Functional and Clinical Impact of CircRNAs in Oral Cancer
Abstract
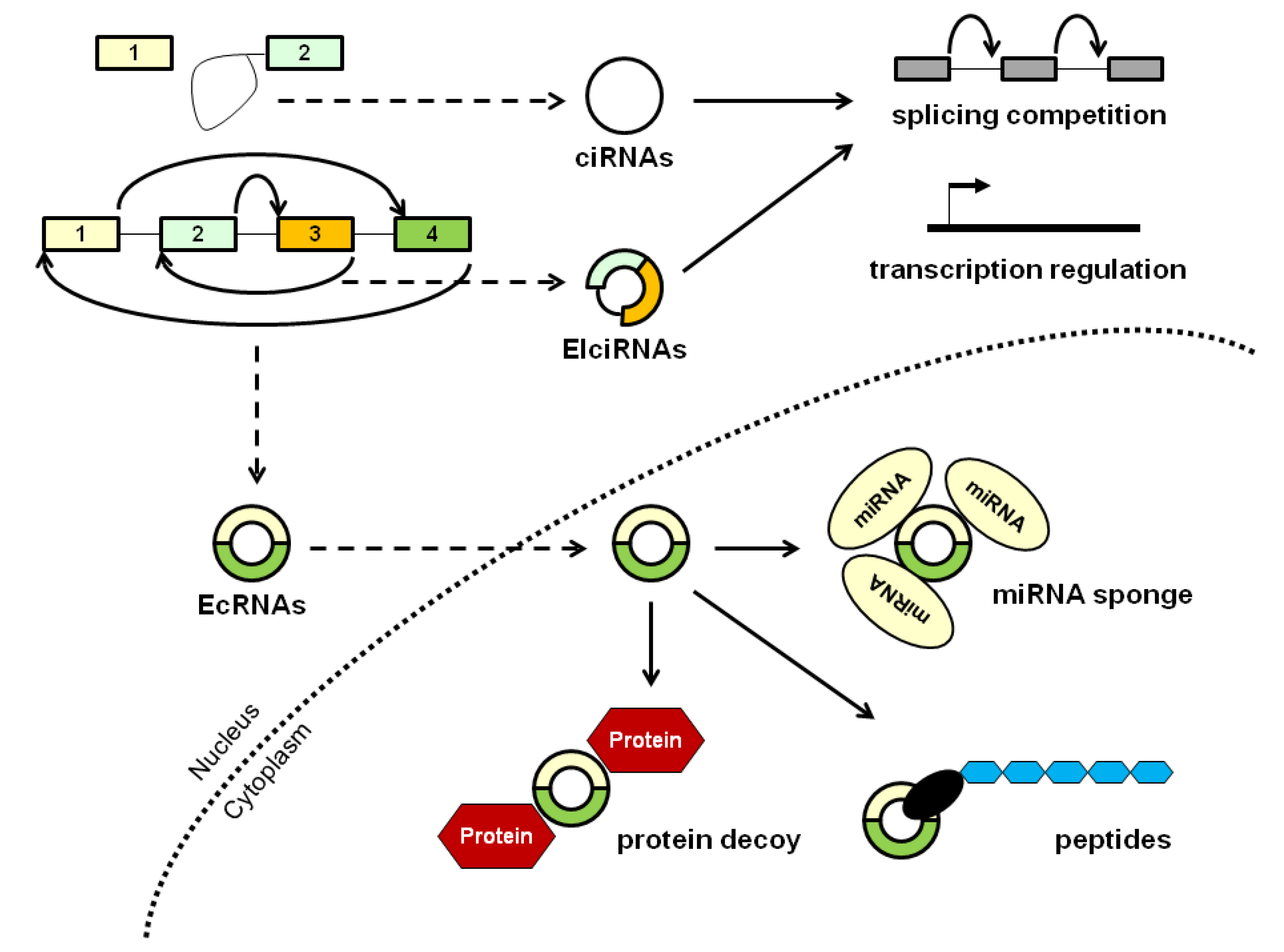
1. Introduction
2. CircRNAs Acting as Oncogenes
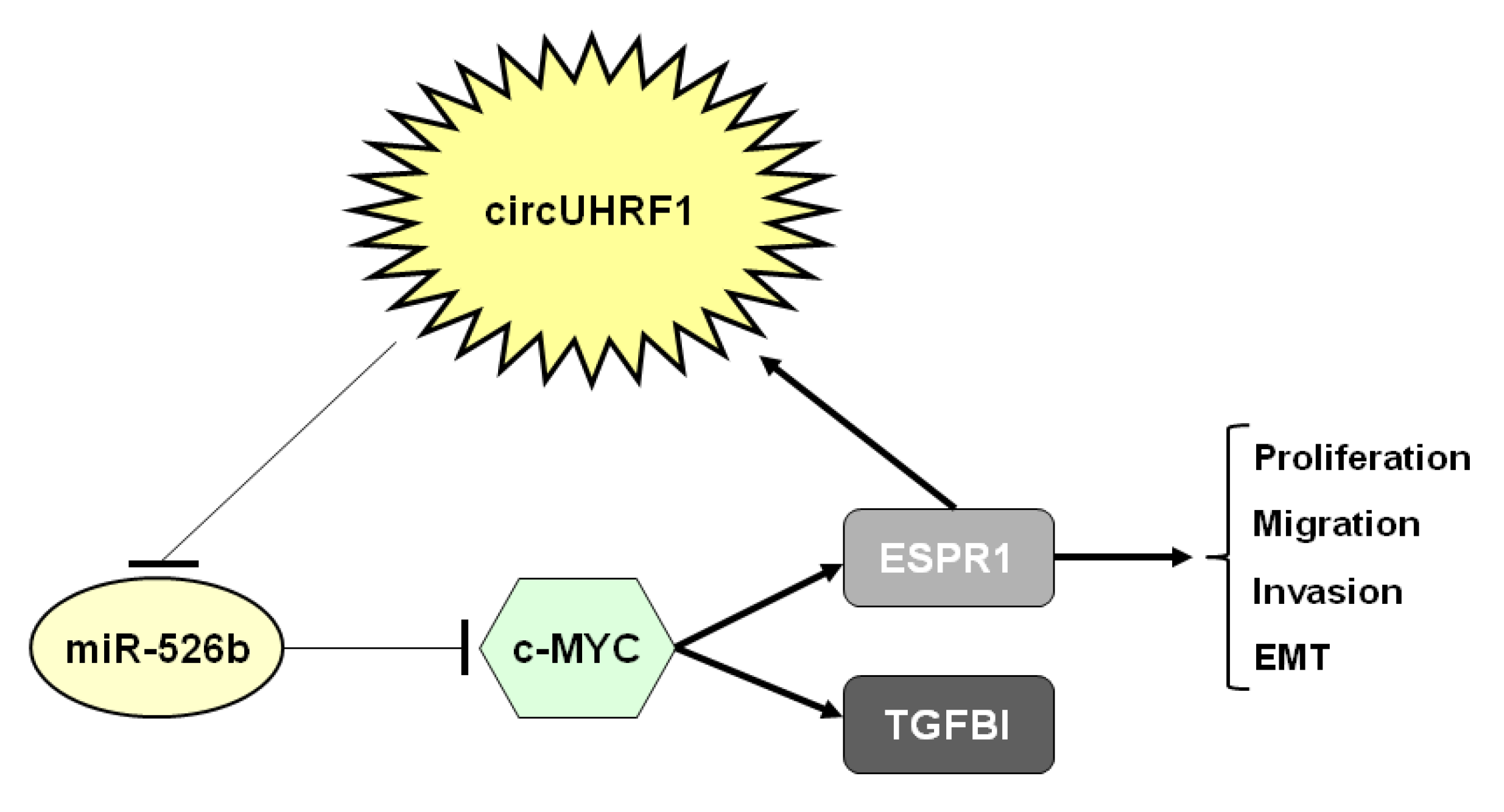
2.1. Circ_0002185
2.2. Circ_0001821
2.3. Circ_100290
2.4. Circ_0001742
2.5. Circ_0059655
2.6. CircHIPK3
2.7. Circ_0001971
2.8. CircDOCK1
3. CircRNAs Acting as Tumor Suppressors
3.1. Circ_0002203
3.2. Circ_0004491
3.3. CircFLNA
3.4. Circ_0063772
3.5. Circ_0070401
3.6. Circ_0005379
3.7. Circ_0007059
3.8. Circ_0012342
4. CircRNAs with Clinical Impact as Novel Biomarkers in Oral Cancer
4.1. Circ_0086414
4.2. Circ_0002185
4.3. Circ_0001821
4.4. Circ_0092125
4.5. CircMAN1A2
4.6. Circ_0072387
4.7. Circ_0008309
4.8. Circ_001242
4.9. Circ_0001874/circ_0001971
5. Conclusions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, N.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2014, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Candia, J.; Fernandez, A.; Somarriva, C.; Horna-Campos, O. Deaths due to oral cancer in Chile in the period 2002–2012. Rev. Med. Chile 2018, 146, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Safi, A.-F.; Grochau, K.; Drebber, U.; Schick, V.; Thiele, O.; Backhaus, T.; Nickenig, H.-J.; Zöller, J.E.; Kreppel, M. A novel histopathological scoring system for patients with oral squamous cell carcinoma. Clin. Oral Investig. 2019, 23, 3759–3765. [Google Scholar] [CrossRef]
- Scott, S.; Grunfeld, E.; McGurk, M. The idiosyncratic relationship between diagnostic delay and stage of oral squamous cell carcinoma. Oral Oncol. 2005, 41, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.; Diel, L.F.; Lamers, M.L. Macrophages and prognosis of oral squamous cell carcinoma: A systematic review. J. Oral Pathol. Med. 2017, 47, 460–467. [Google Scholar] [CrossRef]
- Ribeiro, I.L.A.; De Medeiros, J.J.; Rodrigues, L.V.; Valença, A.M.G.; Neto, E.D.A.L. Factors associated with lip and oral cavity cancer. Rev. Bras. Epidemiol. 2015, 18, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881. [Google Scholar] [CrossRef]
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336. [Google Scholar] [CrossRef]
