Karyopherin-β1 Regulates Radioresistance and Radiation-Increased Programmed Death-Ligand 1 Expression in Human Head and Neck Squamous Cell Carcinoma Cell Lines
Abstract
:1. Introduction
2. Results
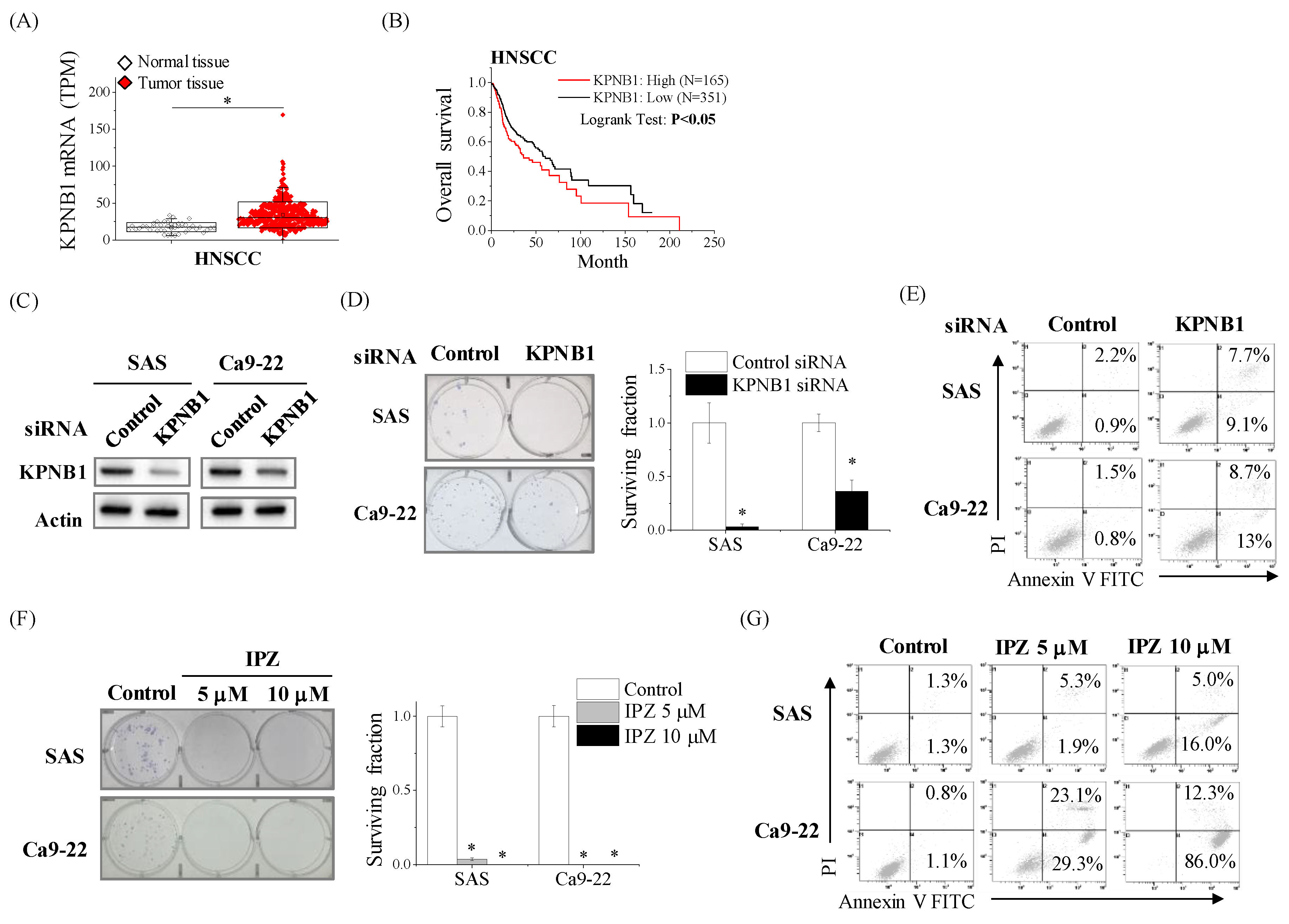
2.1. High Expression Levels of KPNB1 Determines the Proliferation and Apoptosis of HNSCC Cells
2.2. KPNB1 Regulates Radioresistance of HNSCC Cells
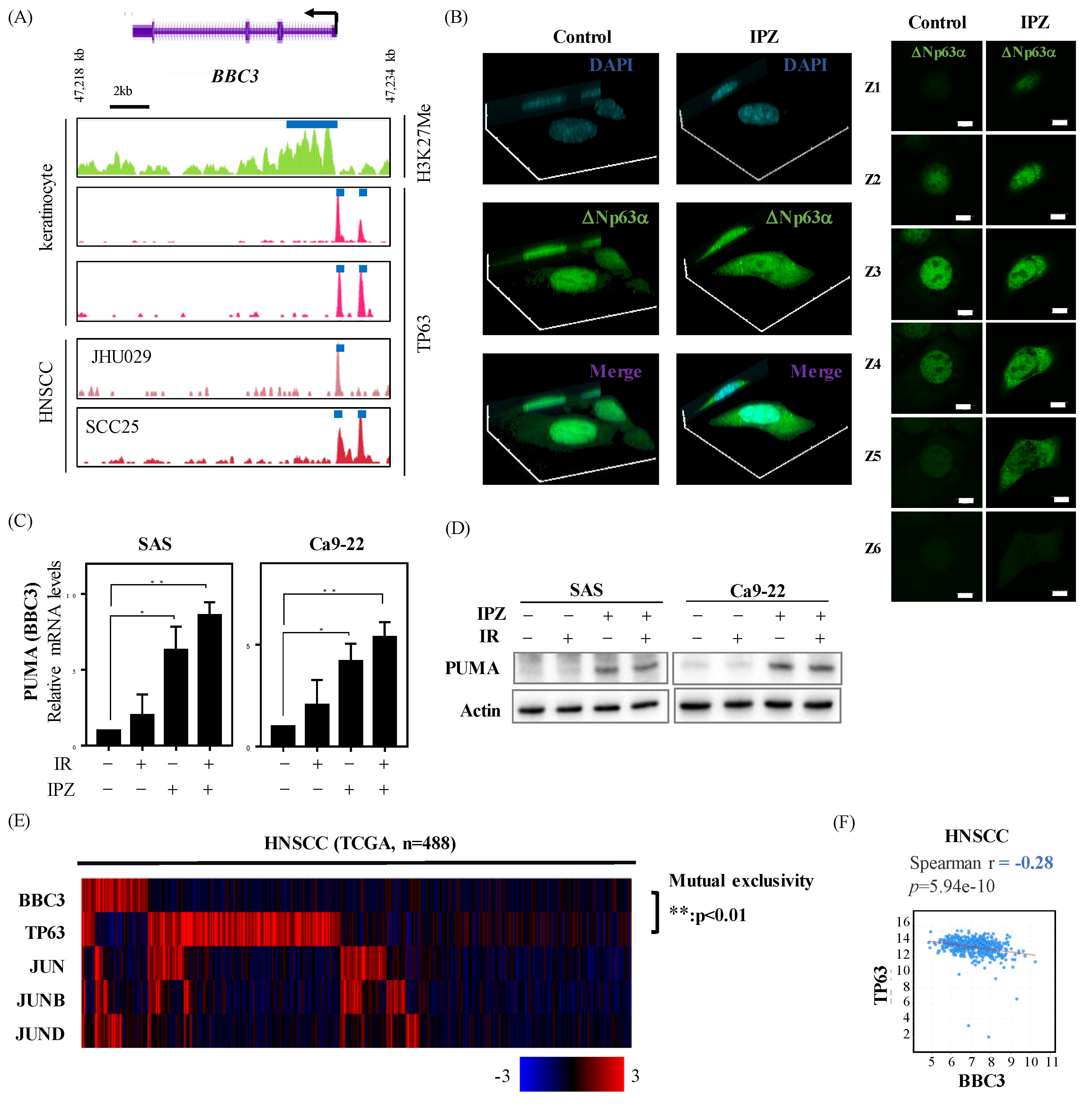
2.3. IPZ Abolishes the Transcriptional Silencing of PUMA by Blocking p63 Nuclear Import in HNSCC Cells
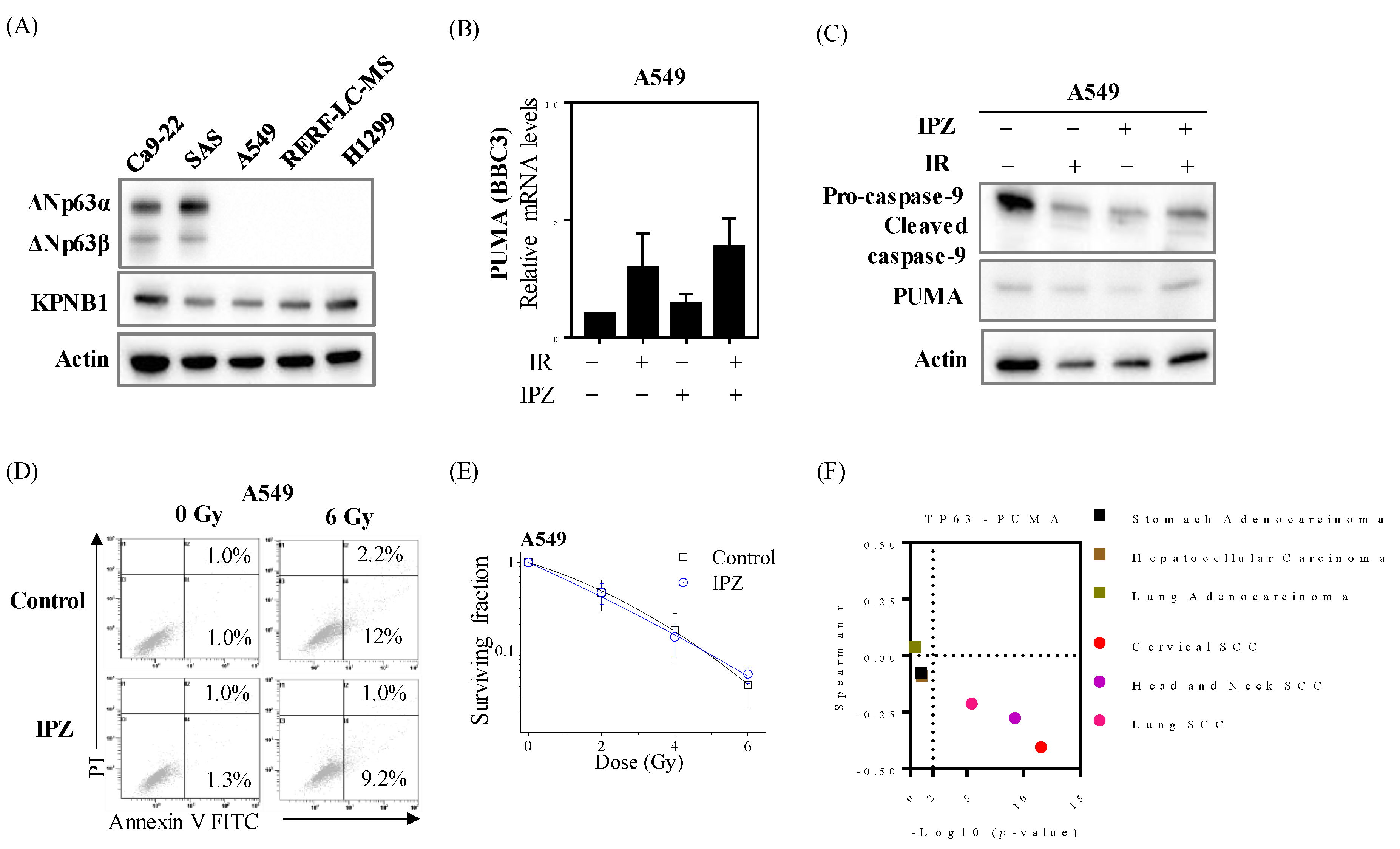
2.4. Radiosensitization Effects through Abolishing ΔNp63α-Mediated PUMA Silencing Is Unique to HNSCC Cells
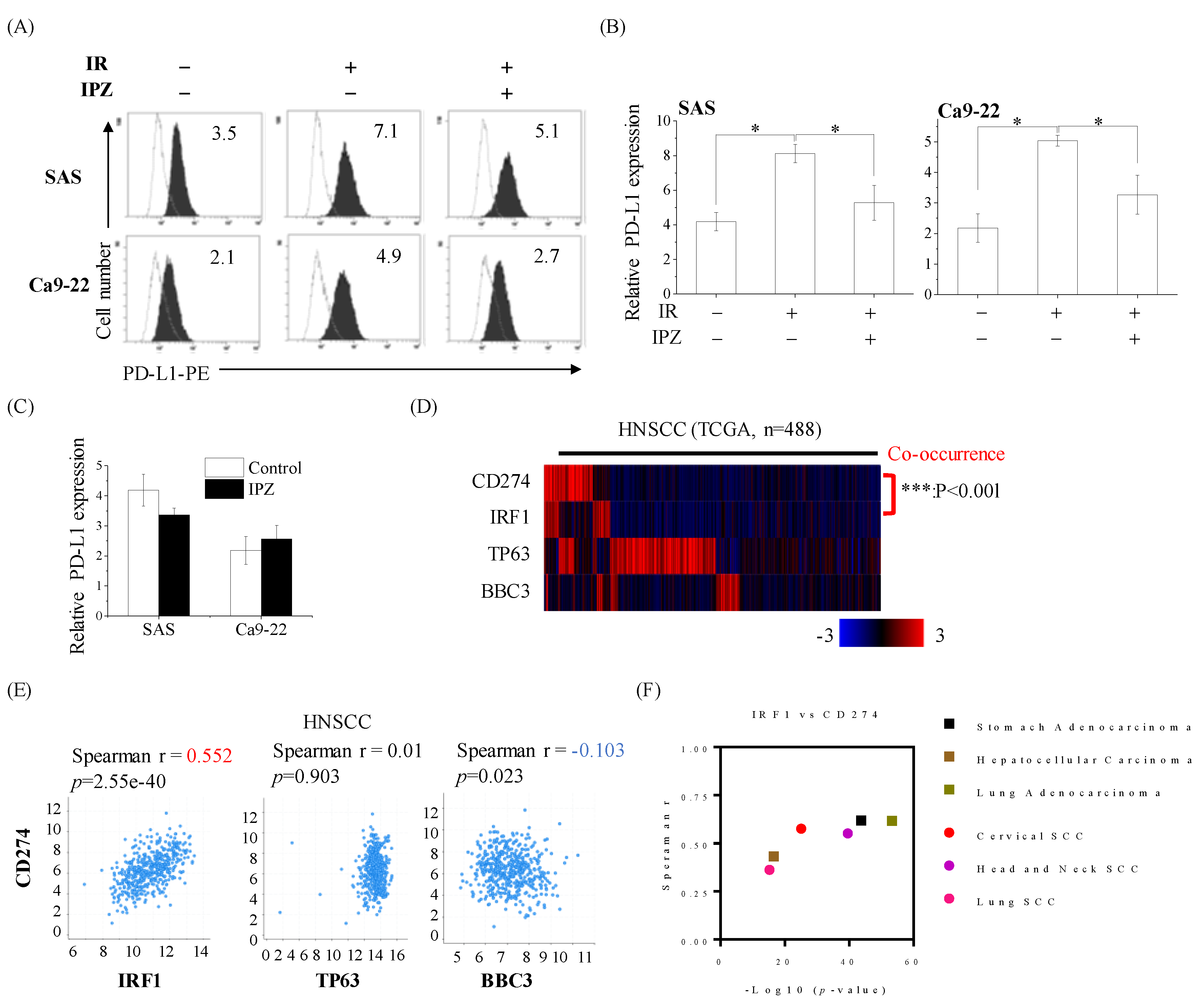
2.5. KPNB1 Is Involved in Radiation-Increased Cell Surface PD-L1 Expression on HNSCC Cells
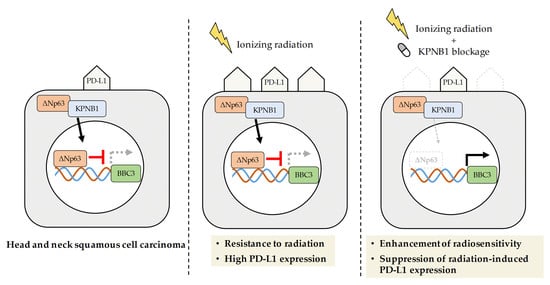
3. Discussion
4. Materials and Methods
4.1. Bioinformatics and Data Analysis
4.2. Reagents
4.3. Cell Culture and Treatment
4.4. siRNA Transfection
4.5. In Vitro X-ray Irradiation
4.6. Clonogenic Survival Assay
4.7. Detection of Apoptosis
4.8. SDS-PAGE and Western Blotting
4.9. Immunofluorescence Analysis
4.10. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.11. Analysis of Cell Surface PD-L1 Expression
4.12. Statistical Analysis
4.13. URLs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Balázs, K.; Kis, E.; Badie, C.; Bogdándi, E.N.; Candéias, S.; Cruz Garcia, L.; Dominczyk, I.; Frey, B.; Gaipl, U.; Jurányi, Z.; et al. Radiotherapy-Induced changes in the systemic immune and inflammation parameters of head and neck cancer patients. Cancers 2019, 11, 1324. [Google Scholar] [CrossRef] [Green Version]
