Abstract
Uveal melanoma (UM) has well-characterised somatic copy number alterations (SCNA) in chromosomes 1, 3, 6 and 8, in addition to mutations in GNAQ, GNA11, CYSLTR2, PLCB4, BAP1, SF3B1 and EIF1AX, most being linked to metastatic-risk. To gain further insight into the molecular landscape of UM, we designed a targeted next-generation sequencing (NGS) panel to detect SCNA and mutations in routine clinical UM samples. We compared hybrid-capture and amplicon-based target enrichment methods and tested a larger cohort of primary UM samples on the best performing panel. UM clinical samples processed either as fresh-frozen, formalin-fixed paraffin embedded (FFPE), small intraocular biopsies or following irradiation were successfully profiled using NGS, with hybrid capture outperforming the PCR-based enrichment methodology. We identified monosomy 3 (M3)-UM that were wild-type for BAP1 but harbored SF3B1 mutations, novel frameshift deletions in SF3B1 and EIF1AX, as well as a PLCB4 mutation outside of the hotspot on exon 20 coinciding with a GNAQ mutation in some UM. We observed samples that harboured mutations in both BAP1 and SF3B1, and SF3B1 and EIF1AX, respectively. Novel mutations were also identified in TTC28, KTN1, CSMD1 and TP53BP1. NGS can simultaneously assess SCNA and mutation data in UM, in a reliable and reproducible way, irrespective of sample type or previous processing. BAP1 and SF3B1 mutations, in addition to 8q copy number, are of added importance when determining UM patient outcome.
1. Introduction
Uveal melanoma (UM), the most common primary intraocular malignancy in adults, has an incidence of 3–8 individuals per million per year in Caucasians [1,2]. Despite successful treatment of the primary tumor with surgery and/or radiotherapy, metastatic death occurs in ~50% of patients [3,4]. Stratifying UM patients on the basis of their metastatic-risk is essential for efficient, personalised care. In Liverpool, UM patients are currently stratified into metastatic-risk groups—i.e., low (LR) or high (HR) risk—using a combination of clinical, histopathological and genetic factors [5,6]. Patients with HR-UM undergo regular liver imaging, using magnetic resonance imaging (MRI) to enable the early detection of metastases, and thereby enhance opportunities for liver resection and enrolment into clinical trials [5]. Liver resection has been shown to prolong the median survival of UM patients by 19 months compared with patients treated palliatively [7]. Conversely, patients with LR-UM can be reassured and avoid long-term surveillance, and there are proven benefits both to them and to health service providers [3].
Distinct somatic copy number alterations (SCNA) occur in UM, the most common being monosomy 3 (M3) [8]. This corresponds with a significantly worse prognosis, especially when accompanied by polysomy chromosome (chr) 8q [9,10]. Increasing copies of chr 8q significantly correlate with reduced survival, in a dose-dependent fashion [11]. SCNA in chr 1p, 6p and 6q have also been linked with survival outcomes [11,12,13,14].
In addition to these well-characterised SCNA, UM has two sets of driver mutations: one which initiates tumorigenesis in the form of mutually exclusive gain-of-function mutations in GNAQ, GNA11, CYSLTR2 or PLCB4, major players in the Gq signalling pathway [15,16,17,18]; and the other consists of mutations in BAP1 [19], SF3B1/SRSF2 [11,20] and EIF1AX [20], which have been correlated with high-, intermediate- and low-metastatic risk groups, respectively [11]. Inactivating mutations in BAP1 are closely associated with HR-M3 UM, with recent data suggesting that the bi-allelic inactivation of BAP1 is required to influence prognosis [21]. Missense mutations in splicing factor SF3B1 are often observed in disomy 3 (D3) UM and have been shown to predispose patients to late-onset metastatic disease [22]. Similarly, mutations in SRSF2, another member of the spliceosome, are observed in D3-UM, suggesting there are some functional similarities between SRSF2- and SF3B1-mutant UM [11]. Mutations in EIF1AX are mainly observed in D3-UM and are associated with LR-UM [23]. Other mutations in FBXW7 [20], DLK2, CSMD1, KTN1, TP53BP1, TTC28 [14] and MAPKAPK5 [11] have also been observed at low frequencies; however, their clinical significance remains unknown.
Recent genomic studies reported that UM could be subdivided into four main groups using unsupervised hierarchical clustering according to genetic alterations (SCNA, mutations and RNA-Seq), which were associated with an increasingly poor prognosis [11,14]. Based on these findings, there have been several efforts to design targeted next-generation sequencing (NGS) panels specifically for UM. In 2017, a bespoke NGS panel was designed to examine mutations in skin melanoma and UM simultaneously; however, this only examined mutations in GNAQ and GNA11, which are not associated with patient prognosis [24]. Another panel combined SCNA analysis of chromosomes 1, 3 and 8 and mutation analysis of GNAQ, GNA11, BAP1, SF3B1 and EIF1AX using the Ion Torrent (Thermofisher Scientific) sequencing platform [25]. More recently, a pan-cancer sequencing panel consisting of 500 genes frequently mutated in cancer (including those frequently mutated in UM) was used to analyse 62 non-irradiated biopsies and fresh resection UM samples [26], and also in another study, 35 matched primary UM and their metastases [27]. The studies reported the successful detection of SCNA and mutations that may enhance survival prognostication. Castle Biosciences have also developed a 7-gene NGS panel ‘DecisionDx-UMSeq’, although to our knowledge this has not been tested with formalin-fixed paraffin embedded (FFPE) or irradiated material.
This study details the largest cohort of UM-patients to be analysed using a targeted NGS panel to date. We examined the ability of NGS to detect both SCNA in chr 1, 3, 6 and 8, and mutations in GNAQ, GNA11, CYSLTR2, PLCB4, BAP1, SF3B1, SRSF2, EIF1AX, FBXW7, DLK2, CSMD1, KTN1, TP53BP1 and TTC28, in irradiated UM, as well as in FFPE tumor samples. Hybrid capture and PCR-based enrichment methods for NGS were initially compared. Following this, the best technology was chosen for the evaluation of a larger UM cohort, and all genetic data were correlated with clinical and histopathological features, and with patient outcome.
2. Results
2.1. Patient and Tumor Demographics
DNA from primary UM samples with a median follow-up of 65 months (range 0–132 months) were from 117 consenting patients treated at the Liverpool Ocular Oncology Centre (LOOC), Liverpool University Hospitals NHS Foundation Trust. Of the UM samples analysed, 27/117 (23%) were biopsies that had residual DNA available (stored at −80 °C), 14/117 (12%) specimens were FFPE and 76/117 (65%) were frozen, resection samples from which DNA could be extracted. Twenty-six cases were selected, as they were taken post-irradiation with either ruthenium plaque radiotherapy (PRXT) or proton beam radiotherapy (PBR) (Figure 1). All samples had previously undergone routine genetic testing by either multiplex ligation dependent probe amplification (MLPA) or microsatellite analysis (MSA).