- Chen, L.-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Boil. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef]
- Sänger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Kolakofsky, D. Isolation and characterization of Sendai virus DI-RNAs. Cell 1976, 8, 547–555. [Google Scholar] [CrossRef]
- Hsu, M.-T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef]
- Jeck, W.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2012, 19, 141–157. [Google Scholar] [CrossRef]
- Ebbesen, K.K.; Hansen, T.; Kjems, J. Insights into circular RNA biology. RNA Boil. 2016, 14, 1035–1045. [Google Scholar] [CrossRef]
- Hansen, T.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Qu, S.; Liu, Z.; Yang, X.; Zhou, J.; Yu, H.; Zhang, R.; Li, H. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018, 414, 301–309. [Google Scholar] [CrossRef] [PubMed]
- El Marabti, E.; Younis, I. The Cancer Spliceome: Reprograming of Alternative Splicing in Cancer. Front. Mol. Biosci. 2018, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.; Pillman, K.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.; Roslan, S.; Schreiber, A.W.; Gregory, P.; Goodall, G.J. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Cui, Y.; Liu, L.; Qi, X.; Liu, J.; Ma, S.; Hu, X.; Zhang, Z.; Wang, Y.; Li, H.; et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2019, 27, 919–933. [Google Scholar] [CrossRef]
- Meyer, K.B.; Maia, A.-T.; O’Reilly, M.; Ghoussaini, M.; Prathalingam, R.; Porter-Gill, P.; Ambs, S.; Prokunina-Olsson, L.; Carroll, J.S.; Ponder, B.A.J. A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with PVT1 Expression. PLoS Genet. 2011, 7, e1002165. [Google Scholar] [CrossRef]
- Lu, D.; Luo, P.; Wang, Q.; Ye, Y.; Wang, B. lncRNA PVT1 in cancer: A review and meta-analysis. Clin. Chim. Acta 2017, 474, 1–7. [Google Scholar] [CrossRef]
- Adhikary, J.; Chakraborty, S.; Dalal, S.; Basu, S.; Dey, A.; Ghosh, A. Circular PVT1: An oncogenic non-coding RNA with emerging clinical importance. J. Clin. Pathol. 2019, 72, 513–519. [Google Scholar] [CrossRef]
- He, T.; Li, X.; Xie, D.; Tian, L. Overexpressed circPVT1 in oral squamous cell carcinoma promotes proliferation by serving as a miRNA sponge. Mol. Med. Rep. 2019, 20, 3509–3518. [Google Scholar] [CrossRef]
