A Disintegrin and Metalloproteinase 9 (ADAM9) in Advanced Hepatocellular Carcinoma and Their Role as a Biomarker During Hepatocellular Carcinoma Immunotherapy
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
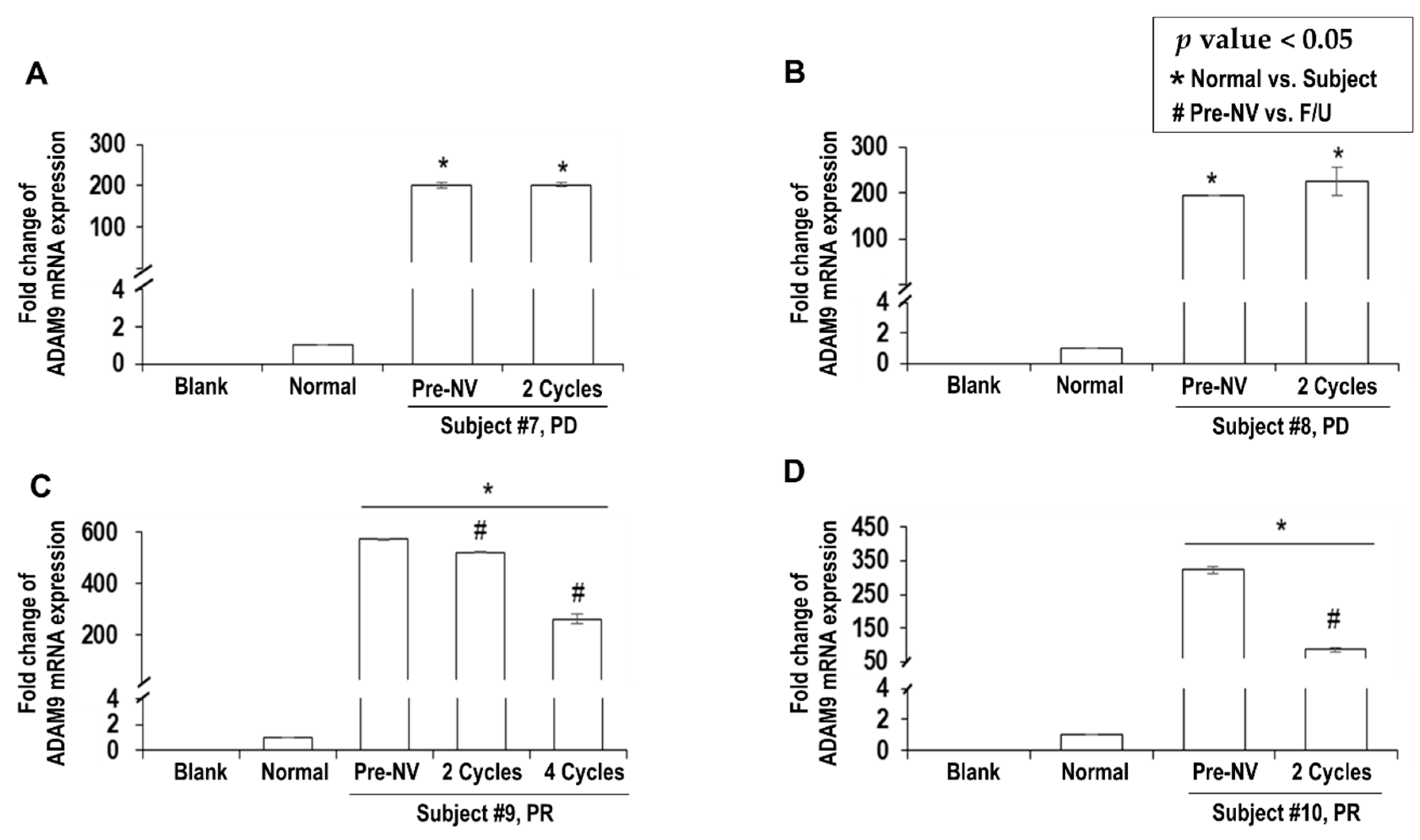
2.2. Overexpression of Plasma ADAM9 mRNA in Untreated HCC Patients
2.3. Decreased ADAM9 mRNA Expression Correlated with Response to Nivolumab
2.4. Immunophenotype Changes following Nivolumab Therapy
2.5. Serum ADAM9 mRNA Expression was Completely Suppressed in Complete Response of HCC
2.6. ADAM9 was Associated with HCC Prognosis in TCGA Database
2.7. ADAM9 Expression is Positively Correlated with PD-1, TIM-3 and BTLA
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Study Design and Protocol
4.3. mRNA Isolation and Real-Time PCR
4.4. Flow Cytometric Analysis
4.5. Statistical Analysis
4.6. In-Silico Analysis with TCGA Database
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chon, Y.E.; Park, H.; Hyun, H.K.; Ha, Y.; Kim, M.N.; Kim, B.K.; Lee, J.H.; Kim, S.U.; Kim, D.Y.; Ahn, S.H.; et al. Development of a New Nomogram Including Neutrophil-to-Lymphocyte Ratio to Predict Survival in Patients with Hepatocellular Carcinoma Undergoing Transarterial Chemoembolization. Cancers 2019, 11, 509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Oh, S.-Y.; Kim, J.Y.; Nishida, N. Cancer immunotherapy for hepatocellular carcinoma. Hepatoma. Res. 2018, 4, 51. Available online: https://hrjournal.net/article/view/2776 (accessed on 20 March 2020). [CrossRef] [Green Version]
- Nishida, N.; Kudo, M. Immune checkpoint blockade for the treatment of human hepatocellular carcinoma. Hepatol. Res. 2018, 48, 622–634. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [Green Version]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef] [Green Version]
- Saffo, S.; Taddei, T.H. Systemic Management for Advanced Hepatocellular Carcinoma: A Review of the Molecular Pathways of Carcinogenesis, Current and Emerging Therapies, and Novel Treatment Strategies. Dig. Dis. Sci. 2019, 64, 1016–1029. [Google Scholar] [CrossRef]
- Ueshima, K.; Nishida, N.; Hagiwara, S.; Aoki, T.; Minami, T.; Chishina, H.; Takita, M.; Minami, Y.; Ida, H.; Takenaka, M.; et al. Impact of Baseline ALBI Grade on the Outcomes of Hepatocellular Carcinoma Patients Treated with Lenvatinib: A Multicenter Study. Cancers 2019, 11, 952. [Google Scholar] [CrossRef] [Green Version]
- Daher, S.; Massarwa, M.; Benson, A.A.; Khoury, T. Current and Future Treatment of Hepatocellular Carcinoma: An Updated Comprehensive Review. J. Clin. Transl. Hepatol. 2018, 6, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef] [Green Version]
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491.e471. [Google Scholar] [CrossRef] [PubMed]
- Bteich, F.; Di Bisceglie, A.M. Current and Future Systemic Therapies for Hepatocellular Carcinoma. Gastroenterol. Hepatol. 2019, 15, 266–272. [Google Scholar]