- Budach, V.; Tinhofer, I. Novel prognostic clinical factors and biomarkers for outcome prediction in head and neck cancer: A systematic review. Lancet Oncol. 2019, 20, e313–e326. [Google Scholar] [CrossRef]
- Perri, F.; Pacelli, R.; Della Vittoria Scarpati, G.; Cella, L.; Giuliano, M.; Caponigro, F.; Pepe, S. Radioresistance in head and neck squamous cell carcinoma: Biological bases and therapeutic implications. Head Neck 2015, 37, 763–770. [Google Scholar] [CrossRef]
- Xie, F.; Xu, M.; Lu, J.; Mao, L.; Wang, S. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol. Cancer 2019, 18, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.T.; Chen, W.C.; Chang, Y.H.; Lin, W.Y.; Chen, M.F. The role of PD-L1 in the radiation response and clinical outcome for bladder cancer. Sci. Rep. 2016, 6, 19740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Chen, P.; Chen, W.; Lu, M.; Lin, P.; Lee, K. The role of PD-L1 in the radiation response and prognosis for esophageal squamous cell carcinoma related to IL-6 and T-cell immunosuppression. Oncotarget 2016, 7, 7913–7924. [Google Scholar] [CrossRef] [PubMed]
- Moudry, P.; Lukas, C.; Macurek, L.; Neumann, B.; Heriche, J.K.; Pepperkok, R.; Ellenberg, J.; Hodny, Z.; Lukas, J.; Bartek, J. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ. 2012, 19, 798–807. [Google Scholar] [CrossRef] [Green Version]
- Chao, C.; Saito, S.; Kang, J.; Anderson, C.W.; Appella, E.; Xu, Y. p53 transcriptional activity is essential for p53-Dependent apoptosis following DNA damage. EMBO J. 2000, 19, 4967–4975. [Google Scholar] [CrossRef] [Green Version]
