Ribosomal Protein S27/Metallopanstimulin-1 (RPS27) in Glioma—A New Disease Biomarker?
Abstract
1. Introduction
2. Results
2.1. Patient Cohort
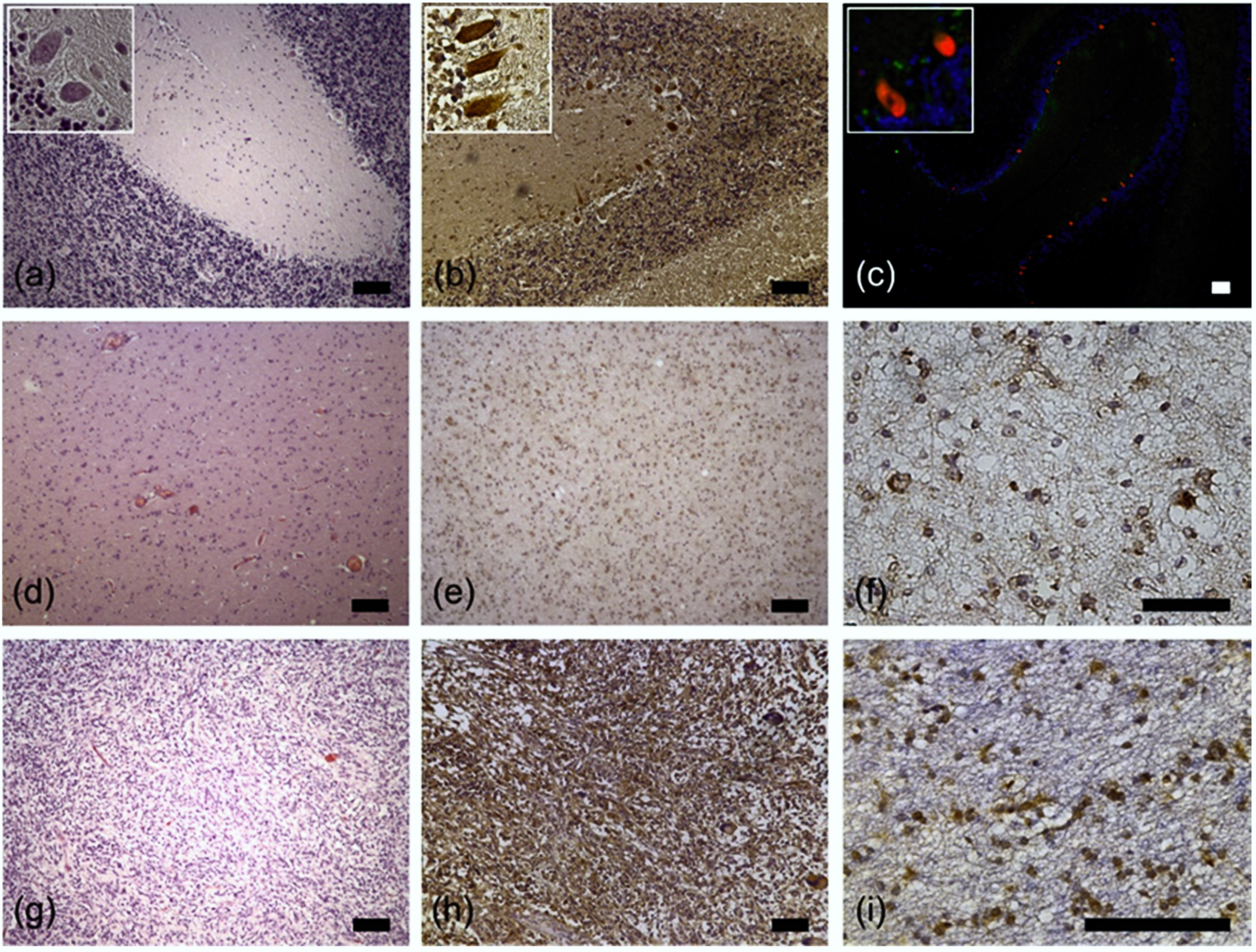
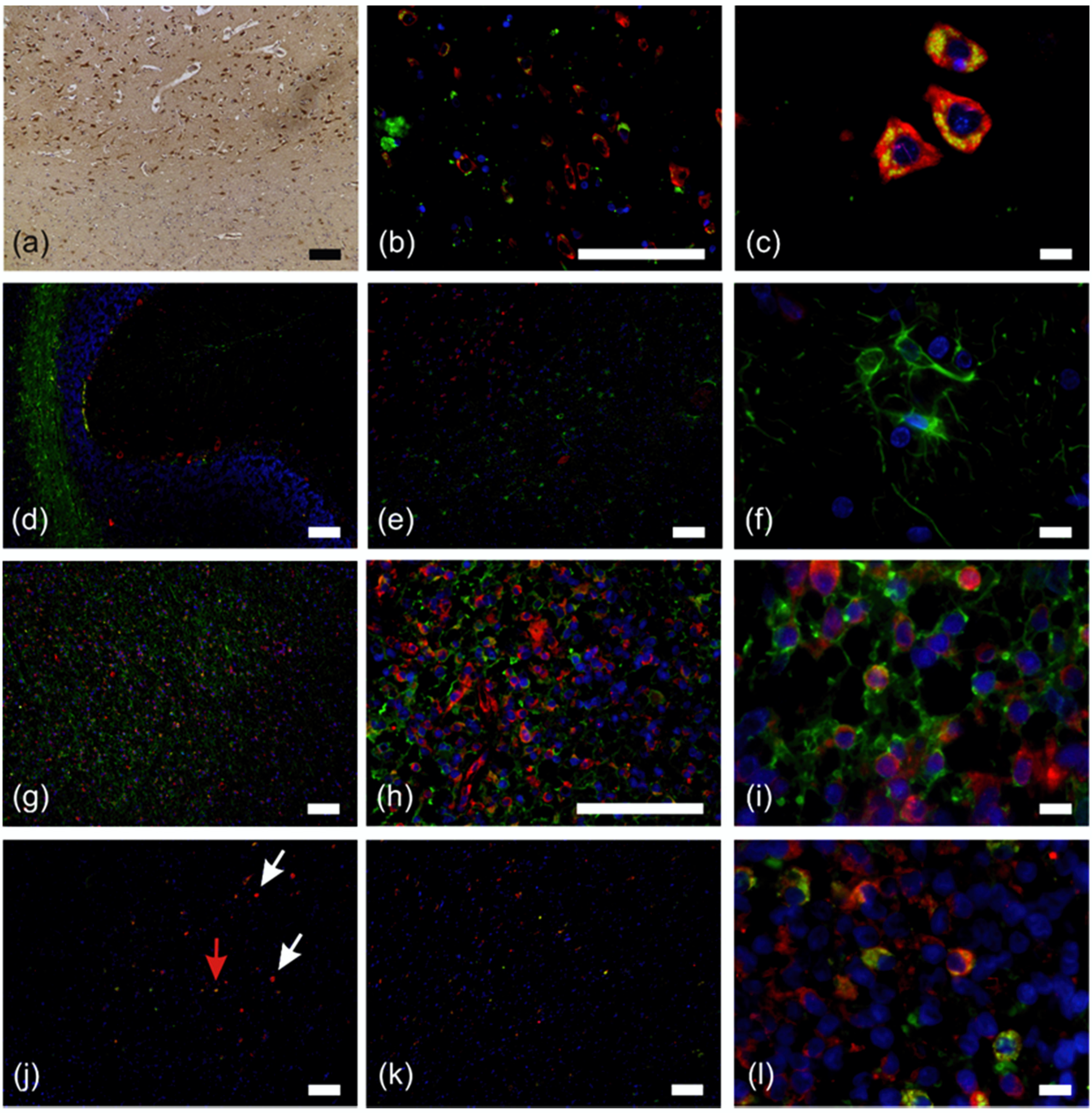
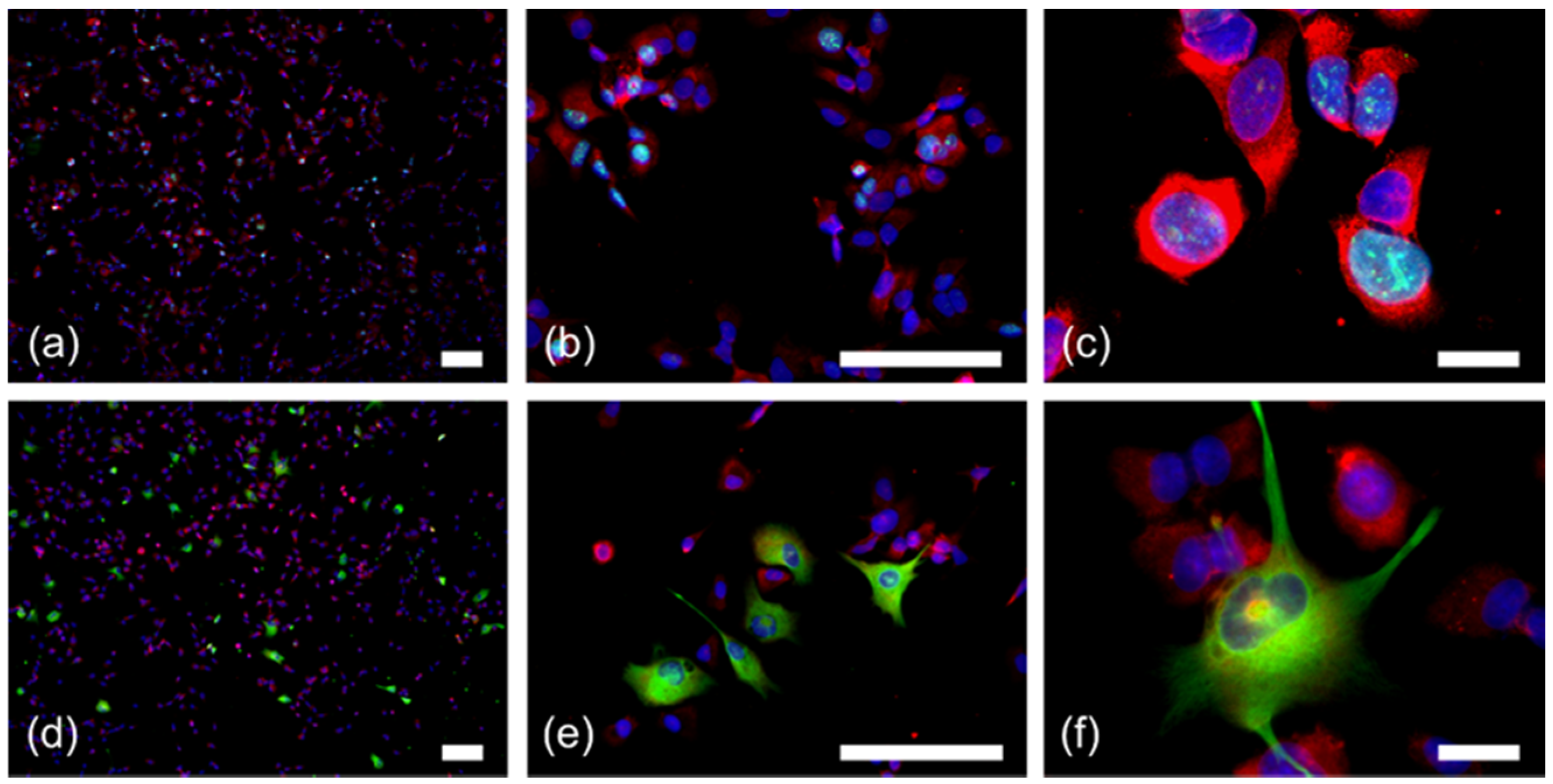
2.2. RPS27 Protein Was Expressed by Neurons, Tumor-Associated Macrophages, Astrocytic Tumor Cells, and Stem-Like GBM Cells
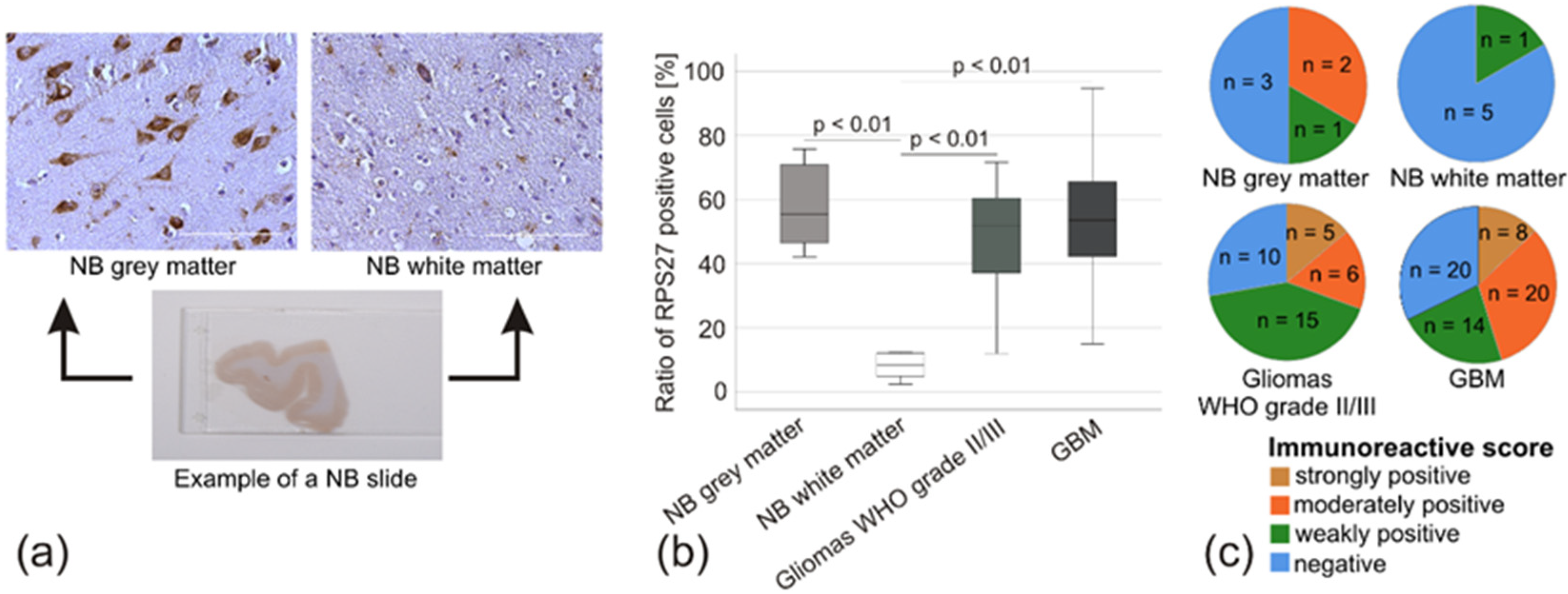
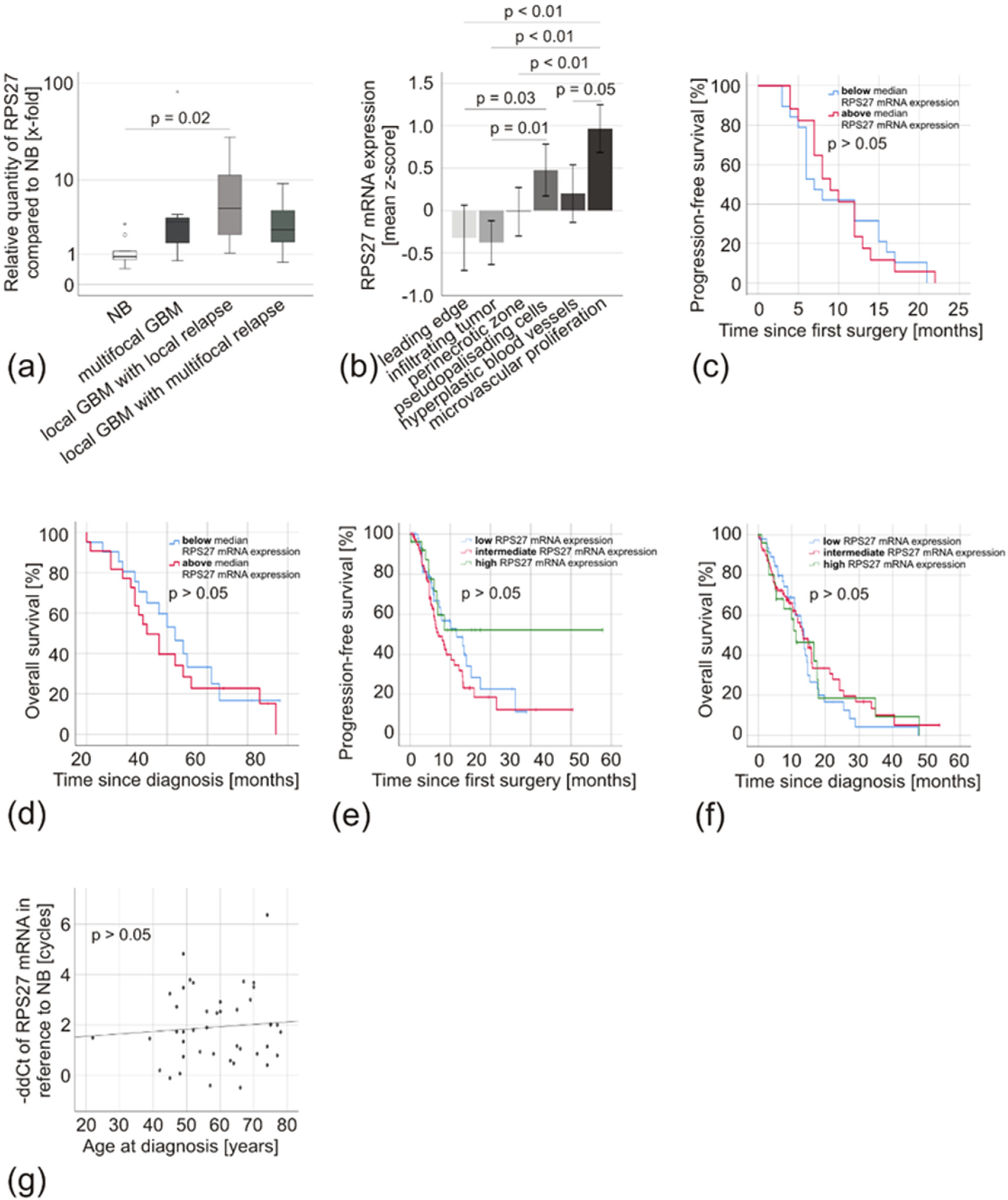
2.3. RPS27 was Overexpressed in Gliomas independently of varying Tumor and Patient Characteristics
3. Discussion
4. Materials and Methods
4.1. Tissue Samples and Clinical Data
4.2. Glioma Stem-Like Cell Culture
4.3. RNA Extraction and Quantitative RT-PCR
4.4. Western Blot Analysis
4.5. Immunohistochemistry (IHC)
4.6. Bioinformatics and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Brandner, S.; Jaunmuktane, Z. Neurological update: Gliomas and other primary brain tumours in adults. J. Neurol. 2018, 265, 717–727. [Google Scholar] [CrossRef]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 1, e15017. [Google Scholar] [CrossRef] [PubMed]
- Hasselblatt, M.; Jaber, M.; Reuss, D.; Grauer, O.; Bibo, A.; Terwey, S.; Schick, U.; Ebel, H.; Niederstadt, T.; Stummer, W.; et al. Diffuse Astrocytoma, IDH-Wildtype: A Dissolving Diagnosis. J. Neuropathol. Exp. Neurol. 2018, 77, 422–425. [Google Scholar] [CrossRef] [PubMed]
- Reuss, D.E.; Kratz, A.; Sahm, F.; Capper, D.; Schrimpf, D.; Koelsche, C.; Hovestadt, V.; Bewerunge-Hudler, M.; Jones, D.T.; Schittenhelm, J.; et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol. 2015, 130, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Wick, W.; Osswald, M.; Wick, A.; Winkler, F. Treatment of glioblastoma in adults. Ther. Adv. Neurol. Disord. 2018, 11, e1756286418790452. [Google Scholar] [CrossRef]
- Hottinger, A.F.; Pacheco, P.; Stupp, R. Tumor treating fields: A novel treatment modality and its use in brain tumors. Neuro-Oncol. 2016, 18, 1338–1349. [Google Scholar] [CrossRef]
