou, W.-C.; Sun, L.; Chen, L.; Suo, J.; Ni, Z.; Zhang, M.; Kong, X.; Hoffman, L.L. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2010, 39, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- D’Errico, M.; de Rinaldis, E.; Blasi, M.F.; Viti, V.; Falchetti, M.; Calcagnile, A.; Sera, F.; Saieva, C.; Ottini, L.; Palli, D. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur. J. Cancer 2009, 45, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Hirao, A.; Cheung, A.; Duncan, G.; Girard, P.-M.; Elia, A.J.; Wakeham, A.; Okada, H.; Sarkissian, T.; Wong, J.A.; Sakai, T. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 2002, 22, 6521–6532. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 2003, 3, 421–429. [Google Scholar] [CrossRef]
- Liu, Z.; Li, J.; Chen, J.; Shan, Q.; Dai, H.; Xie, H.; Zhou, L.; Xu, X.; Zheng, S. MCM family in HCC: MCM6 indicates adverse tumor features and poor outcomes and promotes S/G2 cell cycle progression. BMC Cancer 2018, 18, 200. [Google Scholar] [CrossRef]
- Issac, M.S.M.; Yousef, E.; Tahir, M.R.; Gaboury, L.A. MCM2, MCM4, and MCM6 in Breast Cancer: Clinical Utility in Diagnosis and Prognosis. Neoplasia (N. Y.) 2019, 21, 1015–1035. [Google Scholar] [CrossRef]
- Li, H.; Harrison, M.D.; Avissar, P.L.; Malinowski, D.P. Quantitative Molecular Analysis of MCM6 and MCM7 and Their Association with HPV E6 and E7 mRNA Expression, Viral Load and Physical Status in Invasive Cervical Carcinoma; AACR: Philadelphia, PA, USA, 2006. [Google Scholar]
- Liu, M.; Hu, Q.; Tu, M.; Wang, X.; Yang, Z.; Yang, G.; Luo, R. MCM6 promotes metastasis of hepatocellular carcinoma via MEK/ERK pathway and serves as a novel serum biomarker for early recurrence. J. Exp. Clin. Cancer Res. 2018, 37, 10. [Google Scholar] [CrossRef]
- Liu, Y.-Z.; Wang, B.-S.; Jiang, Y.-Y.; Cao, J.; Hao, J.-J.; Zhang, Y.; Xu, X.; Cai, Y.; Wang, M.-R. MCMs expression in lung cancer: Implication of prognostic significance. J. Cancer 2017, 8, 3641. [Google Scholar] [CrossRef]
- Simon, N.E.; Schwacha, A. The Mcm2-7 replicative helicase: A promising chemotherapeutic target. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Vijayraghavan, S.; Schwacha, A. The eukaryotic Mcm2-7 replicative helicase. In The Eukaryotic Replisome: A Guide to Protein Structure and Function; Springer: Berlin/Heidelberg, Germany, 2012; pp. 113–134. [Google Scholar]
- Forsburg, S.L. Eukaryotic MCM proteins: Beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004, 68, 109–131. [Google Scholar] [CrossRef]
