SOX2 Expression and Transcriptional Activity Identifies a Subpopulation of Cancer Stem Cells in Sarcoma with Prognostic Implications
Abstract
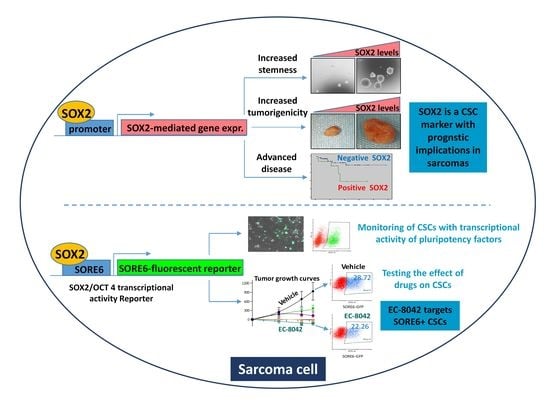
:1. Introduction
2. Results
2.1. SOX2 Expression in Sarcoma Tissue Specimens is Associated to Poor Prognosis and Survival
2.2. SOX2 Is Required to Maintain the Tumorigenic Potential in Sarcoma Cells
2.3. SOX2 Activity Marks a Subpopulation of CSCs in Sarcoma
2.4. SORE6 Response Element is a Valuable Tool to Monitor CSCs Response to Anti-tumor Treatments
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Drugs and Ethics Statement
4.2. Lentiviral Constructions and Cell Transduction
4.3. Flow Cytometry and Cell Sorting
4.4. Western Blotting
4.5. RT-qPCR Assays
4.6. In Vivo Tumor Growth
4.7. Patients and Immunohistochemical Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells: Current status and evolving complexities. Cell Stem Cell 2012, 10, 717–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, J.L.; Loh, Y.H.; Zhang, W.; Chen, X.; Tam, W.L.; Yeap, L.S.; Li, P.; Ang, Y.S.; Lim, B.; Robson, P.; et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005, 25, 6031–6046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granda-Diaz, R.; Menendez, S.T.; Pedregal Mallo, D.; Hermida-Prado, F.; Rodriguez, R.; Suarez-Fernandez, L.; Vallina, A.; Sanchez-Canteli, M.; Rodriguez, A.; Fernandez-Garcia, M.S.; et al. The novel role of SOX2 as an early predictor of cancer risk in patients with laryngeal precancerous lesions. Cancers 2019, 11, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leis, O.; Eguiara, A.; Lopez-Arribillaga, E.; Alberdi, M.J.; Hernandez-Garcia, S.; Elorriaga, K.; Pandiella, A.; Rezola, R.; Martin, A.G. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012, 31, 1354–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piva, M.; Domenici, G.; Iriondo, O.; Rabano, M.; Simoes, B.M.; Comaills, V.; Barredo, I.; Lopez-Ruiz, J.A.; Zabalza, I.; Kypta, R.; et al. Sox2 promotes tamoxifen resistance in breast cancer cells. EMBO Mol. Med. 2014, 6, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Herlyn, M. The emerging roles of Oct4 in tumor-initiating cells. Am. J. Physiol. Cell Physiol. 2015, 309, C709–C718. [Google Scholar] [CrossRef] [PubMed]
- Wuebben, E.L.; Rizzino, A. The dark side of SOX2: Cancer—A comprehensive overview. Oncotarget 2017, 8, 44917–44943. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Qian, W.; Zhang, H.; Liang, Y.; Wu, M.; Zhang, Y.; Zhang, X.; Gao, Q.; Li, Y. SOX2 is a marker for stem-like tumor cells in bladder cancer. Stem Cell Rep. 2017, 9, 429–437. [Google Scholar] [CrossRef] [Green Version]
- Abarrategi, A.; Tornin, J.; Martinez-Cruzado, L.; Hamilton, A.; Martinez-Campos, E.; Rodrigo, J.P.; Gonzalez, M.V.; Baldini, N.; Garcia-Castro, J.; Rodriguez, R. Osteosarcoma: Cells-of-origin, cancer stem cells, and targeted therapies. Stem Cells Int. 2016, 2016, 3631764. [Google Scholar] [CrossRef] [Green Version]
- Brown, H.K.; Tellez-Gabriel, M.; Heymann, D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017, 386, 189–195. [Google Scholar] [CrossRef]