- Stupp, R.; Taillibert, S.; Kanner, A.A.; Kesari, S.; Steinberg, D.M.; Toms, S.A.; Taylor, L.P.; Lieberman, F.; Silvani, A.; Fink, K.L.; et al. Maintenance Therapy with Tumor-Treating Fields Plus Temozolomide vs Temozolomide Alone for Glioblastoma: A Randomized Clinical Trial. Jama 2015, 314, 2535–2543. [Google Scholar] [CrossRef]
- Mehta, M.; Wen, P.; Nishikawa, R.; Reardon, D.; Peters, K. Critical review of the addition of tumor treating fields (TTFields) to the existing standard of care for newly diagnosed glioblastoma patients. Crit. Rev. Oncol. Hematol. 2017, 111, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients with Glioblastoma: A Randomized Clinical Trial. Jama 2017, 318, 2306–2316. [Google Scholar] [CrossRef] [PubMed]
- Herrlinger, U.; Tzaridis, T.; Mack, F.; Steinbach, J.P.; Schlegel, U.; Sabel, M.; Hau, P.; Kortmann, R.D.; Krex, D.; Grauer, O.; et al. Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): A randomised, open-label, phase 3 trial. Lancet 2019, 393, 678–688. [Google Scholar] [CrossRef]
- Osuka, S.; Van Meir, E.G. Overcoming therapeutic resistance in glioblastoma: The way forward. J. Clin. Investig. 2017, 127, 415–426. [Google Scholar] [CrossRef]
- Malmström, A.; Grønberg, B.H.; Marosi, C.; Stupp, R.; Frappaz, D.; Schultz, H.; Abacioglu, U.; Tavelin, B.; Lhermitte, B.; Hegi, M.E.; et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol. 2012, 13, 916–926. [Google Scholar] [CrossRef]
- Wick, W.; Platten, M.; Meisner, C.; Felsberg, J.; Tabatabai, G.; Simon, M.; Nikkhah, G.; Papsdorf, K.; Steinbach, J.P.; Sabel, M.; et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: The NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012, 13, 707–715. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef]
- Feldheim, J.; Kessler, A.F.; Monoranu, C.M.; Ernestus, R.I.; Lohr, M.; Hagemann, C. Changes of O(6)-Methylguanine DNA Methyltransferase (MGMT) Promoter Methylation in Glioblastoma Relapse-A Meta-Analysis Type Literature Review. Cancers 2019, 11. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, 733–745. [Google Scholar] [CrossRef]