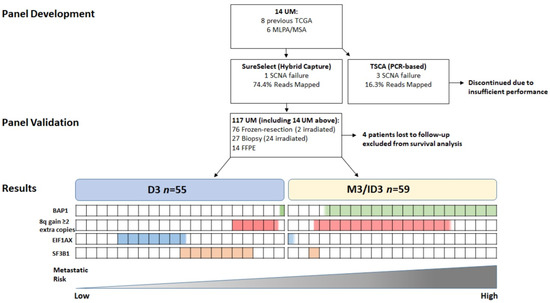
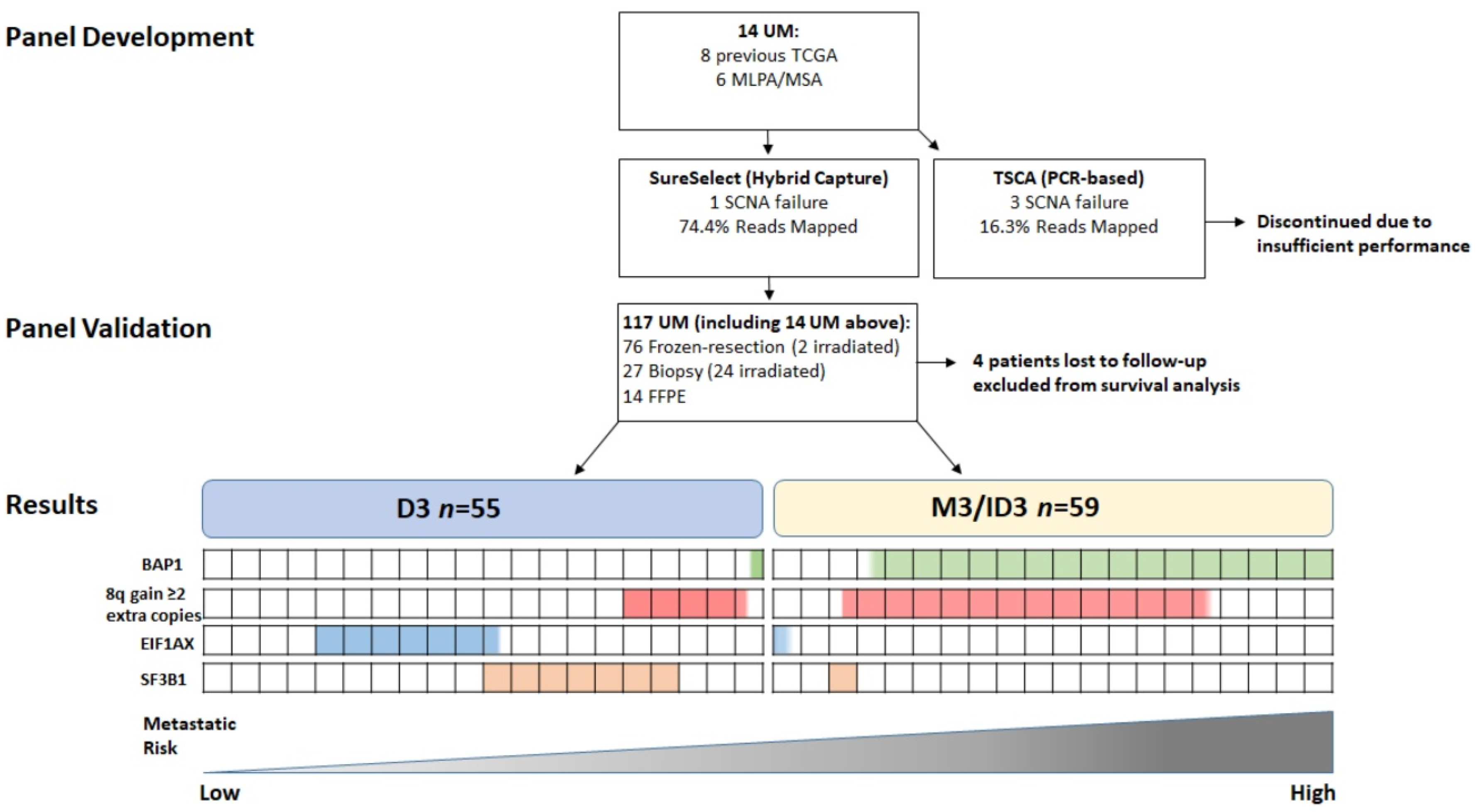
Figure 1.
Flowchart of 117 UM specimens examined in the present study: n = 76 frozen-resection (2 post-irradiation); n = 27 Biopsy (24 post-irradiation); n = 14 FFPE. Four patients were lost to follow-up and excluded from survival analysis. n = 55 were D3, and n = 59 were M3 or ID3. Proportion of cases with the genetic alteration listed are highlighted by the coloured boxes. Each box represents 5% of UM patients examined.
The study consisted of 63 males and 54 females with a median age of 64; range 16–87 years (mean age 62 years) at the time of management of their primary UM. Primary management was enucleation in 78/117 (66%) UM patients; local resection 12/117 (10%); endoresection 1/117 (1%); PRXT 16/117 (14%); and PBR in 10/117 (9%). Secondary treatment was necessary for 4/117 (4%) UM patients, as a result of tumor recurrence (Table 1). Figure 1 describes the flow of patients through this study.

Table 1.
Patient and tumor demographics of n = 117 UM patients treated at Liverpool Ocular Oncology Centre.
The UM median largest basal diameter (LBD) was 15.0; range 4–22 mm (mean 14.6 mm) with a median ultrasound height (UH) 7.5; range 1–15.7 mm (mean 7.5 mm) (Table 1). The American Joint Committee on Cancer (AJCC) stage was: 14/117 (12%) stage 1, 26/117 (22%) stage 2, 57/117 (49%) stage 3 and 20/117 (17%) stage 4. Ciliary body involvement was reported in 36/117 (31%) cases and extraocular UM extension was present in 9/117 (8%) of cases. Epithelioid cells were seen in 50/117 (43%) of cases with the remaining 67/117 (57%) having a spindle cell morphology. Full histological assessment was only undertaken in resection specimens (enucleation or local resection samples; n = 90), which had a mean mitotic count of 7/40 hpf (median 5/40 hpf; range 1–72/hpf). Closed Periodic Acid Schiff (PAS) and connective tissue loops were identified in 47/90 (52%) cases, and focal necrosis was observed in 21/89 (24%) cases. At study closure (23 September 2019), 62/117 (53%) patients were alive without evidence of metastasis, 40/117 (34%) patients had died from metastatic disease, 11/117 (10%) patients died from other causes and 4/117 (3%) patients were lost to follow-up.
2.2. Panel Comparison (14 Samples)
Of the initial 14 UM samples analysed for panel comparison, 1/14 (7%) and 3/14 (21%) failed to produce reportable SCNA data with the SureSelect (SureSelect XT HS using SureDesign, Agilent) and TSCA (TruSeq Custom Amplicon using DesignStudio Illumina) panels, respectively. 13/14 (93%) UM samples had available SCNA data from previous MLPA for chr1, 3, 6 and 8; the remaining sample was tested by MSA for chr3 status only. There was 100% agreement for chr3 status between the MLPA/MSA data and that provided by both NGS tests in this initial sample cohort (Table S1—samples marked by an asterisk). There was 100% concordance for GNAQ, GNA11, BAP1, SF3B1 and EIF1AX mutations between both testing platforms. No false positives were detected in any of the samples. Of note, 6/14 UM test samples had been previously submitted by our group to the TCGA-UM study, and there was also 100% concordance for all mutations identified. The SureSelect panel was chosen to test the larger UM cohort, due to its greater success rate in SCNA analysis and better coverage (Table S2).
2.3. Mutation Frequency
In total, 117 UM samples (including the 14 initial samples analysed) were sequenced using the above bespoke SureSelect NGS panel. This included 26 UM that had previously undergone PBR or PRXT and for which mutation data was successfully obtained. Initiating mutations occurred in 62/117 (53%) for GNAQ; 42/117 (36%) for GNA11; 2/117 (2%) for CYSLTR2 and 1/117 (1%) for PLCB4, which was concomitant with a GNAQ mutation (Table S3). Driver mutations occurred in 50/117 (43%) for BAP1 (1/50 (2%) occurring in a D3-UM); 25/117 (21%) for SF3B1 (3/25 (12%) coincided with a BAP1 mutation 2/25 (8%) coincided with an EIF1AX mutation, 5/25 (20%) had partial loss or M3); 22/117 (19%) for EIF1AX (2/22 (9%) occurring in a M3-UM). Interestingly, two D3-UM were found to have concurrent EIF1AX and SF3B1 mutations.