- Fang, G.; Ye, B.-L.; Hu, B.; Ruan, X.-J.; Shi, Y.-X. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2018, 504, 184–189. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, C.; Zhang, A.; Lu, C.; Liu, L. Overexpression of circRNA_100290 promotes the progression of laryngeal squamous cell carcinoma through the miR-136-5p/RAP2C axis. Biomed. Pharmacother. 2020, 125, 109874. [Google Scholar] [CrossRef]
- Fan, H.; Li, Y.; Liu, C.; Liu, Y.; Bai, J.; Li, W. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem. Biophys. Res. Commun. 2018, 507, 178–184. [Google Scholar] [CrossRef]
- Chen, X.; Yu, J.; Tian, H.; Shan, Z.; Liu, W.; Pan, Z.; Ren, J. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J. Cell. Physiol. 2019, 234, 19130–19140. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; He, L. Hsa_circ_0001742 promotes tongue squamous cell carcinoma progression via modulating miR-634 expression. Biochem. Biophys. Res. Commun. 2019, 513, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-T.; Li, X.-X.; Zeng, L.-W. Circ_0001742 promotes tongue squamous cell carcinoma progression via miR-431-5p/ATF3 axis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10300–10312. [Google Scholar] [PubMed]
- Zhao, F.; Chen, C.-W.; Yang, W.-W.; Xu, L.-H.; Du, Z.-H.; Ge, X.-Y.; Li, S.-L. Hsa_circRNA_0059655 plays a role in salivary adenoid cystic carcinoma by functioning as a sponge of miR-338-3p. Cell. Mol. Boil. 2018, 64, 100–106. [Google Scholar] [CrossRef]
- Xie, Y.; Yuan, X.; Zhou, W.; Kosiba, A.A.; Shi, H.; Gu, J.; Qin, Z. The circular RNA HIPK3 (circHIPK3) and its regulation in cancer progression: Review. Life Sci. 2020, in press. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, S.Y.; Ouyang, S.S.; Huang, Z.K.; Luo, Q.; Liao, L. [Circular RNA circHIPK3 acts as the sponge of microRNA-124 to promote human oral squamous cell carcinoma cells proliferation]. Zhonghua Kou Qiang Yi Xue Za Zhi 2018, 53, 546–551. [Google Scholar]
- Zhao, S.-Y.; Wang, J.; Ouyang, S.-B.; Huang, Z.-K.; Liao, L. Salivary Circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as Novel Biomarkers for the Diagnosis of Oral Squamous Cell Carcinoma. Cell. Physiol. Biochem. 2018, 47, 2511–2521. [Google Scholar] [CrossRef]