- Genadry, K.C.; Pietrobono, S.; Rota, R.; Linardic, C.M. Soft tissue sarcoma cancer stem cells: An overview. Front. Oncol. 2018, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Hatina, J.; Kripnerova, M.; Houfkova, K.; Pesta, M.; Kuncova, J.; Sana, J.; Slaby, O.; Rodriguez, R. Sarcoma stem cell heterogeneity. Adv. Exp. Med. Biol. 2019, 1123, 95–118. [Google Scholar]
- Martins-Neves, S.R.; Corver, W.E.; Paiva-Oliveira, D.I.; Van den Akker, B.E.; Briaire-de-Bruijn, I.H.; Bovee, J.V.; Gomes, C.M.; Cleton-Jansen, A.M. Osteosarcoma stem cells have active WNT/beta-catenin and overexpress SOX2 and KLF4. J. Cell. Physiol. 2016, 231, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Naka, N.; Takenaka, S.; Araki, N.; Miwa, T.; Hashimoto, N.; Yoshioka, K.; Joyama, S.; Hamada, K.; Tsukamoto, Y.; Tomita, Y.; et al. Synovial sarcoma is a stem cell malignancy. Stem Cells 2010, 28, 1119–1131. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Rubio, R.; Menendez, P. Modeling sarcomagenesis using multipotent mesenchymal stem cells. Cell Res. 2012, 22, 62–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiavone, K.; Garnier, D.; Heymann, M.F.; Heymann, D. The Heterogeneity of osteosarcoma: The role played by cancer stem cells. Adv. Exp. Med. Biol. 2019, 1139, 187–200. [Google Scholar]
- Skoda, J.; Nunukova, A.; Loja, T.; Zambo, I.; Neradil, J.; Mudry, P.; Zitterbart, K.; Hermanova, M.; Hampl, A.; Sterba, J.; et al. Cancer stem cell markers in pediatric sarcomas: Sox2 is associated with tumorigenicity in immunodeficient mice. Tumor Biol. 2016, 37, 9535–9548. [Google Scholar] [CrossRef]
- Skoda, J.; Veselska, R. Cancer stem cells in sarcomas: Getting to the stemness core. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2134–2139. [Google Scholar] [CrossRef]
- Basu-Roy, U.; Bayin, N.S.; Rattanakorn, K.; Han, E.; Placantonakis, D.G.; Mansukhani, A.; Basilico, C. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat. Commun. 2015, 6, 6411. [Google Scholar] [CrossRef] [Green Version]
- Basu-Roy, U.; Seo, E.; Ramanathapuram, L.; Rapp, T.B.; Perry, J.A.; Orkin, S.H.; Mansukhani, A.; Basilico, C. Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. Oncogene 2012, 31, 2270–2282. [Google Scholar] [CrossRef] [Green Version]
- Maurizi, G.; Verma, N.; Gadi, A.; Mansukhani, A.; Basilico, C. Sox2 is required for tumor development and cancer cell proliferation in osteosarcoma. Oncogene 2018, 37, 4626–4632. [Google Scholar] [CrossRef] [PubMed]
- Levings, P.P.; McGarry, S.V.; Currie, T.P.; Nickerson, D.M.; McClellan, S.; Ghivizzani, S.C.; Steindler, D.A.; Gibbs, C.P. Expression of an exogenous human Oct-4 promoter identifies tumor-initiating cells in osteosarcoma. Cancer Res. 2009, 69, 5648–5655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suva, M.L.; Riggi, N.; Stehle, J.C.; Baumer, K.; Tercier, S.; Joseph, J.M.; Suva, D.; Clement, V.; Provero, P.; Cironi, L.; et al. Identification of cancer stem cells in Ewing’s sarcoma. Cancer Res. 2009, 69, 