Novel mutations were observed in: PLCB4: 1/117 p.Met549_Gly556delinsIle; KTN1: 2/117 p.Pro195Thr p.Gln86dup; TTC28: 4/117p. Arg21*, p.Pro1216His, p.Ala18Gly and p.lleI1296Val; CCMD1: 2/117 p.Pro1097His, p.Pro108Leu; TP53BP1: 2/117 p.Ile455_Pro456del and p.Glu1529*. These rare variants were confirmed using Integrative Genomics Viewer with a minimum allele frequency of 30%. No mutations were detected in any of the cases for the genes BRAF, DLK2, FBXW7 or SRSF2.
2.4. SCNA Analysis and Comparison with MLPA/MSA
We compared the SCNA datasets to establish whether the SureSelect NGS panel accurately detected SCNA in chr1, 3, 6 and 8 when analysed by MLPA and for chr3 when analysed by MSA. One sample failed to provide clear SCNA data by NGS and was excluded from the concordance data below, as were SCNA deemed ‘unclassifiable’ by MLPA. Concordance was observed with NGS as follows: chr1p—81/98 (83%); chr3—103/112 (92%); chr6p—68/88 (77%); chr6q—77/99 (78%); chr8p—64/102 (63%); and chr8q—72/97 (74%) (Table S1).
SCNA data from the NGS panel was successfully obtained from both non-irradiated and irradiated samples and demonstrated: loss of 1p in 25/116 (22%) with 8/25 (32%) coinciding with a concomitant gain of 1q; gains in 1q in 9/116 (8%); M3 in 55/116 (47%); isodisomy 3 (ID3) in 2/116 (2%); loss of 3p in 1/116 (1%) and loss of 3q in 1/116 (1%), subsequently categorised as partial loss of chr3 (PL3); 6p gain in 46/116 (40%) cases with 37/46 (80%) occurring with D3 and 9/46 (20%) occurring with M3/ ID3/PL3; 6q loss in 25/116 (22%) of samples with 12/25(48%) occurring with M3/isodisomy 3/PL3; 8p loss in 20/116 (17%) each with a concomitant gain of 8q (Table S3). A complete gain of chr8 was seen in 36/116 (31%) UM. Gain of chr8q only occurred in 75/116 (65%) samples; 24/75 (32%) in D3 UM and 51/75 (68%) in M3/ID3/PL3 UM. 8q gain varied with respect to number of extra copies: the median was two extra copies for both M3/ID3/PL3 and D3 UM ranging from 1 to 9 in the former group and from 1 to 4 in D3 UM.
2.5. Cox Regression
Univariate analysis was carried out using a significance level of p < 0.005 after Bonferroni correction.
Factors significantly associated with survival were: epithelioid cytomorphology, LBD, UH, ciliary body involvement, BAP1 and chr3 status (Table 2). These variables were entered into the Cox model and backward selection of covariates was carried out using the likelihood ratio to determine ‘goodness of fit’ of the model. At the 0.01 significance level, chr3 loss was significantly associated with reduced survival (p ≤ 0.001) with a hazard ratio of 5.949 (Table 3).

Table 2.
Univariate analysis of n = 117 UM patients treated at Liverpool Ocular Oncology Centre.

Table 3.
Multivariate analysis of n = 117 UM patients treated at Liverpool Ocular Oncology Centre.
2.6. Survival
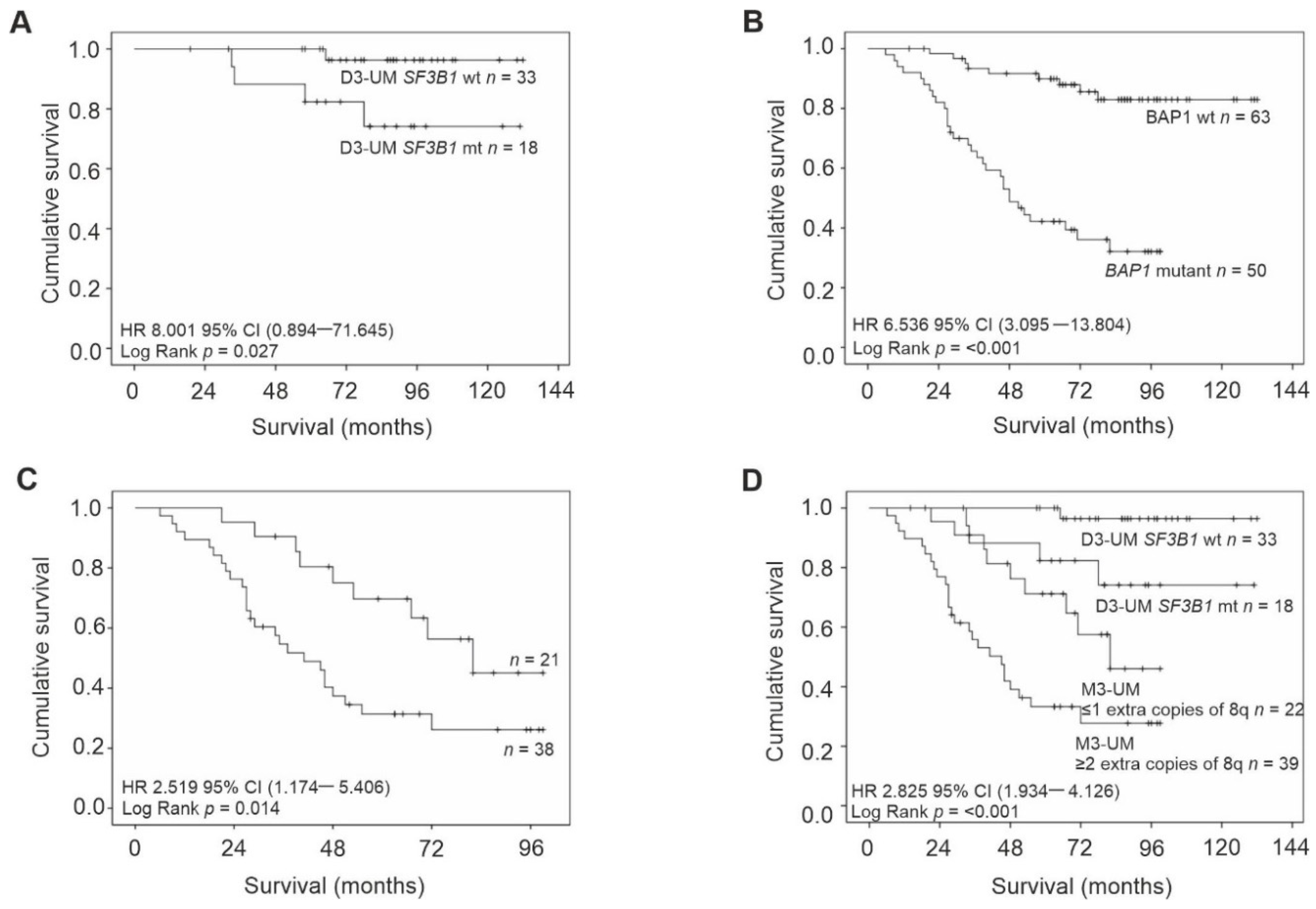
Kaplan–Meier survival curves and tables were examined for all primary UM stratified according to: chr3 status, extra copies of chr8q, and mutations in BAP1 and SF3B1. The following were significantly associated with a reduced survival time: loss of chr3 (Log Rank p < 0.001), BAP1 mutations (Log Rank p < 0.001), M3-UM with more than two copies of 8q (Log Rank p = 0.014) and D3-UM with SF3B1 mutations (Log Rank p = 0.027) (Figure 2).