- Tan, X.; Zhou, C.; Liang, Y.; Lai, Y.F.; Liang, Y. Circ_0001971 regulates oral squamous cell carcinoma progression and chemosensitivity by targeting miR-194/miR-204 in vitro and in vivo. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2470–2481. [Google Scholar]
- Wang, L.; Wei, Y.; Yan, Y.; Wang, H.; Yang, J.; Zheng, Z.; Zha, J.; Bo, P.; Tang, Y.; Guo, X.; et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC. Oncol. Rep. 2017, 39, 951–966. [Google Scholar] [CrossRef]
- Liu, P.; Li, X.; Guo, X.; Chen, J.; Li, C.; Chen, M.; Liu, L.; Zhang, X.; Zu, X. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif. 2019, 52, e12614. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Wang, Y.F.; Wang, F.; Yang, H.J.; Yang, H.Y. Effect of circular RNA hsa_circ_0002203 on the proliferation, migration, invasion, and apoptosis of oral squamous cell carcinoma cells. Hua Xi Kou Qiang Yi Xue Za Zhi 2019, 37, 509–515. [Google Scholar] [PubMed]
- Li, X.; Zhang, H.; Wang, Y.; Sun, S.; Shen, Y.; Yang, H.-Y. Silencing circular RNA hsa_circ_0004491 promotes metastasis of oral squamous cell carcinoma. Life Sci. 2019, 239, 116883. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-X.; Liu, Y.; Jia, X.-J.; Liu, S.-X.; Dong, J.-H.; Ren, X.-M.; Xu, O.; Zhang, H.-Z.; Duan, H.-J.; Shan, C.-G. Upregulation of circFLNA contributes to laryngeal squamous cell carcinoma migration by circFLNA-miR-486-3p-FLNA axis. Cancer Cell Int. 2019, 19, 196. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Y.F.; Su, W.; Yang, H.J. Effect of circular RNA hsa_circ_0063772 on proliferation, migration and invasion of oral squamous cell carcinoma cells. Zhonghua Kou Qiang Yi Xue Za Zhi 2019, 54, 561–567. [Google Scholar]
- Gao, L.; Zhao, C.; Li, S.; Dou, Z.; Wang, Q.; Liu, J.; Ren, W.; Zhi, K.-Q. circ-PKD2 inhibits carcinogenesis via the miR-204-3p/APC2 axis in oral squamous cell carcinoma. Mol. Carcinog. 2019, 58, 1783–1794. [Google Scholar] [CrossRef]
- Su, W.; Wang, Y.; Wang, F.; Sun, S.; Li, M.; Shen, Y.; Yang, H.-Y. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer 2019, 19, 400. [Google Scholar] [CrossRef]