- Liu, X.; Qin, S. Immune Checkpoint Inhibitors in Hepatocellular Carcinoma: Opportunities and Challenges. Oncologist 2019, 24, S3–S10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudo, M. Combination Cancer Immunotherapy with Molecular Targeted Agents/Anti-CTLA-4 Antibody for Hepatocellular Carcinoma. Liver Cancer 2019, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.Y.; Choo, S.P.; Trojan, J.; Welling, T.H.R.; et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017, 389, 2492–2502. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J.H.; Lim, Y.S.; Yeon, J.E.; Song, T.J.; Yu, S.J.; Gwak, G.Y.; Kim, K.M.; Kim, Y.J.; Lee, J.W.; et al. Adjuvant immunotherapy with autologous cytokine-induced killer cells for hepatocellular carcinoma. Gastroenterology 2015, 148, 1383–1391.e1386. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, N.; Neumann-Haefelin, C.; Thimme, R. Cellular immune responses to hepatocellular carcinoma: Lessons for immunotherapy. Dig. Dis. 2012, 30, 483–491. [Google Scholar] [CrossRef]
- Kudo, M. Immune Checkpoint Inhibition in Hepatocellular Carcinoma: Basics and Ongoing Clinical Trials. Oncology 2017, 92 (Suppl. 1), 50–62. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Li, L.; Li, W.; Wang, C.; Yan, X.; Wang, Y.; Niu, C.; Zhang, X.; Li, M.; Tian, H.; Yao, C.; et al. Adoptive transfer of natural killer cells in combination with chemotherapy improves outcomes of patients with locally advanced colon carcinoma. Cytotherapy 2018, 20, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Lee, J.H.; Kwack, K.; Choi, S.W. Natural Killer Cell Therapy: A New Treatment Paradigm for Solid Tumors. Cancers 2019, 11, 1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohga, K.; Takehara, T.; Tatsumi, T.; Ishida, H.; Miyagi, T.; Hosui, A.; Hayashi, N. Sorafenib inhibits the shedding of major histocompatibility complex class I-related chain A on hepatocellular carcinoma cells by down-regulating a disintegrin and metalloproteinase 9. Hepatology 2010, 51, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Arai, J.; Goto, K.; Stephanou, A.; Tanoue, Y.; Ito, S.; Muroyama, R.; Matsubara, Y.; Nakagawa, R.; Morimoto, S.; Kaise, Y.; et al. Predominance of regorafenib over sorafenib: Restoration of membrane-bound MICA in hepatocellular carcinoma cells. J. Gastroenterol. Hepatol. 2018, 33, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Izumi, N.; Ichida, T.; Ku, Y.; Kokudo, N.; Sakamoto, M.; Takayama, T.; Nakashima, O.; Matsui, O.; Matsuyama, Y. Report of the 19th follow-up survey of primary liver cancer in Japan. Hepatol. Res. 2016, 46, 372–390. [Google Scholar] [CrossRef] [PubMed]
- Waldhauer, I.; Goehlsdorf, D.; Gieseke, F.; Weinschenk, T.; Wittenbrink, M.; Ludwig, A.; Stevanovic, S.; Rammensee, H.G.; Steinle, A. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 2008, 68, 6368–6376. [Google Scholar] [CrossRef] [Green Version]