- Fernandez-Pol, J.A.; Klos, D.J.; Hamilton, P.D. Metallopanstimulin gene product produced in a baculovirus expression system is a nuclear phosphoprotein that binds to DNA. Cell Growth Differ. 1994, 5, 811–825. [Google Scholar]
- Fernandez-Pol, J.A.; Fletcher, J.W.; Hamilton, P.D.; Klos, D.J. Expression of metallopanstimulin and oncogenesis in human prostatic carcinoma. Anticancer Res. 1997, 17, 1519–1530. [Google Scholar] [PubMed]
- Ganger, D.R.; Hamilton, P.D.; Klos, D.J.; Jakate, S.; McChesney, L.; Fernandez-Pol, J.A. Differential expression of metallopanstimulin/S27 ribosomal protein in hepatic regeneration and neoplasia. Cancer Detect. Prev. 2001, 25, 231–236. [Google Scholar] [PubMed]
- Atsuta, Y.; Aoki, N.; Sato, K.; Oikawa, K.; Nochi, H.; Miyokawa, N.; Hirata, S.; Kimura, S.; Sasajima, T.; Katagiri, M. Identification of metallopanstimulin-1 as a member of a tumor associated antigen in patients with breast cancer. Cancer Lett. 2002, 182, 101–107. [Google Scholar] [CrossRef]
- Lee, W.J.; Keefer, K.; Hollenbeak, C.S.; Stack, B.C., Jr. A new assay to screen for head and neck squamous cell carcinoma using the tumor marker metallopanstimulin. Otolaryngol. Head Neck Surg. 2004, 131, 466–471. [Google Scholar] [CrossRef]
- Wang, Y.W.; Qu, Y.; Li, J.F.; Chen, X.H.; Liu, B.Y.; Gu, Q.L.; Zhu, Z.G. In vitro and in vivo evidence of metallopanstimulin-1 in gastric cancer progression and tumorigenicity. Clin. Cancer Res. 2006, 12, 4965–4973. [Google Scholar] [CrossRef]
- Fernandez-Pol, J.A. Increased serum level of RPMPS-1/S27 protein in patients with various types of cancer is useful for the early detection, prevention and therapy. Cancer Genom. Proteom. 2012, 9, 203–256. [Google Scholar]
- Fernandez-Pol, J.A. A Novel Marker for Purkinje Cells, Ribosomal Protein MPS1/S27: Expression of MPS1 in Human Cerebellum. Cancer Genom. Proteom. 2016, 13, 47–53. [Google Scholar]