- Su, W.; Wang, Y.; Wang, F.; Zhang, B.; Zhang, H.; Shen, Y.; Yang, H.-Y. Circular RNA hsa_circ_0007059 indicates prognosis and influences malignant behavior via AKT/mTOR in oral squamous cell carcinoma. J. Cell. Physiol. 2019, 234, 15156–15166. [Google Scholar] [CrossRef]
- Lu, H.; Han, N.; Xu, W.; Zhu, Y.; Liu, L.; Liu, S.; Yang, W. Screening and bioinformatics analysis of mRNA, long non-coding RNA and circular RNA expression profiles in mucoepidermoid carcinoma of salivary gland. Biochem. Biophys. Res. Commun. 2018, 508, 66–71. [Google Scholar] [CrossRef]
- Zhang, N.; Gao, L.; Ren, W.; Li, S.; Zhang, D.; Song, X.; Zhao, C.; Zhi, K.-Q. Fucoidan affects oral squamous cell carcinoma cell functions in vitro by regulating FLNA-derived circular RNA. Ann. N. Y. Acad. Sci. 2019, 1462, 65–78. [Google Scholar] [CrossRef]
- Gao, S.; Yu, Y.; Liu, L.; Meng, J.; Li, G. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 2019, 233, 116692. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Z.-T. Hsa_circ_0086414 Might Be a Diagnostic Biomarker of Oral Squamous Cell Carcinoma. Med. Sci. Monit. 2020, 26, e919383. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Wang, Q.-B.; Zhi, Y.; Ren, W.-H.; Li, S.-M.; Zhao, C.-Y.; Xing, X.-M.; Dou, Z.-C.; Liu, J.-C.; Jiang, C.-M.; et al. Down-regulation of hsa_circ_0092125 is related to the occurrence and development of oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2020, 49, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Wang, J.; Tang, Y.; Zhao, J.; He, S.; Xiong, F.; Guo, C.; Xiang, B.; Zhou, M.; Li, X.; et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019, 110, 2180–2188. [Google Scholar] [CrossRef]
- Dou, Z.; Li, S.; Ren, W.; Wang, Q.; Liu, J.; Kong, X.; Gao, L.; Zhi, K.-Q. Decreased expression of hsa_circ_0072387 as a valuable predictor for oral squamous cell carcinoma. Oral Dis. 2019, 25, 1302–1308. [Google Scholar] [CrossRef]
- Li, B.; Wang, F.; Li, X.; Sun, S.; Shen, Y.; Yang, H. Hsa_circ_0008309 May Be a Potential Biomarker for Oral Squamous Cell Carcinoma. Dis. Markers 2018, 2018, 7496890. [Google Scholar] [CrossRef]
- Sun, S.; Li, B.; Wang, Y.; Li, X.; Wang, P.; Wang, F.; Zhang, W.; Yang, H.-Y. Clinical Significance of the Decreased Expression of hsa_circ_001242 in Oral Squamous Cell Carcinoma. Dis. Markers 2018, 2018, 6514795. [Google Scholar] [CrossRef]