- Mazzocca, A.; Giannelli, G.; Antonaci, S. Involvement of ADAMs in tumorigenesis and progression of hepatocellular carcinoma: Is it merely fortuitous or a real pathogenic link? Biochim. Biophys. Acta 2010, 1806, 74–81. [Google Scholar] [CrossRef]
- Seals, D.F.; Courtneidge, S.A. The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 2003, 17, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Xia, C.; Zhang, D.; Li, Y.; Chen, J.; Zhou, H.; Nie, L.; Sun, Y.; Guo, S.; Cao, J.; Zhou, F.; et al. Inhibition of hepatocellular carcinoma cell proliferation, migration, and invasion by a disintegrin and metalloproteinase-17 inhibitor TNF484. J. Res. Med. Sci. 2019, 24, 26. [Google Scholar] [CrossRef]
- Shiu, J.S.; Hsieh, M.J.; Chiou, H.L.; Wang, H.L.; Yeh, C.B.; Yang, S.F.; Chou, Y.E. Impact of ADAM10 gene polymorphisms on hepatocellular carcinoma development and clinical characteristics. Int. J. Med. Sci. 2018, 15, 1334–1340. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Ren, Z.; Wang, Y.; Dang, Y.Z.; Meng, B.X.; Wang, G.D.; Zhang, J.; Wu, J.; Wen, N. ADAM17 promotes cell migration and invasion through the integrin beta1 pathway in hepatocellular carcinoma. Exp. Cell Res. 2018, 370, 373–382. [Google Scholar] [CrossRef]
- Honda, H.; Takamura, M.; Yamagiwa, S.; Genda, T.; Horigome, R.; Kimura, N.; Setsu, T.; Tominaga, K.; Kamimura, H.; Matsuda, Y.; et al. Overexpression of a disintegrin and metalloproteinase 21 is associated with motility, metastasis, and poor prognosis in hepatocellular carcinoma. Sci. Rep. 2017, 7, 15485. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, W.; Liu, S.; Liu, K.; Ji, B.; Wang, Y. miR-365 targets ADAM10 and suppresses the cell growth and metastasis of hepatocellular carcinoma. Oncol. Rep. 2017, 37, 1857–1864. [Google Scholar] [CrossRef]
- Li, S.Q.; Wang, D.M.; Zhu, S.; Ma, Z.; Li, R.F.; Xu, Z.S.; Han, H.M. The important role of ADAM8 in the progression of hepatocellular carcinoma induced by diethylnitrosamine in mice. Hum. Exp. Toxicol. 2015, 34, 1053–1072. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, W.; Liu, K.; Ji, B.; Wang, G. Silencing ADAM10 inhibits the in vitro and in vivo growth of hepatocellular carcinoma cancer cells. Mol. Med. Rep. 2015, 11, 597–602. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Wu, Z.; He, M.; Chen, Y.; Chen, Y.; Shen, X.; Zhao, X.; Zhang, L.; Yuan, B.; Zeng, Z. ADAM9 mediates the interleukin-6-induced Epithelial-Mesenchymal transition and metastasis through ROS production in hepatoma cells. Cancer Lett. 2018, 421, 1–14. [Google Scholar] [CrossRef]
- Hu, D.; Shen, D.; Zhang, M.; Jiang, N.; Sun, F.; Yuan, S.; Wan, K. MiR-488 suppresses cell proliferation and invasion by targeting ADAM9 and lncRNA HULC in hepatocellular carcinoma. Am. J. Cancer Res. 2017, 7, 2070–2080. [Google Scholar]