- Tamatani, T.; Takamaru, N.; Hara, K.; Kinouchi, M.; Kuribayashi, N.; Ohe, G.; Uchida, D.; Fujisawa, K.; Nagai, H.; Miyamoto, Y. Bortezomib-Enhanced radiosensitization through the suppression of radiation-induced nuclear factor-κB activity in human oral cancer cells. Int. J. Oncol. 2013, 42, 935–944. [Google Scholar] [CrossRef]
- Sato, H.; Niimi, A.; Yasuhara, T.; Permata, T.B.M.; Hagiwara, Y.; Isono, M.; Nuryadi, E.; Sekine, R.; Oike, T.; Kakoti, S.; et al. DNA double-Strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 2017, 8, 1751. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, L. PUMA, a potent killer with or without p53. Oncogene 2008, 27, S71–S83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoneda, Y. Nuclear pore-Targeting complex and its role on nuclear protein transport. Arch Histol. Cytol. 1996, 59, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.H.; Clarke, M.F. The nuclear import of p53 is determined by the presence of a basic domain and its relative position to the nuclear localization signal. Oncogene 1999, 18, 2163–2166. [Google Scholar] [CrossRef] [Green Version]
- Hazawa, M.; Sakai, K.; Kobayashi, A.; Yoshino, H.; Iga, Y.; Iwashima, Y.; Lim, K.S.; Chih-Cheng Voon, D.; Jiang, Y.Y.; Horike, S.I.; et al. Disease-Specific alteration of karyopherin-α subtype establishes feed-Forward oncogenic signaling in head and neck squamous cell carcinoma. Oncogene 2019. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Hu, B.; Zhang, Y.; Qiang, S.; Cai, J.; Huang, W.; Gong, C.; Zhang, T.; Zhang, S.; Xu, P.; et al. Suppression of the nuclear transporter-KPNβ1 expression inhibits tumor proliferation in hepatocellular carcinoma. Med. Oncol. 2015, 32, 128. [Google Scholar] [CrossRef]