- Fernandez-Pol, J.A.; Klos, D.J.; Hamilton, P.D. A growth factor-inducible gene encodes a novel nuclear protein with zinc finger structure. J. Biol. Chem. 1993, 268, 21198–21204. [Google Scholar]
- Chen, Z.; Feng, X.; Herting, C.J.; Garcia, V.A.; Nie, K.; Pong, W.W.; Rasmussen, R.; Dwivedi, B.; Seby, S.; Wolf, S.A.; et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017, 77, 2266–2278. [Google Scholar] [CrossRef]
- Remmele, W.; Stegner, H.E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 1987, 8, 138–140. [Google Scholar]
- Gutmann, D.H.; Hedrick, N.M.; Li, J.; Nagarajan, R.; Perry, A.; Watson, M.A. Comparative gene expression profile analysis of neurofibromatosis 1-associated and sporadic pilocytic astrocytomas. Cancer Res. 2002, 62, 2085–2091. [Google Scholar] [PubMed]
- Puchalski, R.B.; Shah, N.; Miller, J.; Dalley, R.; Nomura, S.R.; Yoon, J.G.; Smith, K.A.; Lankerovich, M.; Bertagnolli, D.; Bickley, K.; et al. An anatomic transcriptional atlas of human glioblastoma. Science 2018, 360, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Caracausi, M.; Piovesan, A.; Antonaros, F.; Strippoli, P.; Vitale, L.; Pelleri, M.C. Systematic identification of human housekeeping genes possibly useful as references in gene expression studies. Mol. Med. Rep. 2017, 16, 2397–2410. [Google Scholar] [CrossRef]
- Santa Cruz, D.J.; Hamilton, P.D.; Klos, D.J.; Fernandez-Pol, J.A. Differential expression of metallopanstimulin/S27 ribosomal protein in melanocytic lesions of the skin. J. Cutan. Pathol. 1997, 24, 533–542. [Google Scholar] [CrossRef]
- Wei, J.; Gabrusiewicz, K.; Heimberger, A. The Controversial Role of Microglia in Malignant Gliomas. Clin. Dev. Immunol. 2013, 2013, e12. [Google Scholar] [CrossRef]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef]
- Rong, Y.; Durden, D.L.; Van Meir, E.G.; Brat, D.J. ‘Pseudopalisading’ necrosis in glioblastoma: A familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J. Neuropathol. Exp. Neurol. 2006, 65, 529–539. [Google Scholar] [CrossRef]