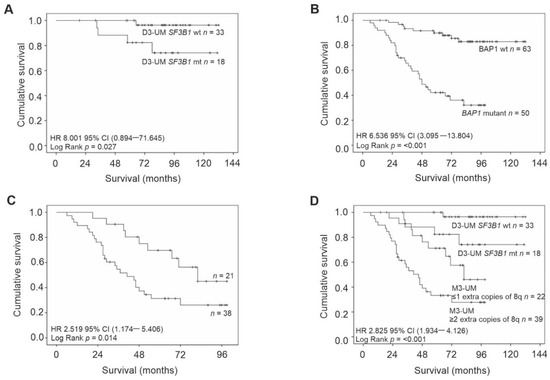
Figure 2.
Kaplan–Meier survival curves estimate survival in UM patients stratified by: (A) SF3B1 wild-type/mutation status in D3-UM n = 51 (Log Rank, p = 0.027); (B) BAP1 wild-type/mutation status n = 113 (Log Rank, p < 0.001); (C) extra copies of chr 8q in M3/ID3-UM n = 59 (Log Rank, p = 0.014) and (D) SF3B1 wild-type/mutation status in D3-UM n = 51 and extra copies of chr 8q in M3/ID3-UM n = 59 (Log Rank, p < 0.001). Number of events indicates the number of deaths due to metastatic melanoma. Log Rank tests were used to compare survival across groups.
2.7. BAP1 IHC
Seventy of the ninety surgical UM samples (enucleation/local resection) had previously undergone routine immunohistochemistry (IHC) to determine nuclear BAP1 (nBAP1) protein expression; the remaining samples did not have enough material for subsequent IHC analysis. nBAP1 protein was absent in 38/70 cases (54%) of which 31 (82%) UM also had mutations in the BAP1 gene. Of the 7/38 (18%) UM with no BAP1 mutations, four patients had M3-UM and three had died from metastatic disease. Furthermore, 3/32 (9%) UM positively expressed nBAP1 protein but had clear mutations in BAP1, all of which were missense alterations (q.Glu31Lys, q.Cys91Gly and q.Ala142Pro).
2.8. SF3B1 Mutations in M3 UM
SF3B1 mutations have previously been associated with D3-UM with late onset metastasis [22]. In our cohort, 5/25 cases (20%) with SF3B1 mutations died of metastatic UM at the time of study closure. Of these five cases, four tumors were D3-UM and one was a M3-UM with a BAP1 mutation. To investigate the prevalence of SF3B1 mutations in M3-UM that lacked mutations in BAP1, we identified 20 additional cases of M3-UM where DNA was available and previous IHC analysis had demonstrated strong nBAP1 positivity, correlating with wild-type BAP1 [28]. This additional UM cohort consisted of 12 males and 8 females with a mean age of 62 years at primary management (median age 62; range 45–80 years). The mean follow-up period was 48 months (median 61 months; range 6–79 months). Primary management was enucleation 17/20 (85%) and local resection 3/20 (15%). The mean LBD was 14.8 mm (median LBD 14.7; range 9.8–22.7 mm) with a mean UH of 8.0 mm (median UH 8.4; range 1.7–12.4 mm). Full histological assessment is detailed in Table S4. Of these additional 20 UM, 5 (25%) had mutations in SF3B1; 3/5 (60%) q.Arg625Cys and 2/5 (40%) q.Arg625His. At study closure, all five patients were alive; of interest, one patient developed liver metastases 40 months after primary management but underwent metastasectomy and is still alive 25 months after surgery.
3. Discussion
This is the largest study to date to profile UM using bespoke targeted NGS panels. It identified chr3 as the most significant factor associated with metastatic death and demonstrated for the first time that irradiated UM samples can be successfully profiled using NGS with no observable differences in quality when compared to non-irradiated UM samples. We identified a subset of M3-UM-patients without nBAP1 loss that demonstrated mutations in SF3B1 and also describe concurrent disruptive frameshift deletions in SF3B1 and EIF1AX. This is consistent with the observation in one case sequenced in TCGA that harboured both an EIF1AX and an atypical SF3B1 (T663P) mutation [11]. We also observed co-occurring mutations in BAP1 and SF3B1 and EIF1AX and SF3B1. Novel mutations were also identified in TTC28, KTN1, CSMD1 and TP53BP1. Of interest, we identified a mutation in PLCB4 that does not fall within the hotspot on exon 20 and coincides with a GNAQ mutation. Furthermore, chr3 results obtained using the NGS panel were comparable to previous MLPA and MSA analyses. We recommend that this bespoke NGS panel ultimately replaces MLPA/MSA testing in routine labs, with the possibility of incorporating molecular data into prognostic tools—e.g., the LUMPO (Liverpool Uveal Melanoma Prognosticator Online), which was recently externally validated in a multicentre study [29].
3.1. Enrichment Comparison
Hybrid capture and PCR-based enrichment methods in NGS vary in how targeted regions are enriched [30]. Hybrid capture methodologies like the SureSelect XT HS used in this study, involve shearing gDNA into smaller fragments, library preparation and hybridisation with targeted biotinylated RNA baits. Using magnetic streptavidin beads, these baits can be separated, and the hybridised library amplified; whilst PCR-based methods hybridise a custom oligo pool flanking-targeted regions on unfragmented gDNA. These are then extended and ligated, and PCR is performed to integrate indexes and sequencing primers. The PCR-based method has the advantages of requiring lower DNA inputs with shorter preparation times. In our study, hybrid capture outperformed the PCR-based enrichment in terms of a larger percentage of reads mapped and a greater mean depth of coverage. Although there were no differences in the ability to call single nucleotide variants (SNV), there was an increased SCNA analysis failure rate for the PCR-based method. Similar comparison investigations in other cancer types found limited sensitivity of PCR-based sequencing, with several variants being missed due to regions of high guanine-cytosine content and suboptimal PCR conditions, yielding a minimal coverage not found when using hybrid capture [31,32,33]. An increased incidence of false positives and missed variants in PCR-based enrichment was also reported when evaluating hybrid capture versus PCR-based methods for whole-exome sequencing [34]. In contrast to our comparison, neither study found differences between the success rates of SCNA analysis.
3.2. Comparison with Previous MLPA
In the current study, we were able to successfully examine both SCNA and SNV using a single NGS assay in fresh, FFPE and also irradiated tissues. Only one sample failed to produce a clear genotype, but this was expected because of a low yield of library post-capture. Furthermore, 10/116 (9%) UM samples were discordant with the original MLPA/MSA analyses for chr3: 2 were isodisomy 3, which had been classified as D3 by MLPA due to its limitations in detecting acquired homozygosity; two were shown to have regions of deletion not identified in previous MLPA, most likely due to an increased number of probes covering chr3 on the NGS panel. Of the remaining six discordant samples, four had been classified as M3 by MLPA but as D3 by NGS; two of these cases had SF3B1 mutations but all patients were alive at the study closure. Two had been classified as D3 by MLPA but M3 by NGS; one had a BAP1 mutation and both patients had died from metastatic disease. For chr1, 6 and 8, the discordance between the MLPA and the NGS SCNA was greater at 17–26% of UM cases, which is likely a result of the low probe coverage for these chromosomes on the MLPA panel. Whilst the median 8q copy number was the same in D3-UM and M3-UM, the 8q copy number burden was generally higher in M3-UM. This was reflected by a reduced survival in M3-UM with an 8q copy number of 4 or more consistent with previous reports that 8q dosage is an important predictor of outcome in UM [11,35].