- Kuusisto, H.V.; Jans, D.A. Hyper-Dependence of breast cancer cell types on the nuclear transporter Importin β1. Biochim. Biophys. Acta 2015, 1853, 1870–1878. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.C.; Hao, J.J.; Nagata, Y.; Xu, L.; Shang, L.; Meng, X.; Sato, Y.; Okuno, Y.; Varela, A.M.; Ding, L.W.; et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 467–473. [Google Scholar] [CrossRef]
- Lee, C.S.; Bhaduri, A.; Mah, A.; Johnson, W.L.; Ungewickell, A.; Aros, C.J.; Nguyen, C.B.; Rios, E.J.; Siprashvili, Z.; Straight, A.; et al. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat. Gene 2014, 46, 1060–1062. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women’s Hospital; Broad Institute, Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University; Greater Poland Cancer Centre, Harvard Medical School; Institute for Systems Biology; et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, M.K.; Kommagani, R.; Payal, V.; Mayo, L.D.; Shamma, H.N.; Kadakia, M.P. ΔNp63α regulates keratinocyte proliferation by controlling PTEN expression and localization. Cell Death Differ. 2011, 18, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D.K.; Carroll, J.S.; Leong, C.O.; Cheng, F.; Brown, M.; Mills, A.A.; Brugge, J.S.; Ellisen, L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006, 8, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Tordella, L.; Koch, S.; Salter, V.; Pagotto, A.; Doondeea, J.B.; Feller, S.M.; Ratnayaka, I.; Zhong, S.; Goldin, R.D.; Lozano, G.; et al. ASPP2 suppresses squamous cell carcinoma via RelA/p65-Mediated repression of p63. Proc. Natl. Acad. Sci. USA 2013, 110, 17969–17974. [Google Scholar] [CrossRef] [Green Version]