- Wan, D.; Shen, S.; Fu, S.; Preston, B.; Brandon, C.; He, S.; Shen, C.; Wu, J.; Wang, S.; Xie, W.; et al. miR-203 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC. Anti-Cancer Agents Med. Chem. 2016, 16, 414–423. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, J.; Li, Y.; Liu, L.; Zhang, X.; Ma, C.Y.; Hua, S.C.; Yang, M.; Yuan, Q. microRNA-1274a, a modulator of sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9) down-regulation in hepatocellular carcinoma. FEBS Lett. 2011, 585, 1828–1834. [Google Scholar] [CrossRef] [Green Version]
- Kohga, K.; Tatsumi, T.; Takehara, T.; Tsunematsu, H.; Shimizu, S.; Yamamoto, M.; Sasakawa, A.; Miyagi, T.; Hayashi, N. Expression of CD133 confers malignant potential by regulating metalloproteinases in human hepatocellular carcinoma. J. Hepatol. 2010, 52, 872–879. [Google Scholar] [CrossRef]
- Itabashi, H.; Maesawa, C.; Oikawa, H.; Kotani, K.; Sakurai, E.; Kato, K.; Komatsu, H.; Nitta, H.; Kawamura, H.; Wakabayashi, G.; et al. Angiotensin II and epidermal growth factor receptor cross-talk mediated by a disintegrin and metalloprotease accelerates tumor cell proliferation of hepatocellular carcinoma cell lines. Hepatol. Res. 2008, 38, 601–613. [Google Scholar] [CrossRef]
- Kohga, K.; Tatsumi, T.; Tsunematsu, H.; Aono, S.; Shimizu, S.; Kodama, T.; Hikita, H.; Yamamoto, M.; Oze, T.; Aketa, H.; et al. Interleukin-1beta enhances the production of soluble MICA in human hepatocellular carcinoma. Cancer Immunol. Immunother. CII 2012, 61, 1425–1432. [Google Scholar] [CrossRef]
- Goto, K.; Kato, N. MICA SNPs and the NKG2D system in virus-induced HCC. J. Gastroenterol. 2015, 50, 261–272. [Google Scholar] [CrossRef]
- Kohga, K.; Takehara, T.; Tatsumi, T.; Ohkawa, K.; Miyagi, T.; Hiramatsu, N.; Kanto, T.; Kasugai, T.; Katayama, K.; Kato, M.; et al. Serum levels of soluble major histocompatibility complex (MHC) class I-related chain A in patients with chronic liver diseases and changes during transcatheter arterial embolization for hepatocellular carcinoma. Cancer Sci. 2008, 99, 1643–1649. [Google Scholar] [CrossRef]
- Jinushi, M.; Takehara, T.; Tatsumi, T.; Kanto, T.; Groh, V.; Spies, T.; Kimura, R.; Miyagi, T.; Mochizuki, K.; Sasaki, Y.; et al. Expression and role of MICA and MICB in human hepatocellular carcinomas and their regulation by retinoic acid. Int. J. Cancer 2003, 104, 354–361. [Google Scholar] [CrossRef]
- Groh, V.; Rhinehart, R.; Secrist, H.; Bauer, S.; Grabstein, K.H.; Spies, T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc. Natl. Acad. Sci. USA 1999, 96, 6879–6884. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Yi Lo, P.H.; Sawai, H.; Kato, N.; Takahashi, A.; Deng, Z.; Urabe, Y.; Mbarek, H.; Tokunaga, K.; Tanaka, Y.; et al. Soluble MICA and a MICA variation as possible prognostic biomarkers for HBV-induced hepatocellular carcinoma. PLoS ONE 2012, 7, e44743. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Kato, N.; Urabe, Y.; Takahashi, A.; Muroyama, R.; Hosono, N.; Otsuka, M.; Tateishi, R.; Omata, M.; Nakagawa, H.; et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat. Genet. 2011, 43, 455–458. [Google Scholar] [CrossRef]