- Das, M.; Prasad, S.B.; Yadav, S.S.; Govardhan, H.; Pandey, L.K.; Singh, S.; Pradhan, S.; Narayan, G. Over expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesis. PLoS ONE 2013, 8, e69607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Keymeulen, S.; Nelson, R.; Tong, T.R.; Yuan, Y.-C.; Yun, X.; Liu, Z.; Lopez, J.; Raz, D.J.; Kim, J.Y. Overexpression of flap endonuclease 1 correlates with enhanced proliferation and poor prognosis of Non–Small-Cell lung cancer. Am. J. Pathol. 2018, 188, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Singh, H.; Srivastava, H.K.; Singh, S.; Kishore, G.; Raghava, G.P. Benchmarking of different molecular docking methods for protein-peptide docking. BMC Bioinform. 2019, 19, 426. [Google Scholar] [CrossRef] [PubMed]
- Bargiela-Iparraguirre, J.; Prado-Marchal, L.; Fernandez-Fuente, M.; Gutierrez-González, A.; Moreno-Rubio, J.; Muñoz-Fernandez, M.; Sereno, M.; Sanchez-Prieto, R.; Perona, R.; Sanchez-Perez, I. CHK1 expression in Gastric Cancer is modulated by p53 and RB1/E2F1: Implications in chemo/radiotherapy response. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Dai, Y.; Grant, S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin. Cancer Res. 2010, 16, 376–383. [Google Scholar] [CrossRef]
- Smits, V.A.; Gillespie, D.A. DNA damage control: Regulation and functions of checkpoint kinase 1. FEBS J. 2015, 282, 3681–3692. [Google Scholar] [CrossRef]
- Tapia-Alveal, C.; Calonge, T.M.; O’Connell, M.J. Regulation of chk1. Cell Div. 2009, 4, 8. [Google Scholar] [CrossRef]
- Aha, S.K.; Islam, S.R.; Kwak, K.-S.; Rahman, M.S.; Cho, S.-G. PROM1 and PROM2 expression differentially modulates clinical prognosis of cancer: A multiomics analysis. Cancer Gene Ther. 2019, 1. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Wang, F.; Wang, X.; Cheng, B.; Ye, F.; Xie, X.; Zhou, C.; Lu, W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene 2013, 32, 976. [Google Scholar] [CrossRef]
- Cole, K.A.; Huggins, J.; Laquaglia, M.; Hulderman, C.E.; Russell, M.R.; Bosse, K.; Diskin, S.J.; Attiyeh, E.F.; Sennett, R.; Norris, G. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc. Natl. Acad. Sci. USA 2011, 108, 3336–3341. [Google Scholar] [CrossRef]
- Yao, H.; Yang, Z.; Li, Y. Expression of checkpoint kinase 1 and polo-like kinase 1 and its clinicopathological significance in benign and malignant lesions of the stomach. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2010, 35, 1080–1084. [Google Scholar] [PubMed]
- Zhou, J.; Chen, Z.; Malysa, A.; Li, X.; Oliveira, P.; Zhang, Y.; Bepler, G. A kinome screen identifies checkpoint kinase 1 (CHK1) as a sensitizer for RRM1-dependent gemcitabine efficacy. PLoS ONE 2013, 8, e58091. [Google Scholar]