3.3. Irradiated Samples
This is the first study to examine irradiated UM samples using a NGS panel. No diminished quality or ability to genotype these tumors was observed amongst these samples. This is consistent with our findings using MSA/MLPA to genotype irradiated UM [36,37,38].
3.4. BAP1 Mutations
The frequency of BAP1 mutations in the present study was 43% in total, occurring in 82% of M3-UM; these data are consistent with the findings of others [11,14,19,25]. The presence of a BAP1 mutation in UM was associated with a worse survival. We have previously reported that nBAP1+ M3-UM have a better prognosis as compared with nBAP1− M3-UM [21]; however, interestingly in this current study, M3-UM that were wild-type for BAP1 (10/57; 18%) did not correlate with an increased survival time as compared with M3-UM with BAP1 mutations. This may be due to either the observation that BAP1 mutations do not always correlate with loss of nBAP1 protein expression, or to the smaller cohort of patients in the present study [28,39].
3.5. SF3B1 Mutations
The frequency of SF3B1 mutations in UM ranges in the literature from 11–34% [14,25], and in this study SF3B1 mutations occurred in 21% of cases. SF3B1 mutations are reported to occur mainly in D3-UM associated with late onset metastasis and decreased survival (22). This is consistent with our study in which 20/25 (80%) SF3B1 mutations occurred in D3-UM with a significantly reduced survival time as compared with D3/SF3B1wt UM (p = 0.027).
A novel disruptive frameshift deletion in SF3B1 of 15 nucleotides was observed in p.Lys653_Ser657del on heat domain 4, outside the hotspot region of codon 625; the significance of this is unclear. Of particular interest in our study are five M3-UM or UM with PL of chromosome 3 with SF3B1 mutations. Two of these UM harboured BAP1 mutations, previously described in one other study (11); one patient succumbed to metastatic disease 12 months after primary management, and the second patient died of other causes 99 months (8.25 years) later. Three SF3B1 mutations were recorded in M3-BAP1wt UM, a phenomenon only observed in one other study to date [11]. To examine this further, we tested an additional 20 cases of M3-UM with nBAP1 positivity and identified five cases with SF3B1 mutations; at the time of study closure, all five patients were alive. Additional cases and longer follow-up are required to fully understand the clinical relevance of SF3B1 mutations in M3-UM.
3.6. EIF1AX Mutations
EIF1AX mutations were detected in the present study in 19% of UM, which is consistent with that reported by other groups [11,14,18,25]. Interestingly, two UM demonstrated mutations in both EIF1AX and SF3B1 despite previous reports describing that these occur in a mutually exclusive manner [11,25]. Of note, both patients died from metastatic disease at 34 and 58 months, respectively, after primary treatment. EIF1AX mutations are typically associated with D3-UM; however, we identified two M3-UM that displayed mutations in this gene. A novel disruptive frameshift deletion of 6 nucleotides from the coding sequence was also identified in p.Arg14_Gly15del of EIF1AX.
3.7. Initiating Mutations
Mutations in GNAQ and GNA11 occurred in 89% of UM in a mutually exclusive manner (53% and 39%, respectively), consistent with the literature [11,14,25]. Mutations predominantly occurred in exon 5 for GNAQ and GNA11, and two UM had mutations in exon 4. One sample contained two unusual mutations in exon 4 of GNA11 p.R214K and p.R214S. These regions do not lie within any of the known functional domains of GNA11 and have not been previously described; their effect on GNA11 protein function is unknown. Mutations in CYSLTR2 were found in two UM in the hot spot region p.L129Q in exon 1 and occurred in a mutually exclusive manner to mutations in GNAQ and GNA11, as previously reported [17]. Consistent with our general understanding of the function of these mutations, there were no differences in survival outcome based on the mutational status of the driver mutations GNAQ, GNA11 and CYSLTR2.
Disruptive frameshift deletions in p.M549_G556delinsI and M561_G568delinsI mutations were observed in PLCB4 in a single UM sample. These cases also showed a p.R183Q mutation in GNAQ. Previous studies identified recurrent mutations in PLCB4 in a hot-spot region p.D630Y and p.D630N on exon 20 [18]. The mutation identified in our study occurred in exon 18 and is the first mutation in this region to be described in UM. Though it was initially thought that PLCB4 mutations occurred in a mutually exclusive manner to GNAQ, GNA11 and CYSLTR2, our study and that of Robertson et al. [11] demonstrate PLCB4 mutations concurrent to GNAQ and GNA11 mutations.
3.8. Other Mutations
We observed low frequency (3%) somatic mutations in genes originally identified by Royer-Bertrand et al. (6%), namely in TTC28, CSMD1, KTN1 and TP53BP1 [14]. Most of these genes are involved in various cellular processes, e.g., cell cycle regulation [40], cell migration and proliferation [41,42], kinesin binding [43] and DNA double-strand break repair [44]. Our NGS panel was custom-designed to have full coverage of the TTC28, CSMD1, KTN1 and TP53BP1 genes, and because of its targeted nature had greater coverage in comparison to whole-exome sequencing methodologies. Due to their low frequency in this study, no association could be made between the mutations in TTC28, CSMD1, KTN1 or TP53BP1 and UM with particular clinical or morphological features. It is worth noting that previously described mutations in SRSF2, DLK2 or FBXW7 were not detected in this large study [14,20,45].
4. Materials and Methods
4.1. Patients
In this retrospective cohort study, primary UM samples were collected from 117 patients who were treated at the Liverpool Ocular Oncology Centre (LOOC), Liverpool University Hospitals NHS Foundation Trust, between January 2008 and May 2015. This time period was chosen to allow sufficient follow-up (median, 65 months). The follow-up period was calculated from the date of primary management to either study end (23 September 2019) or to death from metastatic disease or other causes. Patients were treated either by radiotherapy or surgical resection, and their UM was genotyped using either MLPA or MSA, as described below.
4.2. Specimen Characteristics
Specimens consisted of DNA (stored at −80 °C) previously extracted from fresh biopsies all preserved in CytoLyt (Cytyc Corp) and stored at 4 °C, fresh-tumor tissue snap-frozen in liquid nitrogen and stored at −80 °C, and FFPE UM samples stored at room temperature. Twenty-six of the DNA samples analysed were post-irradiation specimens.
4.3. Study Design
The clinical endpoint examined in this study was death from metastatic disease. Patients who died from causes other than those relating to UM were included in the study, and data for these records were treated as right-censored cases for evaluation purposes. This study conformed to the principles of the Declaration of Helsinki and Good Clinical Practice guidelines. Approval for the study was obtained from the Health Research Authority South Central - Hampshire B Research Ethics Committee (REC ref 15/SC/0611). All samples and data were provided by the Ocular Oncology Biobank (REC ref 16/NW/0380). All patients had provided informed consent for the use of their samples and data in research.
4.4. Assay Methods
4.4.1. Morphological/Histological Studies
All samples underwent routine histopathological and cytological workup assessing cell type, mitotic count, and presence of PAS+ connective tissue loops where possible (28). Furthermore, 90/117 enucleation and local resection specimens had a full histological workup, whilst 27/117 biopsies and endoresection specimens underwent cytological examination only. Additionally, IHC analysis of nBAP1 expression was undertaken in 70/117 cases, as described previously [21].
4.4.2. DNA Extraction and Quantification
Methods for DNA extraction from FFPE and frozen UM have been published elsewhere [46]. DNA integrity of FFPE samples was qualified by performing a qPCR using the Agilent NGS FFPE QC Kit. (Agilent Technologies Inc., UK).