- Floristan, A.; Morales, L.; Hanniford, D.; Martinez, C.; Castellano-Sanz, E.; Dolgalev, I.; Ulloa-Morales, A.; Vega-Saenz de Miera, E.; Moran, U.; Darvishian, F.; et al. Functional analysis of RPS27 mutations and expression in melanoma. Pigment Cell Melanoma Res. 2019. [Google Scholar] [CrossRef]
- Stupp, R.; Hegi, M.E.; Gorlia, T.; Erridge, S.C.; Perry, J.; Hong, Y.K.; Aldape, K.D.; Lhermitte, B.; Pietsch, T.; Grujicic, D.; et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014, 15, 1100–1108. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Godard, S.; Dietrich, P.Y.; Regli, L.; Ostermann, S.; Otten, P.; Van Melle, G.; de Tribolet, N.; Stupp, R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004, 10, 1871–1874. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Zhao, Y.; He, H.; Sun, Y. Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene 2011, 30, 1798–1811. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tan, M.; Liu, X.; Xiong, X.; Sun, Y. Inactivation of ribosomal protein S27-like confers radiosensitivity via the Mdm2-p53 and Mdm2-MRN-ATM axes. Cell Death Dis. 2018, 9, e145. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Pierson, S.; Dudney, C.; Zeng, Y.; Macleod, V.; Shaughnessy, J.D.; Stack, B.C., Jr. Ribosomal protein metallopanstimulin-1 impairs multiple myeloma CAG cells growth and inhibits fibroblast growth factor receptor 3. Clin. Lymphoma Myeloma Leukemia 2011, 11, 490–497. [Google Scholar] [CrossRef][Green Version]
- Valdivieso, A.G.; Mori, C.; Clauzure, M.; Massip-Copiz, M.; Santa-Coloma, T.A. CFTR modulates RPS27 gene expression using chloride anion as signaling effector. Arch. Biochem. Biophys. 2017, 633, 103–109. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Qu, Y.; Zhang, Q.; Wei, M.; Liu, C.X.; Chen, X.H.; Yan, M.; Zhu, Z.G.; Liu, B.Y.; Chen, G.Q.; et al. Knockdown of metallopanstimulin-1 inhibits NF-kappaB signaling at different levels: The role of apoptosis induction of gastric cancer cells. Int. J. Cancer 2012, 130, 2761–2770. [Google Scholar] [CrossRef]