- Xu, N.; Libertini, S.; Black, E.; Lao, Y.; Hegarat, N.; Walker, M.; Gillespie, D. Cdk-mediated phosphorylation of Chk1 is required for efficient activation and full checkpoint proficiency in response to DNA damage. Oncogene 2012, 31, 1086. [Google Scholar] [CrossRef] [PubMed]
- Fadaka, A.O.; Pretorius, A.; Klein, A. MicroRNA Assisted Gene Regulation in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4899. [Google Scholar] [CrossRef] [PubMed]
- Cittelly, D.M.; Das, P.M.; Spoelstra, N.S.; Edgerton, S.M.; Richer, J.K.; Thor, A.D.; Jones, F.E. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer 2010, 9, 317. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Baran, J.; Cros, A.; Guberman, J.M.; Haider, S.; Hsu, J.; Liang, Y.; Rivkin, E.; Wang, J.; Whitty, B. International Cancer Genome Consortium Data Portal—A one-stop shop for cancer genomics data. Database 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Klonowska, K.; Czubak, K.; Wojciechowska, M.; Handschuh, L.; Zmienko, A.; Figlerowicz, M.; Dams-Kozlowska, H.; Kozlowski, P. Oncogenomic portals for the visualization and analysis of genome-wide cancer data. Oncotarget 2016, 7, 176. [Google Scholar] [CrossRef]
- Albertson, D.G.; Collins, C.; McCormick, F.; Gray, J.W. Chromosome aberrations in solid tumors. Nat. Genet. 2003, 34, 369. [Google Scholar] [CrossRef]
- Albertson, D.G. Gene amplification in cancer. Trends Genet. 2006, 22, 447–455. [Google Scholar] [CrossRef]
- Matsui, A.; Ihara, T.; Suda, H.; Mikami, H.; Semba, K. Gene amplification: Mechanisms and involvement in cancer. Biomol. Concepts 2013, 4, 567–582. [Google Scholar] [CrossRef]
- Prince, E.W.; Balakrishnan, I.; Shah, M.; Mulcahy Levy, J.M.; Griesinger, A.M.; Alimova, I.; Harris, P.S.; Birks, D.K.; Donson, A.M.; Davidson, N.; et al. Checkpoint kinase 1 expression is an adverse prognostic marker and therapeutic target in MYC-driven medulloblastoma. Oncotarget 2016, 7, 53881–53894. [Google Scholar] [CrossRef] [PubMed]
- van Harten, A.M.; Buijze, M.; van der Mast, R.; Rooimans, M.A.; Martens-de Kemp, S.R.; Bachas, C.; Brink, A.; Stigter-van Walsum, M.; Wolthuis, R.M.F.; Brakenhoff, R.H. Targeting the cell cycle in head and neck cancer by Chk1 inhibition: A novel concept of bimodal cell death. Oncogenesis 2019, 8, 38. [Google Scholar] [CrossRef]