4.4.3. Chromosomal SCNA Analysis
MLPA (MRC Holland, The Netherlands) and MSA were used to assess SCNA, and subsequent comparison with NGS data were undertaken during routine genetic testing of patient samples, as previously described [47,48]. Cases yielding >100 ng of DNA were tested using MLPA, whilst MSA was undertaken for UM samples with lower DNA yields.
4.4.4. Next-Generation Sequencing
Two custom NGS panels were designed: SureSelect XT HS using SureDesign (Agilent) and TruSeq Custom Amplicon (TSCA) using DesignStudio (Illumina). Both panels were designed to cover mutations in GNAQ (exons 4 & 5), GNA11 (exons 4 & 5), SF3B1 (exons 12 & 14), EIF1AX (exons 1 and 2), and all exons of BAP1, FBXW7, DLK2, CSMD1, CYSLTR2, KTN1, TP53BP1, SRSF2, PLCB4, TTC28 and BRAF (negative control). Both enrichment methods included the incorporation of unique molecular identifiers or barcodes to reduce errors and quantitative bias introduced by the amplification process. For the SureSelect XT HS, additional probes were included to examine SCNA in chr1: 1541 probes; chr3: 1287 probes; chr6: 1094 probes; chr8: 933. The TSCA panel included additional probes to examine SCNA in chr3: 83 amplicons; chr6: 76 amplicons and chr8: 67 amplicons. Chr1 was not included in the TSCA NGS panel due to tiling limitations. As the panels were worked up on larger resection samples, the DNA input was 50 ng for both panels. Libraries were constructed using either the SureSelect XT HS Reagent and Capture Library Kit (Agilent Technologies Inc., United Kingdom) or TruSeq Custom Amplicon Low Input Kit (Illumina Inc., United Kingdom), according to the manufacturer’s instructions. The two panels were tested and compared using 14 frozen UM samples, 8 of which had been previously profiled by The Cancer Genome Atlas (TCGA) UM study [11], and 6 had available data from previous genotyping plus an additional two reference samples (Genome In A Bottle, HDx).
The SureSelect XT HS was subsequently selected to test a larger cohort of 95 fresh and 13 FFPE UM samples with reference samples included in each sequencing run. The DNA input varied (5 ng–25 ng) depending upon the sample type.
4.4.5. Sanger Sequencing
Exon 14 of SF3B1 was sequenced using PCR-based capillary Sanger sequencing in an additional twenty M3-UM with unusual nBAP1+ protein expression [21]. Oligonucleotides were constructed by Eurofins Genomics; forward 5’-GGCCGAGAGATCATTTCT-3, reverse 5’-AAGAAGGGCAATAAAGAAGGA-3’, product size 289bp. PCR was performed in a reaction volume of 50 μL containing 100 ng of genomic DNA, 0.25 μL of Thermo-Start Taq DNA Polymerase (Thermo Scientific), 5 μL of HP Buffer, 4 μL of 25 mM MgCl2, 2 μL of dNTP (2 mM each), 31.25 μL Nuclease Free water and 1 μL of each of the primers. The thermal cycling profile was as follows: initial denaturation at 95 °C for 15 min and 35 rounds of amplification at 95 °C for 15 s, 55 °C for 30 s and 72 °C for 1 min. A final extension step at 72 °C for 5 min was added. PCR products were purified using the QIAquick PCR purification kit (Qiagen, United Kingdom) according to the manufacturer’s protocol. Sequencing of PCR products was carried out by GATC at Eurofins Genomics in accordance with ISO 17025. Sequencing data were analysed using Chromas Lite (2.1.1., Technelysium Pty Ltd.).
4.5. NGS Data Analysis
NGS libraries were sequenced on the Illumina MiSeq platform (2 × 250 bp paired-end) by the Centre for Genomic Research (www.cgr.liv.ac.uk), University of Liverpool, UK. Base-calling and de-multiplexing of indexed reads were performed by CASAVA version 1.8.2 (Illumina) to produce the raw sequence data in FASTQ format. The raw FASTQ reads were trimmed to remove Illumina adapter sequences using Cutadapt version 1.2, and low-quality bases using Sickle version 1.200.
Trimmed reads were aligned to the human GRCh37 reference genome (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/phase2_reference_assembly_sequence/hs37d5.fa.gz) with the short-read alignment tool, BWA-MEM (version 0.7.5a-r405). Following alignment, PCR and optical duplicate reads were identified and removed with UMI-tools (https://github.com/CGATOxford/UMI-tools). Subsequently, the Genome Analysis Toolkit (GATK) (version 3.7) Indel Re-Aligner module was used to locally realign reads around the putative insertion and deletion sites. GATK BaseRecalibrator module was used for recalibrating the base calls. The aligned data were then analysed using tCoNut (https://github.com/tgen/tCoNuT) to detect SCNAs. The variants were called by GATK and annotated by SNPeff.
4.6. Statistical Analysis Methods
Survival time (months) was calculated from the date of primary management until death from metastases or study closure on 23 September 2019. Median survival time was estimated using the Kaplan–Meier product limit method. Univariate associations between survival time, clinical, histological and genetic features were examined using Cox proportional hazards regression models. Analyses were undertaken using SPSS Statistics v.24 (IBM), Microsoft R 3.5.1 and the packages rms, cmprsk and mstate. Cut-offs for SCNA used established values based on previous clustering analysis carried out at our centre: log rank < 0.85 loss, > 1.15 amplification [49]. The allelic frequency threshold to call a mutation was 10%.
5. Conclusions
Our bespoke UM NGS panel enables detailed SCNA and mutational information to be obtained from small UM biopsies, FFPE material and previously irradiated UM. This is in distinct contrast to some current methodologies, which, when applied to biopsies, can only determine chr3 status due to the low DNA yield. Moreover, consistent with other reports, BAP1 and SF3B1 mutations in addition to 8q copy number are of added importance when determining patient outcome and moves UM stratification away from a binary genetic classification based on chr3 copy number only. Identifying metastatic risk groups with greater precision than is currently possible with SCNA assessment alone will have implications on the frequency at which patients are followed up for subsequent liver imaging, and the imaging techniques applied, as well as on patient selection for clinical trials. Although at present, mutations in UM are not therapeutically actionable, it is hoped that continued advances in our understanding of this disease will result in the use of these biomarkers to predict response to emerging therapies.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/4/1039/s1, Table S1: Concordance Data, Table S2: NGS Quality Comparison, Table S3: Clinical, Molecular and Histopathological data 117 patients, Table S4: Monosomy 3 SF3B1 Cohort Data.
Author Contributions
Conceptualization S.E.C., H.K., S.T., H.H. and R.H.; methodology, S.T., L.O., J.S.S. and N.K.; software, X.L. and S.H.; validation, S.T.; formal analysis, S.T. and A.T.; investigation, S.T.; resources, S.E.C., H.K., C.H.-F. and J.G.K.; data curation, X.L., S.H., and S.T.; writing—original draft preparation, S.T.; writing—review and editing, S.E.C., H.K., S.T., L.O., J.S.S., J.G.K., C.H.-F., A.T., H.H., R.H., N.K., X.L. and S.H.; visualization, S.T. and H.K.; supervision, S.E.C., H.K. and J.G.K.; project administration, S.E.C. and H.K.; funding acquisition, S.T., S.E.C., H.K. and C.H.-F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Eye Tumour Research Charitable Funds, Liverpool University Hospitals NHS Foundation Trust grant number [A091/CF], who funded the PhD studentship of Sophie Thornton; and The Liverpool Health Genomics Healthcare Laboratory, Centre for Genomic Research, University of Liverpool, which funded part of the sequencing work.