- Cao, D.; Luo, Y.; Qin, S.; Yu, M.; Mu, Y.; Ye, G.; Yang, N.; Cong, Z.; Chen, J.; Qin, J.; et al. Metallopanstimulin-1 (MPS-1) mediates the promotion effect of leptin on colorectal cancer through activation of JNK/c-Jun signaling pathway. Cell Death Dis. 2019, 10, e655. [Google Scholar] [CrossRef]
- Sundblad, A.S.; Ricci, L.; Xynos, F.P.; Fernandez-Pol, J.A. Metallopanstimulin/S27 Ribosomal Antigen Expression in Stages I and II Breast Cancer: Its Relationship with Clinicopathologic Factors. Cancer Genom. Proteom. 2005, 2, 53–60. [Google Scholar]
- Fernandez-Pol, J.A. Metallopanstimulin as a novel tumor marker in sera of patients with various types of common cancers: Implications for prevention and therapy. Anticancer Res. 1996, 16, 2177–2185. [Google Scholar]
- Fernandez-Pol, J.A.; Hamilton, P.D.; Klos, D.J. Genomics, Proteomics and Cancer: Specific Ribosomal, Mitochondrial, and Tumor Reactive Proteins Can Be Used as Biomarkers for Early Detection of Breast Cancer in Serum. Cancer Genom. Proteom. 2005, 2, 1–24. [Google Scholar]
- Stack, B.C., Jr.; Hollenbeak, C.S.; Lee, C.M.; Dunphy, F.R.; Lowe, V.J.; Hamilton, P.D. Metallopanstimulin as a marker for head and neck cancer. World J. Surg. Oncol. 2004, 2, e45. [Google Scholar] [CrossRef] [PubMed]
- Feldheim, J.; Kessler, A.F.; Schmitt, D.; Wilczek, L.; Linsenmann, T.; Dahlmann, M.; Monoranu, C.M.; Ernestus, R.I.; Hagemann, C.; Lohr, M. Expression of activating transcription factor 5 (ATF5) is increased in astrocytomas of different WHO grades and correlates with survival of glioblastoma patients. Onco. Targets Ther. 2018, 11, 8673–8684. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, T.P.; Kricka, L.J.; Carter, T.J.; Thorpe, G.H. Analytical luminescence: Its potential in the clinical laboratory. Clin. Chem. 1979, 25, 1531–1546. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ruifrok, A.C.; Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Patients’ Characteristics | IDHmut Glioma | IDHwt Glioma |
|---|---|---|
| Sex | female: 10/35.7% male: 18/64.3% | female: 4/30.8% male: 9/69.2% |