- Lee, H.-J.; Cao, Y.; Pham, V.; Blackwood, E.; Wilson, C.; Evangelista, M.; Klijn, C.; Stokoe, D.; Settleman, J. Ras–MEK Signaling Mediates a Critical Chk1-Dependent DNA Damage Response in Cancer Cells. Mol. Cancer Ther. 2017, 16, 694. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Chen, F.; Jiang, J.; Zhang, H.; Zhou, M. Screening key genes and signaling pathways in colorectal cancer by integrated bioinformatics analysis. Mol. Med. Rep. 2019, 20, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Zhang, W.-Y.; Xia, Q.-J.; Li, H.-G.; Wang, R.; Yang, L.; Sun, X.-F.; Zhou, Z.-G. mRNA expression of minichromosome maintenance 2 in colonic adenoma and adenocarcinoma. Eur. J. Cancer Prev. 2009, 18, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Mensink, R.G.; Verlind, E.; Sijmons, R.H.; Buys, C.H.; Hofstra, R.M.; Berends, M.J.; Kleibeuker, J.H.; Post, J.G.; Kempinga, C. Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology 2001, 120, 1580–1587. [Google Scholar] [CrossRef]
- Chhabra, G.; Sharma, P.; Anant, A.; Deshmukh, S.; Kaushik, H.; Gopal, K.; Srivastava, N.; Sharma, N.; Garg, L.C. Identification and modeling of a drug target for Clostridium perfringens SM101. Bioinformation 2010, 4, 278. [Google Scholar] [CrossRef]
- Elkayam, E.; Kuhn, C.-D.; Tocilj, A.; Haase, A.D.; Greene, E.M.; Hannon, G.J.; Joshua-Tor, L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012, 150, 100–110. [Google Scholar] [CrossRef]
- Fadaka, A.O.; Ojo, B.A.; Adewale, O.B.; Esho, T.; Pretorius, A. Effect of dietary components on miRNA and colorectal carcinogenesis. Cancer Cell Int. 2018, 18, 130. [Google Scholar] [CrossRef]
- Fadaka, A.O.; Pretorius, A.; Klein, A. Biomarkers for stratification in colorectal cancer: MicroRNAs. Cancer Control 2019, 26. [Google Scholar] [CrossRef]
- Fadaka, A.O.; Klein, A.; Pretorius, A. In silico identification of microRNAs as candidate colorectal cancer biomarkers. Tumor Biol. 2019, 41. [Google Scholar] [CrossRef] [PubMed]
- James, A.B.; Fadaka, A.O.; Magbagbeola, O.A.; Oturu, A.; Kolawole, O.O.; Ogunjimi, A.; Oshodi, T.; Habeebu, M.; Onawoga, F.O.; Ajogbeje, E.O. Haematological and miRNAs (let-7g, miR-21, miR-141) expression modulation profile in serum samples of Human prostate cancer. FASEB J. 2017, 31. [Google Scholar] [CrossRef]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, Y.; Ma, X.; Pertsemlidis, A. miR-195 potentiates the efficacy of microtubule-targeting agents in non-small cell lung cancer. Cancer Lett. 2018, 427, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, Y.; Cavazos, D.; Ma, X.; Zhao, Z.; Du, L.; Pertsemlidis, A. miR-195 targets cyclin D3 and survivin to modulate the tumorigenesis of non-small cell lung cancer. Cell Death Dis. 2018, 9, 193. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liang, X.; Li, X.; Zhang, Y.; Sun, Z.; Liu, Y.; Wang, J. MicroRNA-195: A review of its role in cancers. Oncotargets Ther. 2018, 11, 7109–7123. [Google Scholar] [CrossRef]