1776–1781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermida-Prado, F.; Villaronga, M.A.; Granda-Diaz, R.; Del-Rio-Ibisate, N.; Santos, L.; Hermosilla, M.A.; Oro, P.; Allonca, E.; Agorreta, J.; Garmendia, I.; et al. The SRC inhibitor dasatinib induces stem cell-like properties in head and neck cancer cells that are effectively counteracted by the mithralog EC-8042. J. Clin. Med. 2019, 8, 1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iglesias, J.M.; Gumuzio, J.; Martin, A.G. Linking pluripotency reprogramming and cancer. Stem Cells Transl. Med. 2017, 6, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, J.M.; Leis, O.; Perez Ruiz, E.; Gumuzio Barrie, J.; Garcia-Garcia, F.; Aduriz, A.; Beloqui, I.; Hernandez-Garcia, S.; Lopez-Mato, M.P.; Dopazo, J.; et al. The activation of the Sox2 RR2 pluripotency transcriptional reporter in human breast cancer cell lines is dynamic and labels cells with higher tumorigenic potential. Front. Oncol. 2014, 4, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, S.; Furuhashi, M.; Nakane, R.; Nakazawa, S.; Goudarzi, H.; Hamada, J.; Iizasa, H. Isolation and characterization of human breast cancer cells with SOX2 promoter activity. Biochem. Biophys. Res. Commun. 2013, 437, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Stoltz, K.; Sinyuk, M.; Hale, J.S.; Wu, Q.; Otvos, B.; Walker, K.; Vasanji, A.; Rich, J.N.; Hjelmeland, A.B.; Lathia, J.D. Development of a Sox2 reporter system modeling cellular heterogeneity in glioma. Neuro Oncol. 2015, 17, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.; Raviv, A.; Esposito, D.; Flanders, K.C.; Daniel, C.; Nghiem, B.T.; Garfield, S.; Lim, L.; Mannan, P.; Robles, A.I.; et al. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Rep. 2015, 4, 155–169. [Google Scholar] [CrossRef] [Green Version]
- Vaddi, P.K.; Stamnes, M.A.; Cao, H.; Chen, S. Elimination of SOX2/OCT4-associated prostate cancer stem cells blocks tumor development and enhances therapeutic response. Cancers 2019, 11, 1331. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhang, J.; Wang, P.; Ye, X.; Jung, K.; Bone, K.M.; Pearson, J.D.; Ingham, R.J.; McMullen, T.P.; Ma, Y.; et al. Identification of two novel phenotypically distinct breast cancer cell subsets based on Sox2 transcription activity. Cell. Signal. 2012, 24, 1989–1998. [Google Scholar] [CrossRef] [PubMed]
- Funes, J.M.; Quintero, M.; Henderson, S.; Martinez, D.; Qureshi, U.; Westwood, C.; Clements, M.O.; Bourboulia, D.; Pedley, R.B.; Moncada, S.; et al. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc. Natl. Acad. Sci. USA 2007, 104, 6223–6228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, R.; Rosu-Myles, M.; Arauzo-Bravo, M.; Horrillo, A.; Pan, Q.; Gonzalez-Rey, E.; Delgado, M.; Menendez, P. Human bone marrow stromal cells lose immunosuppressive and anti-inflammatory properties upon oncogenic transformation. Stem Cell Rep. 2014, 3, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, R.; Tornin, J.; Suarez, C.; Astudillo, A.; Rubio, R.; Yauk, C.; Williams, A.; Rosu-Myles, M.; Funes, J.M.; Boshoff, C.; et al. Expression of FUS-CHOP fusion protein in immortalized/transformed human mesenchymal stem cells drives mixoid liposarcoma