- Liu, X.-X.; Yang, Y.-E.; Zhang, M.-Y.; Li, R.; Yin, Y.-H.; Qu, Y.-Q.; Liu, X. A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J. Transl. Med. 2019, 17, 50. [Google Scholar] [CrossRef]
- Wang, C.; Tan, S.; Liu, W.-R.; Lei, Q.; Qiao, W.; Wu, Y.; Liu, X.; Cheng, W.; Wei, Y.; Peng, Y.; et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol. Cancer 2019, 18, 134–136. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.; Yang, Q.; Tang, C.; Wang, T.; Shen, X.; Ju, S. Identification of hsa_circ_0001821 as a Novel Diagnostic Biomarker in Gastric Cancer via Comprehensive Circular RNA Profiling. Front. Genet. 2019, 10, 878. [Google Scholar] [CrossRef]
| CircRNA | Function | Tumor | Pathway | References |
|---|---|---|---|---|
| Circ_0002185 | Oncogene | OSCC 1 | circUHRF1/miR-526-5p/c-Myc/TGFB1/ESRP1 loop | [24] |
| Circ_0001821 | Oncogene | OSCC | circPVT1/miR-125b/STAT3 | [28] |
| Circ_100290 | Oncogene | OSCC | circ_100290/miR-378a/GLUT1 | [32] |
| Circ_0001742 | Oncogene | TSCC 2 | circ_0001742/miR-431-5p/ATF3 | [34] |
| Circ_0059655 | Oncogene | SACC 3 | circ_0059655/miR-338-3p/CCND1 | [35] |
| CircHIPK3 | Oncogene | OSCC | circHIPK3/miR-124 | [37] |
| Circ_0001971 | Oncogene | OSCC | circ_0001971/miR-194/miR-204 | [39] |
| CircDOCK1 | Oncogene | OSCC | circDOCK1/miR-196-5p/BIRC3 | [41] |
| Circ_0002203 | TS 4 | OSCC | Unknown | [42] |
| Circ_0004491 | TS | OSCC | Unknown | [43] |
| CircFLNA | TS | OSCC | circFLNA/miR-486-3p | [44] |
| Circ_0063772 | TS | OSCC | Unknown | [45] |
| Circ_0070401 | TS | OSCC | circPDK2/miR-204-3p/APC2 | [46] |
| Circ_0005379 | TS | OSCC | EGFR pathway | [47] |
| Circ_0007059 | TS | OSCC | AKT/mTOR pathway | [48] |
| Circ_0012342 | TS | MECSG 5 | Unknown | [49] |
| Functional Role | CircRNAs |
|---|---|
| Proliferation | circUHRF1, circPVT1, circ_100290, circ_0001742, circ_0059655, circHIPK3, circ_0001971, circ_0002203, circFLNA, circ_0063772, circ_0070401, circ_0005379, circ_0007059 |
| Migration | circUHRF1, circ_0059655, circ_0001971, circ_0002203, circ_0004491, circFLNA, circ_0063772, circ_0070401, circ_0005379, circ_0007059 |