- Cai, C.; Chen, Q.-B.; Han, Z.-D.; Zhang, Y.-Q.; He, H.-C.; Chen, J.-H.; Chen, Y.-R.; Yang, S.-B.; Wu, Y.-D.; Zeng, Y.-R.; et al. miR-195 Inhibits Tumor Progression by Targeting RPS6KB1 in Human Prostate Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 4922–4934. [Google Scholar] [CrossRef]
- Liu, C.; Guan, H.; Wang, Y.; Chen, M.; Xu, B.; Zhang, L.; Lu, K.; Tao, T.; Zhang, X.; Huang, Y. miR-195 Inhibits EMT by Targeting FGF2 in Prostate Cancer Cells. PLoS ONE 2015, 10, e0144073. [Google Scholar] [CrossRef]
- Lin, X.; Wang, S.; Sun, M.; Zhang, C.; Wei, C.; Yang, C.; Dou, R.; Liu, Q.; Xiong, B. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019, 12, 20. [Google Scholar] [CrossRef]
- Sun, M.; Song, H.; Wang, S.; Zhang, C.; Zheng, L.; Chen, F.; Shi, D.; Chen, Y.; Yang, C.; Xiang, Z.; et al. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J. Hematol. Oncol. 2017, 10, 79. [Google Scholar] [CrossRef]
- Liu, H.; Chen, Y.; Li, Y.; Li, C.; Qin, T.; Bai, M.; Zhang, Z.; Jia, R.; Su, Y.; Wang, C. miR-195 suppresses metastasis and angiogenesis of squamous cell lung cancer by inhibiting the expression of VEGF. Mol. Med. Rep. 2019, 20, 2625–2632. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, Y.; Wu, B.; Kurie, J.M.; Pertsemlidis, A. The miR-195 Axis Regulates Chemoresistance through TUBB and Lung Cancer Progression through BIRC5. Mol. Oncolytics 2019, 14, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Chae, D.-K.; Park, J.; Cho, M.; Ban, E.; Jang, M.; Yoo, Y.S.; Kim, E.E.; Baik, J.-H.; Song, E.J. MiR-195 and miR-497 suppress tumorigenesis in lung cancer by inhibiting SMURF2-induced TGF-β receptor I ubiquitination. Mol. Oncol. 2019, 13, 2663–2678. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, F.; Zhang, J.; Sun, R.; Li, F.; Li, Y.; Chang, S.E.; Wang, L.; Wang, X.; Liu, L.; et al. EGR1 interacts with DNMT3L to inhibit the transcription of miR-195 and plays an anti-apoptotic role in the development of gastric cancer. J. Cell. Mol. Med. 2019, 23, 7372–7381. [Google Scholar] [CrossRef]
- Zhong, J.; Yuan, H.; Xu, X.; Kong, S. MicroRNA-195 inhibits cell proliferation, migration and invasion by targeting defective in cullin neddylation 1 domain containing 1 in cervical cancer. Int. J. Mol. Med. 2018, 42, 779–788. [Google Scholar] [CrossRef]