- Hazawa, M.; Lin, D.C.; Kobayashi, A.; Jiang, Y.Y.; Xu, L.; Dewi, F.; Mohamed, M.S.; Nakada, M.; Meguro-Horike, M.; Horike, S.I.; et al. ROCK-Dependent phosphorylation of NUP62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 2018, 19, 73–88. [Google Scholar] [CrossRef] [Green Version]
- Moergel, M.; Abt, E.; Stockinger, M.; Kunkel, M. Overexpression of p63 is associated with radiation resistance and prognosis in oral squamous cell carcinoma. Oral Oncol. 2010, 46, 667–671. [Google Scholar] [CrossRef]
- Rocco, J.W.; Leong, C.O.; Kuperwasser, N.; DeYoung, M.P.; Ellisen, L.W. p63 mediates survival in squamous cell carcinoma by suppression of p73-Dependent apoptosis. Cancer Cell 2006, 9, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Stelma, T.; Leaner, V.D. KPNB1-Mediated nuclear import is required for motility and inflammatory transcription factor activity in cervical cancer cells. Oncotarget 2017, 8, 32833–32847. [Google Scholar] [CrossRef] [Green Version]
- Stelma, T.; Chi, A.; van der Watt, P.J.; Verrico, A.; Lavia, P.; Leaner, V.D. Targeting nuclear transporters in cancer: Diagnostic, prognostic and therapeutic pohurtential. IUBMB Life 2016, 68, 268–280. [Google Scholar] [CrossRef] [Green Version]
- Green, D.R.; Llambi, F. Cell death signaling. Cold Spring Harb. Perspect Biol. 2015, 7, a006080. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, X.; Li, B.; Lin, F.; Dong, K.; Gao, P.; Zhang, H.Z. Tumor-Specific adenovirus-Mediated PUMA gene transfer using the survivin promoter enhances radiosensitivity of breast cancer cells in vitro and in vivo. Breast Cancer Res. Treat. 2009, 117, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yue, W.; Wu, B.; Zhang, L. PUMA sensitizes lung cancer cells to chemotherapeutic agents and irradiation. Clin. Cancer Res. 2006, 12, 2928–2936. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Zhang, B.; Lu, Y.; Sun, C.; Zhao, W.; Jiao, X.; Hu, J.; Mu, P.; Lu, H.; Zhou, C. Slug inhibition upregulates radiation-Induced PUMA activity leading to apoptosis in cholangiocarcinomas. Med. Oncol. 2011, 28, S301–S309. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Jung, S.Y.; Park, J.I.; Ahn, J.; Park, J.K.; Um, H.D.; Park, I.C.; Hwang, S.G.; Ha, H.; Song, J.Y. Inhibition of karyopherin-α2 augments radiation-Induced cell death by perturbing BRCA1-mediated DNA repair. Int. J. Mol. Sci. 2019, 20, 2843. [Google Scholar] [CrossRef] [Green Version]
- Teng, S.C.; Wu, K.J.; Tseng, S.F.; Wong, C.W.; Kao, L. Importin KPNA2, NBS1, DNA repair and tumorigenesis. J. Mol. Histol. 2006, 37, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Senoo, M.; Pinto, F.; Crum, C.P.; McKeon, F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 2007, 129, 523–536. [Google Scholar] [CrossRef] [Green Version]
- Gatti, V.; Fierro, C.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. ΔNp63 in squamous cell carcinoma: Defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol. Oncol. 2019, 13, 981–1001. [Google Scholar] [CrossRef] [Green Version]
- Azad, A.; Yin Lim, S.; D′Costa, Z.; Jones, K.; Diana, A.; Sansom, O.J.; Kruger, P.; Liu, S.; McKenna, W.G.; Dushek, O.; et al. PD-L1 blockade enhances response of pancreatic ductal adenocarcinoma to radiotherapy. EMBO Mol. Med. 2017, 9, 167–180. [Google Scholar] [CrossRef]
- Gong, X.; Li, X.; Jiang, T.; Xie, H.; Zhu, Z.; Zhou, F.; Zhou, C. Combined radiotherapy and anti-PD-L1 antibody synergistically enhances antitumor effect in non-small cell lung cancer. J. Thorac. Oncol. 2017, 12, 1085–1097. [Google Scholar] [CrossRef] [Green Version]