| Age (median, quartiles) | 38.0 years (33.0–46.0 years) | 45.0 years (28.5–57.5 years) |
| OS (median, quartiles) | 31.0 months (8.0–41.0 months) | 32.0 months (12.0–49.0 months) |
| Growth pattern | usual: 11/39.3% highly diffuse: 17/60.7% | usual: 6/46.2% highly diffuse: 7/53.5% |
| Patients’ Characteristics | |
|---|---|
| Sex | |
| female | 18/41.9% |
| male | 25/58.1% |
| Age (median, quartiles) | 59.0 years (49.0–70.0 years) |
| ECOG | |
| 0 | 22/51.2% |
| 1 | 17/39.5% |
| >1 | 4/9.3% |
| Tumor Characteristics | |
| Volume (median, quartiles) | 30.1 cm3 (16.2–54.3 cm3) |
| Localization | |
| left hemisphere | 24/55.8% |
| right hemisphere | 16/37.2% |
| both hemispheres | 3/7.0% |
| Localization (lobe) | |
| frontal | 13/30.2% |
| occipital | 5/11.6% |
| temporal | 7/16.3% |
| parietal | 5/11.6% |
| multiple | 13/30.2% |
| MGMT promoter methylation 1 | |
| unmethylated | 10/31.3% |
| methylated | 22/68.7% |
| Ki67 staining (median, quartiles) | 25% (20–30) |
| Therapy | |
| Time from diagnosis to therapy | |
| 0–7 days | 27/62.9% |
| 8–14 days | 9/20.9% |
| >14 days | 7/16.2% |
| Surgical intervention | |
| biopsy | 6/14.0% |
| complete resection | 9/20.9% |
| incomplete resection | 28/65.1% |
| Radiation therapy | |
| yes | 40/93.0% |
| no | 3/7.0% |
| TMZ chemotherapy | |
| yes | 35/81.4% |
| no | 8/18.6% |
| Relapse and Outcome | |
| PFS 2 | |
| 0–6 months | 2/5.6% |
| >6 months | 34/94.4% |
| Relapse | |
| primarily multifocal | 7/16.3% |
| local | 24/55.8% |
| multifocal | 12/27.9% |
| OS | |
| 0–6 months | 4/9.3% |
| >6 months | 39/90.7% |
| Patient Characteristics | Correlation Coefficient r | p-Value | Statistical Test |
|---|---|---|---|
| Sex (female/male) | n.d. | 0.676 | ANOVA |
| Age | 0.519 | <0.01 | Non-parametric correlation |
| OS | −0.214 | 0.294 | Non-parametric correlation |
| Growth pattern (usual/highly diffuse) | n.d. | 0.520 | ANOVA |
| Patient and Tumor Characteristics | Correlation Coefficient r | p-Value | Statistical Test |
|---|---|---|---|
| Sex (female/male) | n.d. | 0.47 | ANOVA |