- Feitelson, M.A.; Bonamassa, B.; Arzumanyan, A. The roles of hepatitis B virus-encoded X protein in virus replication and the pathogenesis of chronic liver disease. Expert Opin. Ther. Targets 2014, 18, 293–306. [Google Scholar] [CrossRef]
- Ou, D.P.; Tao, Y.M.; Tang, F.Q.; Yang, L.Y. The hepatitis B virus X protein promotes hepatocellular carcinoma metastasis by upregulation of matrix metalloproteinases. Int. J. Cancer 2007, 120, 1208–1214. [Google Scholar] [CrossRef]
- Lunemann, S.; Malone, D.F.; Hengst, J.; Port, K.; Grabowski, J.; Deterding, K.; Markova, A.; Bremer, B.; Schlaphoff, V.; Cornberg, M.; et al. Compromised function of natural killer cells in acute and chronic viral hepatitis. J. Infect. Dis. 2014, 209, 1362–1373. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zhang, X.J.; Shi, K.Q.; Chen, Y.P.; Ren, Y.F.; Song, Y.J.; Li, G.; Xue, Y.F.; Fang, Y.X.; Deng, Z.J.; et al. Hepatitis B surface antigen inhibits MICA and MICB expression via induction of cellular miRNAs in hepatocellular carcinoma cells. Carcinogenesis 2014, 35, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Chotiyaputta, W.; Lok, A.S. Hepatitis B virus variants. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 453–462. [Google Scholar] [CrossRef]
- Abu-Amara, M.; Feld, J.J. Does antiviral therapy for chronic hepatitis B reduce the risk of hepatocellular carcinoma? Semin. Liver Dis. 2013, 33, 157–166. [Google Scholar] [CrossRef]
- Morgan, R.L.; Baack, B.; Smith, B.D.; Yartel, A.; Pitasi, M.; Falck-Ytter, Y. Eradication of hepatitis C virus infection and the development of hepatocellular carcinoma: A meta-analysis of observational studies. Ann. Intern. Med. 2013, 158, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, J.J.; Gao, S.; Liu, Q.; Bai, J.; Zhao, X.Q.; Hao, Y.H.; Ding, H.H.; Zhu, F.; Yang, D.L.; et al. Decreased peripheral natural killer cells activity in the immune activated stage of chronic hepatitis B. PLoS ONE 2014, 9, e86927. [Google Scholar] [CrossRef]
- Jinushi, M.; Takehara, T.; Tatsumi, T.; Kanto, T.; Groh, V.; Spies, T.; Suzuki, T.; Miyagi, T.; Hayashi, N. Autocrine/paracrine IL-15 that is required for type I IFN-mediated dendritic cell expression of MHC class I-related chain A and B is impaired in hepatitis C virus infection. J. Immunol. 2003, 171, 5423–5429. [Google Scholar] [CrossRef]
- Goto, K.; Arai, J.; Stephanou, A.; Kato, N. Novel therapeutic features of disulfiram against hepatocellular carcinoma cells with inhibitory effects on a disintegrin and metalloproteinase 10. Oncotarget 2018, 9, 18821–18831. [Google Scholar] [CrossRef] [Green Version]
- Takayama, T.; Makuuchi, M.; Sekine, T.; Terui, S.; Shiraiwa, H.; Kosuge, T.; Yamazaki, S.; Hasegawa, H.; Suzuki, K.; Yamagata, M.; et al. Distribution and therapeutic effect of intraarterially transferred tumor-infiltrating lymphocytes in hepatic malignancies. A preliminary report. Cancer 1991, 68, 2391–2396. [Google Scholar] [CrossRef]