- Purohit, P.K.; Edwards, R.; Tokatlidis, K.; Saini, N. MiR-195 regulates mitochondrial function by targeting mitofusin-2 in breast cancer cells. RNA Biol. 2019, 16, 918–929. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Von Mering, C. STRING v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape stringApp: Network analysis and visualization of proteomics data. J. Proteome Res. 2018, 18, 623–632. [Google Scholar] [CrossRef]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data; AACR: Philadelphia, PA, USA, 2012. [Google Scholar]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef] [PubMed]
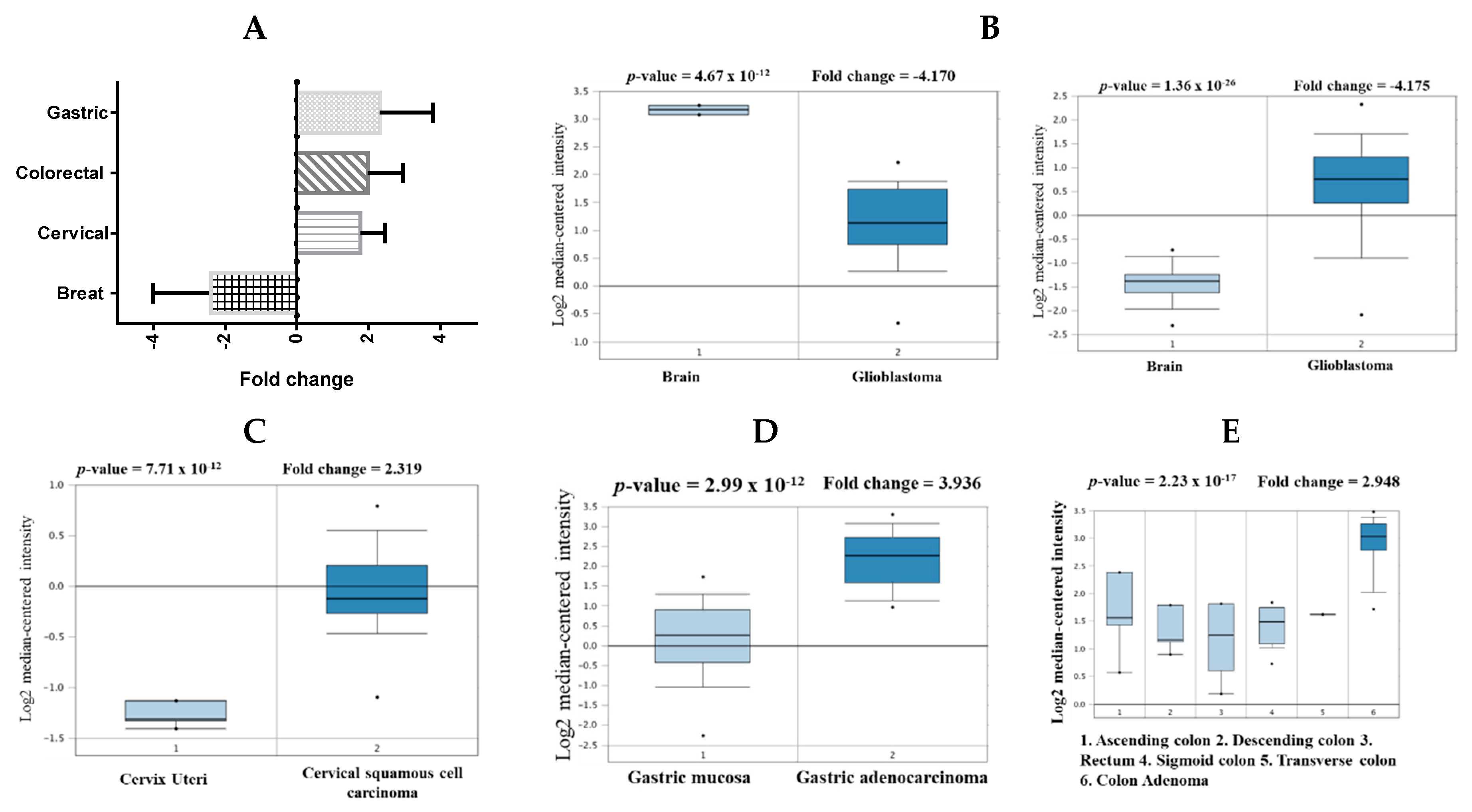



| Cancer | Subtype | p-Value | Fold Change | Sample | Ref. |
|---|---|---|---|---|---|
| Brain | Glioblastoma | 1.36 × 10−26 | 4.157 | 180 | [27] |
| Glioblastoma | 4.67 × 10−12 | −4.170 | 101 | [28] | |
| Cervical | Cervical Squamous Cell Carcinoma | 7.71 × 10−12 | 2.319 | 45 | [29] |
| Colorectal | Colon Adenoma | 2.23 × 10−12 | 2.948 | 64 | [30] |
| Gastric | Gastric cancer | 2.23 × 10−9 | 2.409 | 180 | [31] |
| Gastric Intestinal Type Adenocarcinoma | 2.99 × 10−12 | 3.936 | 69 | [32] |
| Cancer | Dataset | Endpoint | PROBE ID | N | Cox p-value | HR |
|---|---|---|---|---|---|---|
| Bladder | GSE13507 | Overall Survival | ILMN_1664630 | 165 | 0.040924 | 1.35 |
| GSE13507 | Disease Specific Survival | ILMN_1664630 | 165 | 0.00706943 | 1.75 | |
| Brain | GSE4271-GPL96 | Overall Survival | 205394_at | 77 | 0.000296208 | 1.92 |
| Breast | GSE12276 | Relapse Free Survival | 205393_s_at | 204 | 0.000166722 | 1.46 |
| GSE9195 | Distant Metastasis Free Survival | 205393_s_at | 77 | 0.00319767 | 3.57 | |
| GSE1456-GPL96 | Relapse Free Survival | 205394_at | 159 | 0.00259906 | 2.11 | |
| GSE1456-GPL96 | Overall Survival | 205393_s_at | 159 | 0.0298328 | 1.78 | |
| GSE1456-GPL96 | Disease Specific Survival | 205394_at | 159 | 0.00442993 | 2.23 | |
| GSE1456-GPL96 | Overall Survival | 205394_at | 159 | 0.00383569 | 2.02 | |
| GSE1456-GPL96 | Relapse Free Survival | 205393_s_at | 159 | 0.000924772 | 2.44 | |
| GSE1456-GPL96 | Disease Specific Survival | 205393_s_at | 159 | 0.00145812 | 2.60 | |
| GSE4922-GPL97 | Disease Free Survival | 238075_at | 249 | 0.0101338 | 1.77 | |
| Colorectal | GSE14333 | Disease Free Survival | 205394_at | 226 | 0.0121161 | 0.56 |
| Lung | GSE13213 | Overall Survival | A_23_P116123 | 117 | 0.00199884 | 1.40 |
| GSE31210 | Overall Survival | 238075_at | 204 | 0.001305 | 2.39 | |
| GSE31210 | Relapse Free Survival | 205393_s_at | 204 | 0.000142 | 1.91 | |
| GSE31210 | Overall Survival | 205394_at | 204 | 0.000872076 | 1.93 | |
| Ovarian | DUKE-OC | Overall Survival | 205394_at | 133 | 0.0130251 | 0.71 |
| GSE8841 | Overall Survival | 2515 | 81 | 0.017998 | 1.91 |
| Cancer | CHEK1 | ATM | CDC25A | CDC25C | EXO1 | CLSPN | RAD51 | ATR | TOPBP1 | MCM4 |
|---|---|---|---|---|---|---|---|---|---|---|