formation. Stem Cells 2013, 31, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Cruzado, L.; Tornin, J.; Santos, L.; Rodriguez, A.; Garcia-Castro, J.; Moris, F.; Rodriguez, R. Aldh1 expression and activity increase during tumor evolution in sarcoma cancer stem cell populations. Sci. Rep. 2016, 6, 27878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tornin, J.; Martinez-Cruzado, L.; Santos, L.; Rodriguez, A.; Nunez, L.E.; Oro, P.; Hermosilla, M.A.; Allonca, E.; Fernandez-Garcia, M.T.; Astudillo, A.; et al. Inhibition of SP1 by the mithramycin analog EC-8042 efficiently targets tumor initiating cells in sarcoma. Oncotarget 2016, 7, 30935–30950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garofalo, A.; Naumova, E.; Manenti, L.; Ghilardi, C.; Ghisleni, G.; Caniatti, M.; Colombo, T.; Cherrington, J.M.; Scanziani, E.; Nicoletti, M.I.; et al. The combination of the tyrosine kinase receptor inhibitor SU6668 with paclitaxel affects ascites formation and tumor spread in ovarian carcinoma xenografts growing orthotopically. Clin. Cancer Res. 2003, 9, 3476–3485. [Google Scholar]
- Martinez-Cruzado, L.; Tornin, J.; Rodriguez, A.; Santos, L.; Allonca, E.; Fernandez-Garcia, M.T.; Astudillo, A.; Garcia-Pedrero, J.M.; Rodriguez, R. Trabectedin and campthotecin synergistically eliminate cancer stem cells in cell-of-origin sarcoma models. Neoplasia 2017, 19, 460–470. [Google Scholar] [CrossRef]
- Li, J.; Shen, J.; Wang, K.; Hornicek, F.; Duan, Z. The roles of sox family genes in sarcoma. Curr. Drug Targets 2016, 17, 1761–1772. [Google Scholar] [CrossRef]
- Ren, C.; Ren, T.; Yang, K.; Wang, S.; Bao, X.; Zhang, F.; Guo, W. Inhibition of SOX2 induces cell apoptosis and G1/S arrest in Ewing’s sarcoma through the PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2016, 35, 44. [Google Scholar] [CrossRef] [Green Version]
- Riggi, N.; Suva, M.L.; De Vito, C.; Provero, P.; Stehle, J.C.; Baumer, K.; Cironi, L.; Janiszewska, M.; Petricevic, T.; Suva, D.; et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010, 24, 916–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slemmons, K.K.; Crose, L.E.S.; Riedel, S.; Sushnitha, M.; Belyea, B.; Linardic, C.M. A novel notch-YAP circuit drives stemness and tumorigenesis in embryonal rhabdomyosarcoma. Mol. Cancer Res. 2017, 15, 1777–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sannino, G.; Marchetto, A.; Ranft, A.; Jabar, S.; Zacherl, C.; Alba-Rubio, R.; Stein, S.; Wehweck, F.S.; Kiran, M.M.; Holting, T.L.B.; et al. Gene expression and immunohistochemical analyses identify SOX2 as major risk factor for overall survival and relapse in Ewing sarcoma patients. EBioMedicine 2019, 47, 156–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zayed, H.; Petersen, I. Stem cell transcription factor SOX2 in synovial sarcoma and other soft tissue tumors. Pathol. Res. Pract. 2018, 214, 1000–1007. [Google Scholar] [CrossRef] [Green Version]
- Rey, V.; Menendez, S.T.; Estupinan, O.; Rodriguez, A.; Santos, L.; Tornin, J.; Martinez-Cruzado, L.; Castillo, D.; Ordonez, G.R.; Costilla, S.; et al. New chondrosarcoma cell lines with preserved stem cell properties to study the genomic drift during in vitro/in vivo growth. J. Clin. Med. 2019, 8, 455. [Google Scholar] [CrossRef] [Green Version]