- Kawata, A.; Une, Y.; Hosokawa, M.; Wakizaka, Y.; Namieno, T.; Uchino, J.; Kobayashi, H. Adjuvant chemoimmunotherapy for hepatocellular carcinoma patients. Adriamycin, interleukin-2, and lymphokine-activated killer cells versus adriamycin alone. Am. J. Clin. Oncol. 1995, 18, 257–262. [Google Scholar] [CrossRef]
- Takayama, T.; Sekine, T.; Makuuchi, M.; Yamasaki, S.; Kosuge, T.; Yamamoto, J.; Shimada, K.; Sakamoto, M.; Hirohashi, S.; Ohashi, Y.; et al. Adoptive immunotherapy to lower postsurgical recurrence rates of hepatocellular carcinoma: A randomised trial. Lancet 2000, 356, 802–807. [Google Scholar] [CrossRef]
- Liu, D.; Staveley-O’Carroll, K.F.; Li, G. Immune-based therapy clinical trials in hepatocellular carcinoma. J. Clin. Cell. Immunol. 2015, 6, 376. [Google Scholar] [CrossRef] [Green Version]
- Ueno, S.; Tanabe, G.; Nuruki, K.; Hamanoue, M.; Komorizono, Y.; Oketani, M.; Hokotate, H.; Inoue, H.; Baba, Y.; Imamura, Y.; et al. Prognostic performance of the new classification of primary liver cancer of Japan (4th edition) for patients with hepatocellular carcinoma: A validation analysis. Hepatol. Res. 2002, 24, 395–403. [Google Scholar] [CrossRef]
- Lencioni, R.; Llovet, J.M. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin. Liver Dis. 2010, 30, 52–60. [Google Scholar] [CrossRef] [Green Version]



| Subject No. | Age | Sex | Etiology | Antiviral Therapy | Serum AFP (ng/mL) | Child-Pugh Class | Tumor Size (cm) | PV Invasion (Vp) * | mUICC Stage | Extra-Hepatic Metastasis | Type of Therapy |
|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | 58 | M | HBV | TDF | 97,387 | A | 8 | 4 | IVb | Yes | Sorafenib |
| #2 | 61 | M | HBV | ETV | >200,000 | B | 22 | 2 | Ivb | Yes | Sorafenib |
| #3 | 61 | M | HBV | ETV | 190 | B | 5.2 | 3 | Ivb | Yes | Sorafenib |
| #4 | 55 | M | HBV | TDF | 71.7 | A | 3 | 0 | III | No | TACE, Sorafenib |
| #5 | 58 | M | HBV | ETV | 14.7 | A | 10 | 2 | Ivb | Yes | Sorafenib, Nivolumab |
| #6 | 59 | M | HBV | ETV | 82.4 | A | 4 | 2 | Iva | No | Sorafenib, Nivolumab + NK cell therapy, Regorafenib + NK cell therapy |
| #7 | 45 | M | HBV | TDF | 154.7 | B | 11 | 4 | Iva | No | Sorafenib, Nivolumab |
| #8 | 44 | F | HCV | DAC/SUN | 6519.4 | A | 4.5 | 0 | Ivb | Yes | Sorafenib, Regorafenib, Nivolumab |
| #9 | 58 | F | HBV | TDF | 66 | A | 9 | 2 | Ivb | Yes | Sorafenib, Nivolumab Regorafenib |
| #10 | 76 | M | NASH | none | 4594.1 | A | 5.2 | 1 | Ivb | Yes | Sorafenib, Nivolumab |
| Cell Type | Phenotype Marker | Non-Responders | Responders | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Subject #7 | Subject #8 | Subject #9 | Subject #10 | ||||||
| Pre-NV | 3 Cycles | Pre-NV | 4 Cycles | Pre-NV | 4 Cycles | Pre-NV | 4 Cycles | ||
| T cells | CD3+ | 63.62 | 52.98 | 74.53 | 84.57 | 6.18 | 19.02 | 45.01 | 53.65 |