| Bladder | 2% | 14% | 2.2% | 1% | 2% | 4% | 2.5% | 9% | 4% | 7% |
| Colon | 4% | 13% | 7% | 4% | 7% | 10% | 5% | 6% | 6% | 1% |
| Breast | 0.4% | 2.9% | 0.2% | 0.4% | 5% | 0.6% | 0.8% | 1.2% | 1.5% | 5% |
| Brain | 2.6% | 1.4% | 1.6% | 0.2% | 0.6% | 0,4% | 0.2% | 0.8% | 0.4% | 0.4% |
| Breast | 0.4% | 0.5% | 0.2% | 0.6% | 23% | 0.7% | 0.7% | 5% | 0.9% | 12% |
| Cervical | 4% | 6% | 1.4% | 0.4% | 2.2% | 3% | 1.4% | 10% | 9% | 1.4% |
| Stomach | 2.3% | 12% | 2.5% | 2.1% | 6% | 3% | 0.7% | 8% | 4% | 6% |
| Liver | 1.4% | 5% | 0.3% | 1.4% | 8% | 0.6% | 0.8% | 4% | 1.1% | 7% |
| Lung | 3% | 10% | 0.8% | 2.2% | 7% | 2.4% | 2.6% | 4% | 2.6% | 6% |
| Ovarian | 7% | 11% | 7% | 4% | 9% | 12% | 5% | 21% | 22% | 8% |
| Prostate | 16% | 19% | 22% | 21% | 28% | 10% | 9% | 28% | 27% | 38% |
| Pancreas | 9% | 10% | 4% | 6% | 11% | 7% | 6% | 11% | 8% | 12% |
| Ago Complex | Score | Area | ACE |
|---|---|---|---|
| miRNA-AGO | 20558 | 3146.10 | −386.43 |
| miRNA-mRNA-AGO | 21006 | 3504.30 | −275.84 |
| AGO Complex | Hydrophobic AA | Aromatic AA |
|---|---|---|
| miR-195-5p | (41) Ala221, Ile353, Ile365, Ile756, Val797 | Tyr804, Phe811, Tyr815 |
| miR-195-5p-CHEK1 | (43) Val70, Val154, Ile159, Val347 | Phe156 |
| Ago Complex | Atoms | AA Residues | Distance | NA Residues |
|---|---|---|---|---|
| MiR-195-5p | ARG351 LYS709 ARG761 ARG761 ASP358 THR361 ARG635 ASP436 HIS753 | HH21—OP1 HZ2—OP2 H—OP1 HE—OP2 O—HO2′ HB—O2′ HD2—O3′ OD2—H5′2 ND1—H4′ | 2.0 1.4 1.9 1.3 1.3 1.5 1.5 1.8 1.9 | C37 C34 C34 U35 G3 G3 U38 A12 G33 |
| miRNA-CHEK1 | ARG72 ARG69 LYS278 HIS56 ARG635 GLU157 | HH12—OP1 HH21—O2′ O—H42 HE1—OP2 HD2—OP1 OE2—H5′ | 2.0 1.7 2.0 1.9 1.7 1.9 | G20 G3 C4 U17 C9 A16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fadaka, A.O.; Bakare, O.O.; Sibuyi, N.R.S.; Klein, A. Gene Expression Alterations and Molecular Analysis of CHEK1 in Solid Tumors. Cancers 2020, 12, 662. https://doi.org/10.3390/cancers12030662
Fadaka AO, Bakare OO, Sibuyi NRS, Klein A. Gene Expression Alterations and Molecular Analysis of CHEK1 in Solid Tumors. Cancers. 2020; 12(3):662. https://doi.org/10.3390/cancers12030662
Chicago/Turabian StyleFadaka, Adewale Oluwaseun, Olalekan Olanrewaju Bakare, Nicole Remaliah Samantha Sibuyi, and Ashwil Klein. 2020. "Gene Expression Alterations and Molecular Analysis of CHEK1 in Solid Tumors" Cancers 12, no. 3: 662. https://doi.org/10.3390/cancers12030662
APA StyleFadaka, A. O., Bakare, O. O., Sibuyi, N. R. S., & Klein, A. (2020). Gene Expression Alterations and Molecular Analysis of CHEK1 in Solid Tumors. Cancers, 12(3), 662. https://doi.org/10.3390/cancers12030662