Acknowledgments
The authors would like to thank and acknowledge the Biomedical Scientists of the Ophthalmic Pathology team at Liverpool Clinical Laboratories, namely Simon Biddolph and Anna Ikin; Gary Cheetham for maintaining the database of the Liverpool Ocular Oncology Centre; and Antonio Eleuteri for his statistical expertise and advice. A special appreciation to all the patients who kindly donated tissue used in this study. The second (SEC) and senior (HK) authors are associated with the UMCure2020 project (https://www.umcure2020.org/en/).
Conflicts of Interest
The authors declare no potential conflicts of interest.
References
- Virgili, G.; Gatta, G.; Ciccolallo, L.; Capocaccia, R.; Biggeri, A.; Crocetti, E.; Lutz, J.M.; Paci, E. Incidence of uveal melanoma in Europe. Ophthalmology 2007, 114, 2309–2315. [Google Scholar] [CrossRef] [PubMed]
- Yonekawa, Y.; Kim, I.K. Epidemiology and management of uveal melanoma. Hematol./Oncol. Clin. N. Am. 2012, 26, 1169–1184. [Google Scholar] [CrossRef] [PubMed]
- Damato, B. Progress in the management of patients with uveal melanoma. the 2012 Ashton Lecture. Eye 2012, 26, 1157–1172. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, R.D.; Schwartz, G.K.; Tezel, T.; Marr, B.; Francis, J.H.; Nathan, P.D. Metastatic disease from uveal melanoma: Treatment options and future prospects. Br. J. Ophthalmol. 2017, 101, 38–44. [Google Scholar] [CrossRef]
- Damato, B.; Eleuteri, A.; Taktak, A.F.G.; Coupland, S.E. Estimating prognosis for survival after treatment of choroidal melanoma. Prog. IN Retin. Eye Res. 2011, 30, 285–295. [Google Scholar] [CrossRef]
- DeParis, S.W.; Taktak, A.; Eleuteri, A.; Enanoria, W.; Heimann, H.; Coupland, S.E.; Damato, B. External Validation of the Liverpool Uveal Melanoma Prognosticator Online. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6116–6122. [Google Scholar] [CrossRef]
- Gomez, D.; Wetherill, C.; Cheong, J.; Jones, L.; Marshall, E.; Damato, B.; Coupland, S.E.; Ghaneh, P.; Poston, G.J.; Malik, H.Z.; et al. The Liverpool uveal melanoma liver metastases pathway: Outcome following liver resection. J. Surg. Oncol. 2014, 109, 542–547. [Google Scholar] [CrossRef]
- Prescher, G.; Bornfeld, N. Prognostic implications of monosomy 3 in uveal melanoma. Lancet 1996, 347, 1222. [Google Scholar]
- Horsman, D.E.; Rootman, J.; White, V.A.; Sroka, H. Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer Genet. Cytogenet. 1990, 45, 249–253. [Google Scholar] [CrossRef]
- Sisley, K.; Cottam, D.W.; Rennie, I.G.; Parsons, M.A.; Potter, A.M.; Potter, C.W.; Rees, R.C. Non-random abnormalities of chromosomes 3, 6, and 8 associated with posterior uveal melanoma. Genes Chromosomes Cancer 1992, 5, 197–200. [Google Scholar] [CrossRef]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220. [Google Scholar] [CrossRef]
- Kilic, E.; Naus, N.C.; van Gils, W.; Klaver, C.C.; van Til, M.E.; Verbiest, M.M.; Stijnen, T.; Mooy, C.M.; Paridaens, D.; Beverloo, H.B.; et al. Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2253–2257. [Google Scholar] [CrossRef] [PubMed]
- Damato, B.; Dopierala, J.; Klaasen, A.; van Dijk, M.; Sibbring, J.; Coupland, S.E. Multiplex ligation-dependent probe amplification of uveal melanoma: Correlation with metastatic death. Investig. Ophthalmol. Vis. Sci. 2009, 50, 3048–3055. [Google Scholar] [CrossRef] [PubMed]
- Royer-Bertrand, B.; Torsello, M.; Rimoldi, D.; El Zaoui, I.; Cisarova, K.; Pescini-Gobert, R.; Raynaud, F.; Zografos, L.; Schalenbourg, A.; Speiser, D.; et al. Comprehensive Genetic Landscape of Uveal Melanoma by Whole-Genome Sequencing. Am. J. Hum. Genet. 2016, 99, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009, 457, 599–602. [Google Scholar] [CrossRef]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef]
- Moore, A.R.; Ceraudo, E.; Sher, J.J.; Guan, Y.; Shoushtari, A.N.; Chang, M.T.; Zhang, J.Q.; Walczak, E.G.; Kazmi, M.A.; Taylor, B.S.; et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016, 48, 675–680. [Google Scholar] [CrossRef]
- Johansson, P.; Aoude, L.G.; Wadt, K.; Glasson, W.J.; Warrier, S.K.; Hewitt, A.W.; Kiilgaard, J.F.; Heegaard, S.; Isaacs, T.; Franchina, M.; et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 2015, 7, 4624–4631. [Google Scholar] [CrossRef]
- Harbour, J.W.; Onken, M.D.; Worley, L.A.; Matatall, K.A.; Roberson, E.D.O.; Duan, S.; Cao, L.; Council, M.L.; Helms, C.; Bowcock, A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef]
- Martin, M.; Maßhöfer, L.; Temming, P.; Rahmann, S.; Metz, C.; Bornfeld, N.; van de Nes, J.; Klein-Hitpass, L.; Hinnebusch, A.G.; Horsthemke, B.; et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013, 45, 933–936. [Google Scholar] [CrossRef]
- Farquhar, N.; Thornton, S.; Coupland, S.E.; Coulson, J.M.; Sacco, J.J.; Krishna, Y.; Heimann, H.; Taktak, A.; Cebulla, C.M.; Abdel-Rahman, M.H.; et al. Patterns of BAP1 protein expression provide insights into prognostic significance and the biology of uveal melanoma. J. Pathol. Clin. Res. 2017, 4, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Yavuzyigitoglu, S.; Koopmans, A.E.; Verdijk, R.M.; Vaarwater, J.; Eussen, B.; van Bodegom, A.; Paridaens, D.; Kilic, E.; de Klein, A. Uveal Melanomas with SF3B1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology 2016, 123, 1118–1128. [Google Scholar] [CrossRef]
- Dono, M.; Angelini, G.; Cecconi, M.; Amaro, A.; Esposito, A.I.; Mirisola, V.; Maric, I.; Lanza, F.; Nasciuti, F.; Viaggi, S.; et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: Detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br. J. Cancer 2014, 110, 1058–1065. [Google Scholar] [CrossRef]
- Reiman, A.; Kikuchi, H.; Scocchia, D.; Smith, P.; Tsang, Y.W.; Snead, D.; Cree, I.A. Validation of an NGS mutation detection panel for melanoma. BMC Cancer 2017, 17, 150. [Google Scholar] [CrossRef]