| Helper T cells | CD3+CD4+ | 27.46 | 26.36 | 59.91 | 52.43 | 5.51 | 10.58 | 30.57 | 35.58 |
| Cytotoxic T cells | CD3+CD8+ | 34.15 | 20.03 | 13.95 | 30.7 | 0.73 | 7.41 | 14.57 | 18.75 |
| B cells | CD19+ | 10.15 | 7.27 | 1.88 | 1.06 | 3.21 | 60.7 | 19.1 | 5.84 |
| NK cells | CD3−CD56+ | 10.77 | 5 | 0.21 | 5.52 | 0.61 | 7.49 | 19.87 | 18.88 |
| NKT cells | CD3+CD56+ | 4 | 2.37 | 2.03 | 2.59 | 0.23 | 0.78 | 6.68 | 4.63 |
| Cytotoxic T cells | CD3+CD8+PD-1+ | 3.95 | 6.48 | 22.34 | 2.87 | ND | 0.12 | 24.65 | 7.99 |
| CD3+CD8+TIM3+ | 21.05 | 52.11 | 15.06 | 5.3 | ND | 45.32 | 11.57 | 6.73 | |
| CD3+CD8+LAG3+ | 6.58 | 16.47 | 38.57 | 37.64 | ND | 3.33 | 29.95 | 30.63 | |
| CD3+CD8+BTLA+ | 14.47 | 11.32 | 29.87 | 54.32 | ND | 6.7 | 2.7 | 1.79 | |
| Helper T cells | CD3+CD4+PD-1+ | 0 | 1.22 | 15.44 | 2.15 | 7.69 | 7.34 | 13.09 | 3.3 |
| CD3+CD4+TIM3+ | 19.81 | 37.3 | 3.42 | 11.89 | 69.74 | 45.52 | 10.2 | 4.01 | |
| CD3+CD4+LAG3+ | 0.94 | 16.51 | 35.16 | 25.04 | 8.21 | 2.42 | 6.93 | 4.53 | |
| CD3+CD4+BTLA+ | 13.16 | 9.4 | 24.81 | 29.19 | 0.51 | 2 | 1.94 | 11.14 | |
| NK cells | CD3−CD56+PD-1+ | 8.7 | 1.24 | ND | 1.58 | ND | 1.19 | 4.39 | 3.94 |
| CD3−CD56+TIM-3+ | 4.35 | 22.39 | ND | 21.45 | ND | 33.6 | 25.45 | 14.6 | |
| CD3−CD56+LAG3+ | 0 | 2.74 | ND | 14.51 | ND | 5.06 | 46.1 | 25.15 | |
| CD3−CD56+BTLA+ | 47.83 | 26.18 | ND | 9.15 | ND | 3.45 | 1.63 | 1.47 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, S.; Park, Y.; Lee, H.-J.; Lee, J.; Lee, S.-H.; Baek, Y.-S.; Chun, S.-K.; Lee, S.-M.; Kim, M.; Chon, Y.-E.; et al. A Disintegrin and Metalloproteinase 9 (ADAM9) in Advanced Hepatocellular Carcinoma and Their Role as a Biomarker During Hepatocellular Carcinoma Immunotherapy. Cancers 2020, 12, 745. https://doi.org/10.3390/cancers12030745
Oh S, Park Y, Lee H-J, Lee J, Lee S-H, Baek Y-S, Chun S-K, Lee S-M, Kim M, Chon Y-E, et al. A Disintegrin and Metalloproteinase 9 (ADAM9) in Advanced Hepatocellular Carcinoma and Their Role as a Biomarker During Hepatocellular Carcinoma Immunotherapy. Cancers. 2020; 12(3):745. https://doi.org/10.3390/cancers12030745
Chicago/Turabian StyleOh, Sooyeon, YoungJoon Park, Hyun-Jung Lee, Jooho Lee, Soo-Hyeon Lee, Young-Seok Baek, Su-Kyung Chun, Seung-Min Lee, Mina Kim, Young-Eun Chon, and et al. 2020. "A Disintegrin and Metalloproteinase 9 (ADAM9) in Advanced Hepatocellular Carcinoma and Their Role as a Biomarker During Hepatocellular Carcinoma Immunotherapy" Cancers 12, no. 3: 745. https://doi.org/10.3390/cancers12030745
APA StyleOh, S., Park, Y., Lee, H.-J., Lee, J., Lee, S.-H., Baek, Y.-S., Chun, S.-K., Lee, S.-M., Kim, M., Chon, Y.-E., Ha, Y., Cho, Y., Kim, G. J., Hwang, S.-G., & Kwack, K. (2020). A Disintegrin and Metalloproteinase 9 (ADAM9) in Advanced Hepatocellular Carcinoma and Their Role as a Biomarker During Hepatocellular Carcinoma Immunotherapy. Cancers, 12(3), 745. https://doi.org/10.3390/cancers12030745