| Invasion | circUHRF1, circ_0001742, circ_0059655, circ_0001971, circ_0002203, circ_0004491, circFLNA, circ_0063772, circ_0070401, circ_0005379, circ_0007059 |
| Apoptosis | circ_100290, circ_0001742, circ_0001971, circDOCK1, circFLNA, circ_0070401, circ_0005379, circ_0007059 |
| EMT 1 | circUHRF1, circ_0001742, circ_0004491, circ_0005379 |
| Tumor growth in vivo | circUHRF1, circ_0001971, circ_0002203, circ_0063772, circ_0070401, circ_0005379, circ_0007059 |
| Drug resistance | circ_0001971 (cisplatin), circ_0005379 (cetuximab) |
| CircRNA | Expression | Sample | Clinical Value | References |
|---|---|---|---|---|
| Circ_0086414 | Low | Tumor | Diagnostic | [52] |
| Circ_0002185 | High | Tumor | Prognostic | [24] |
| Circ_0001821 | High | Tumor | Diagnostic | [28] |
| Circ_0092125 | Low | Tumor | Prognostic | [53] |
| CircMAN1A2 | High | Serum | Diagnostic | [54] |
| Circ_0072387 | Low | Tumor | Diagnostic | [55] |
| Circ_0008309 | Low | Tumor | Diagnostic | [56] |
| Circ_001242 | Low | Tumor | Diagnostic | [57] |
| Circ_0001874/circ_0001971 | High | Saliva | Diagnostic | [38] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cristóbal, I.; Caramés, C.; Rubio, J.; Sanz-Alvarez, M.; Luque, M.; Madoz-Gúrpide, J.; Rojo, F.; García-Foncillas, J. Functional and Clinical Impact of CircRNAs in Oral Cancer. Cancers 2020, 12, 1041. https://doi.org/10.3390/cancers12041041
Cristóbal I, Caramés C, Rubio J, Sanz-Alvarez M, Luque M, Madoz-Gúrpide J, Rojo F, García-Foncillas J. Functional and Clinical Impact of CircRNAs in Oral Cancer. Cancers. 2020; 12(4):1041. https://doi.org/10.3390/cancers12041041
Chicago/Turabian StyleCristóbal, Ion, Cristina Caramés, Jaime Rubio, Marta Sanz-Alvarez, Melani Luque, Juan Madoz-Gúrpide, Federico Rojo, and Jesús García-Foncillas. 2020. "Functional and Clinical Impact of CircRNAs in Oral Cancer" Cancers 12, no. 4: 1041. https://doi.org/10.3390/cancers12041041
APA StyleCristóbal, I., Caramés, C., Rubio, J., Sanz-Alvarez, M., Luque, M., Madoz-Gúrpide, J., Rojo, F., & García-Foncillas, J. (2020). Functional and Clinical Impact of CircRNAs in Oral Cancer. Cancers, 12(4), 1041. https://doi.org/10.3390/cancers12041041