| Age | -0.07 | 0.66 | Non-parametric correlation |
| ECOG (median) | n.d. | 0.42 | ANOVA |
| Tumor volume | 0.01 | 0.93 | Non-parametric correlation |
| Tumor localization (left/right/both hemispheres) | n.d. | 0.90 | ANOVA |
| Tumor localization (lobe: frontal/occipital/temporal/parietal/multifocal) | n.d. | 0.27 | ANOVA |
| MGMT promoter methylation (unmethylated/methylated) | n.d. | 0.43 | ANOVA |
| Ki67 staining | 0.33 | 0.04 | Non-parametric correlation |
| Surgical intervention | n.d. | 0.79 | ANOVA |
| (biopsy/incomplete resection/complete resection) Tumor growth pattern (multifocal/local relapse [primary tumor/relapse]/multifocal relapse [primary tumor/relapse]) | n.d. | 0.84 | ANOVA |
| PFS | 0.04 | 0.83 | Non-parametric correlation |
| OS | -0.15 | 0.33 | Non-parametric correlation |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feldheim, J.; Kessler, A.F.; Schmitt, D.; Salvador, E.; Monoranu, C.M.; Feldheim, J.J.; Ernestus, R.-I.; Löhr, M.; Hagemann, C. Ribosomal Protein S27/Metallopanstimulin-1 (RPS27) in Glioma—A New Disease Biomarker? Cancers 2020, 12, 1085. https://doi.org/10.3390/cancers12051085
Feldheim J, Kessler AF, Schmitt D, Salvador E, Monoranu CM, Feldheim JJ, Ernestus R-I, Löhr M, Hagemann C. Ribosomal Protein S27/Metallopanstimulin-1 (RPS27) in Glioma—A New Disease Biomarker? Cancers. 2020; 12(5):1085. https://doi.org/10.3390/cancers12051085
Chicago/Turabian StyleFeldheim, Jonas, Almuth F. Kessler, Dominik Schmitt, Ellaine Salvador, Camelia M. Monoranu, Julia J. Feldheim, Ralf-Ingo Ernestus, Mario Löhr, and Carsten Hagemann. 2020. "Ribosomal Protein S27/Metallopanstimulin-1 (RPS27) in Glioma—A New Disease Biomarker?" Cancers 12, no. 5: 1085. https://doi.org/10.3390/cancers12051085
APA StyleFeldheim, J., Kessler, A. F., Schmitt, D., Salvador, E., Monoranu, C. M., Feldheim, J. J., Ernestus, R.-I., Löhr, M., & Hagemann, C. (2020). Ribosomal Protein S27/Metallopanstimulin-1 (RPS27) in Glioma—A New Disease Biomarker? Cancers, 12(5), 1085. https://doi.org/10.3390/cancers12051085