- Dovedi, S.J.; Adlard, A.L.; Lipowska-Bhalla, G.; McKenna, C.; Jones, S.; Cheadle, E.J.; Stratford, I.J.; Poon, E.; Morrow, M.; Stewart, R.; et al. Acquired resistance to fractionated radiotherapy can be overcome by concurrent PD-L1 blockade. Cancer Res. 2014, 74, 5458–5468. [Google Scholar] [CrossRef] [Green Version]
- Heinrich, P.C.; Behrmann, I.; Müller-Newen, G.; Schaper, F.; Graeve, L. Interleukin-6-Type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998, 334 Pt 2, 297–314. [Google Scholar] [CrossRef] [Green Version]
- Umegaki, N.; Tamai, K.; Nakano, H.; Moritsugu, R.; Yamazaki, T.; Hanada, K.; Katayama, I.; Kaneda, Y. Differential regulation of karyopherin alpha 2 expression by TGF-beta1 and IFN-Gamma in normal human epidermal keratinocytes: Evident contribution of KPNA2 for nuclear translocation of IRF-1. J. Invest. Dermatol. 2007, 127, 1456–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, H.; Iwabuchi, M.; Kazama, Y.; Furukawa, M.; Kashiwakura, I. Effects of retinoic acid-Inducible gene-I-Like receptors activations and ionizing radiation cotreatment on cytotoxicity against human non-Small cell lung cancer in vitro. Oncol. Lett. 2018, 15, 4697–4705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, H.; Konno, H.; Ogura, K.; Sato, Y.; Kashiwakura, I. Relationship between the regulation of caspase-8-Mediated apoptosis and radioresistance in human THP-1-Derived macrophages. Int. J. Mol. Sci. 2018, 19, 3154. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, H.; Kumai, Y.; Kashiwakura, I. Effects of endoplasmic reticulum stress on apoptosis induction in radioresistant macrophages. Mol. Med. Rep. 2017, 15, 2867–2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, H.; Kashiwakura, I. Involvement of reactive oxygen species in ionizing radiation-Induced upregulation of cell surface Toll-Like receptor 2 and 4 expression in human monocytic cells. J. Radiat. Res. 2017, 58, 626–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Sequence (5′→3′) | |
|---|---|
| PUMA F | GACGACCTCAACGCACAGTA |
| PUMA R | CACCTAATTGGGCTCCATCT |
| GAPDH F | GTCAGTGGTGGACCTGACCT |
| GAPDH R | AGGGGTCTACATGGCAACTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hazawa, M.; Yoshino, H.; Nakagawa, Y.; Shimizume, R.; Nitta, K.; Sato, Y.; Sato, M.; Wong, R.W.; Kashiwakura, I. Karyopherin-β1 Regulates Radioresistance and Radiation-Increased Programmed Death-Ligand 1 Expression in Human Head and Neck Squamous Cell Carcinoma Cell Lines. Cancers 2020, 12, 908. https://doi.org/10.3390/cancers12040908
Hazawa M, Yoshino H, Nakagawa Y, Shimizume R, Nitta K, Sato Y, Sato M, Wong RW, Kashiwakura I. Karyopherin-β1 Regulates Radioresistance and Radiation-Increased Programmed Death-Ligand 1 Expression in Human Head and Neck Squamous Cell Carcinoma Cell Lines. Cancers. 2020; 12(4):908. https://doi.org/10.3390/cancers12040908
Chicago/Turabian StyleHazawa, Masaharu, Hironori Yoshino, Yuta Nakagawa, Reina Shimizume, Keisuke Nitta, Yoshiaki Sato, Mariko Sato, Richard W. Wong, and Ikuo Kashiwakura. 2020. "Karyopherin-β1 Regulates Radioresistance and Radiation-Increased Programmed Death-Ligand 1 Expression in Human Head and Neck Squamous Cell Carcinoma Cell Lines" Cancers 12, no. 4: 908. https://doi.org/10.3390/cancers12040908