- Smit, K.N.; van Poppelen, N.M.; Vaarwater, J.; Verdijk, R.; van Marion, R.; Kalirai, H.; Coupland, S.E.; Thornton, S.; Farquhar, N.; Dubbink, H.J.; et al. Combined mutation and copy-number variation detection by targeted next-generation sequencing in uveal melanoma. Mod. Pathol. Off. J. USA Can. Acad. Pathol. Inc. 2018, 31, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Afshar, A.R.; Damato, B.E.; Stewart, J.M.; Zablotska, L.B.; Roy, R.; Olshen, A.B.; Joseph, N.M.; Bastian, B.C. Next-Generation Sequencing of Uveal Melanoma for Detection of Genetic Alterations Predicting Metastasis. Transl. Vis. Sci. Technol. 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Bagger, M.M.; Yu, R.; Chang, D.; Liu, S.; Vemula, S.; Weier, J.F.; Wadt, K.; Heegaard, S.; Bastian, B.C.; et al. The genetic evolution of metastatic uveal melanoma. Nat. Genet. 2019, 51, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, A.E.; Verdijk, R.M.; Brouwer, R.W.; van den Bosch, T.P.; van den Berg, M.M.; Vaarwater, J.; Kockx, C.E.; Paridaens, D.; Naus, N.C.; Nellist, M.; et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod. Pathol. Off. J. USA Can. Acad. Pathol. Inc. 2014, 27, 1321–1330. [Google Scholar] [CrossRef]
- Cunha Rola, A.; Taktak, A.; Eleuteri, A.; Kalirai, H.; Heimann, H.; Hussain, R.; Bonnett, L.J.; Hill, C.J.; Traynor, M.; Jager, M.J.; et al. Multicenter External Validation of the Liverpool Uveal Melanoma Prognosticator Online: An OOG Collaborative Study. Cancers 2020, 12, 477. [Google Scholar] [CrossRef]
- Kozarewa, I.; Armisen, J.; Gardner, A.F.; Slatko, B.E.; Hendrickson, C.L. Overview of Target Enrichment Strategies. Curr. Protoc. Mol. Biol. 2015, 112, 7–21. [Google Scholar] [CrossRef]
- Mamedov, T.G.; Pienaar, E.; Whitney, S.E.; TerMaat, J.R.; Carvill, G.; Goliath, R.; Subramanian, A.; Viljoen, H.J. A fundamental study of the PCR amplification of GC-rich DNA templates. Comput. Biol. Chem. 2008, 32, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Nolan, T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. JBT 2004, 15, 155–166. [Google Scholar] [PubMed]
- Hung, S.S.; Meissner, B.; Chavez, E.A.; Ben-Neriah, S.; Ennishi, D.; Jones, M.R.; Shulha, H.P.; Chan, F.C.; Boyle, M.; Kridel, R.; et al. Assessment of Capture and Amplicon-Based Approaches for the Development of a Targeted Next-Generation Sequencing Pipeline to Personalize Lymphoma Management. J. Mol. Diagn. JMD 2018, 20, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Samorodnitsky, E.; Jewell, B.M.; Hagopian, R.; Miya, J.; Wing, M.R.; Lyon, E.; Damodaran, S.; Bhatt, D.; Reeser, J.W.; Datta, J.; et al. Evaluation of Hybridization Capture Versus Amplicon-Based Methods for Whole-Exome Sequencing. Hum. Mutat. 2015, 36, 903–914. [Google Scholar] [CrossRef]
- Versluis, M.; de Lange, M.J.; van Pelt, S.I.; Ruivenkamp, C.A.; Kroes, W.G.; Cao, J.; Jager, M.J.; Luyten, G.P.; van der Velden, P.A. Digital PCR validates 8q dosage as prognostic tool in uveal melanoma. PLoS ONE 2015, 10, e0116371. [Google Scholar] [CrossRef]
- Coupland, S.E.; Kalirai, H.; Ho, V.; Thornton, S.; Damato, B.E.; Heimann, H. Concordant chromosome 3 results in paired choroidal melanoma biopsies and subsequent tumour resection specimens. Br. J. Ophthalmol. 2015, 99, 1444–1450. [Google Scholar] [CrossRef]
- Hussain, R.N.; Kalirai, H.; Groenewald, C.; Kacperek, A.; Errington, R.D.; Coupland, S.E.; Heimann, H.; Damato, B. Prognostic Biopsy of Choroidal Melanoma after Proton Beam Radiation Therapy. Ophthalmology 2016, 123, 2264–2265. [Google Scholar] [CrossRef]
- Thornton, S.; Coupland, S.E.; Heimann, H.; Hussain, R.; Groenewald, C.; Kacperek, A.; Damato, B.; Taktak, A.; Eleuteri, A.; Kalirai, H. Effects of plaque brachytherapy and proton beam radiotherapy on prognostic testing: A comparison of uveal melanoma genotyped by microsatellite analysis. Br. J. Ophthalmol. 2020. [Google Scholar] [CrossRef]
- van de Nes, J.A.; Nelles, J.; Kreis, S.; Metz, C.H.; Hager, T.; Lohmann, D.R.; Zeschnigk, M. Comparing the Prognostic Value of BAP1 Mutation Pattern, Chromosome 3 Status, and BAP1 Immunohistochemistry in Uveal Melanoma. Am. J. Surg. Pathol. 2016, 40, 796–805. [Google Scholar] [CrossRef]
- Izumiyama, T.; Minoshima, S.; Yoshida, T.; Shimizu, N. A novel big protein TPRBK possessing 25 units of TPR motif is essential for the progress of mitosis and cytokinesis. Gene 2012, 511, 202–217. [Google Scholar] [CrossRef]
- Tang, M.-R.; Wang, Y.-X.; Guo, S.; Han, S.-Y.; Wang, D. CSMD1 exhibits antitumor activity in A375 melanoma cells through activation of the Smad pathway. Apoptosis 2012, 17, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Quesnelle, K.M.; Sparano, A.; Rao, S.; Park, M.S.; Cohen, M.A.; Wang, Y.; Samanta, M.; Kumar, M.S.; Aziz, M.U.; et al. Characterization CSMD1 in a large set of primary lung, head and neck, breast and skin cancer tissues. Cancer Biol. Ther. 2009, 8, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Machleidt, T.; Geller, P.; Schwandner, R.; Scherer, G.; Kronke, M. Caspase 7-induced cleavage of kinectin in apoptotic cells. FEBS Lett. 1998, 436, 51–54. [Google Scholar] [CrossRef]
- Panier, S.; Boulton, S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef] [PubMed]
- van Poppelen, N.M.; Drabarek, W.; Smit, K.N.; Vaarwater, J.; Brands, T.; Paridaens, D.; Kilic, E.; de Klein, A. SRSF2 Mutations in Uveal Melanoma: A Preference for In-Frame Deletions? Cancers 2019, 11, 1200. [Google Scholar] [CrossRef]
- Lake, S.L.; Kalirai, H.; Dopierala, J.; Damato, B.E.; Coupland, S.E. Comparison of formalin-fixed and snap-frozen samples analyzed by multiplex ligation-dependent probe amplification for prognostic testing in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2647–2652. [Google Scholar] [CrossRef]
- Dopierala, J.; Lake, S.L.; Coupland, S.E.; Damato, B.E.; Taktak, A.F.G. Genetic heterogeneity in uveal melanoma assessed by multiplex ligation-dependent probe amplification. Investig. Ophthalmol. Vis. Sci. 2010, 51, 4898–4905. [Google Scholar] [CrossRef]
- Thomas, S.; Putter, C.; Weber, S.; Bornfeld, N.; Lohmann, D.R.; Zeschnigk, M. Prognostic significance of chromosome 3 alterations determined by microsatellite analysis in uveal melanoma: A long-term follow-up study. Br. J. Cancer 2012, 106, 1171–1176. [Google Scholar] [CrossRef]
- Caines, R.; Eleuteri, A.; Kalirai, H.; Fisher, A.C.; Heimann, H.; Damato, B.E.; Coupland, S.E.; Taktak, A.F. Cluster analysis of multiplex ligation-dependent probe amplification data in choroidal melanoma. Mol. Vis. 2015, 21, 1–11. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).