- Golan, H.; Shukrun, R.; Caspi, R.; Vax, E.; Pode-Shakked, N.; Goldberg, S.; Pleniceanu, O.; Bar-Lev, D.D.; Mark-Danieli, M.; Pri-Chen, S.; et al. In vivo expansion of cancer stemness affords novel cancer stem cell targets: Malignant rhabdoid tumor as an example. Stem Cell Rep. 2018, 11, 795–810. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.K.; Kollipara, R.K.; Vemireddy, V.; Yang, X.L.; Sun, Y.; Regmi, N.; Klingler, S.; Hatanpaa, K.J.; Raisanen, J.; Cho, S.K.; et al. Oncogenes activate an autonomous transcriptional regulatory circuit that drives glioblastoma. Cell Rep. 2017, 18, 961–976. [Google Scholar] [CrossRef]
- Vanner, R.J.; Remke, M.; Gallo, M.; Selvadurai, H.J.; Coutinho, F.; Lee, L.; Kushida, M.; Head, R.; Morrissy, S.; Zhu, X.; et al. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 2014, 26, 33–47. [Google Scholar] [CrossRef] [Green Version]
- Nunez, L.E.; Nybo, S.E.; Gonzalez-Sabin, J.; Perez, M.; Menendez, N.; Brana, A.F.; Shaaban, K.A.; He, M.; Moris, F.; Salas, J.A.; et al. A novel mithramycin analogue with high antitumor activity and less toxicity generated by combinatorial biosynthesis. J. Med. Chem. 2012, 55, 5813–5825. [Google Scholar] [CrossRef] [Green Version]
- Rudin, C.M.; Durinck, S.; Stawiski, E.W.; Poirier, J.T.; Modrusan, Z.; Shames, D.S.; Bergbower, E.A.; Guan, Y.; Shin, J.; Guillory, J.; et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 2012, 44, 1111–1116. [Google Scholar] [CrossRef]
- Tornin, J.; Hermida-Prado, F.; Padda, R.S.; Gonzalez, M.V.; Alvarez-Fernandez, C.; Rey, V.; Martinez-Cruzado, L.; Estupinan, O.; Menendez, S.T.; Fernandez-Nevado, L.; et al. FUS-CHOP Promotes Invasion in Myxoid Liposarcoma through a SRC/FAK/RHO/ROCK-Dependent Pathway. Neoplasia 2018, 20, 44–56. [Google Scholar] [CrossRef] [PubMed]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menendez, S.T.; Rey, V.; Martinez-Cruzado, L.; Gonzalez, M.V.; Morales-Molina, A.; Santos, L.; Blanco, V.; Alvarez, C.; Estupiñan, O.; Allonca, E.; et al. SOX2 Expression and Transcriptional Activity Identifies a Subpopulation of Cancer Stem Cells in Sarcoma with Prognostic Implications. Cancers 2020, 12, 964. https://doi.org/10.3390/cancers12040964
Menendez ST, Rey V, Martinez-Cruzado L, Gonzalez MV, Morales-Molina A, Santos L, Blanco V, Alvarez C, Estupiñan O, Allonca E, et al. SOX2 Expression and Transcriptional Activity Identifies a Subpopulation of Cancer Stem Cells in Sarcoma with Prognostic Implications. Cancers. 2020; 12(4):964. https://doi.org/10.3390/cancers12040964
Chicago/Turabian StyleMenendez, Sofia T., Veronica Rey, Lucia Martinez-Cruzado, M. Victoria Gonzalez, Alvaro Morales-Molina, Laura Santos, Verónica Blanco, Carlos Alvarez, Oscar Estupiñan, Eva Allonca, and et al. 2020. "SOX2 Expression and Transcriptional Activity Identifies a Subpopulation of Cancer Stem Cells in Sarcoma with Prognostic Implications" Cancers 12, no. 4: 964. https://doi.org/10.3390/cancers12040964