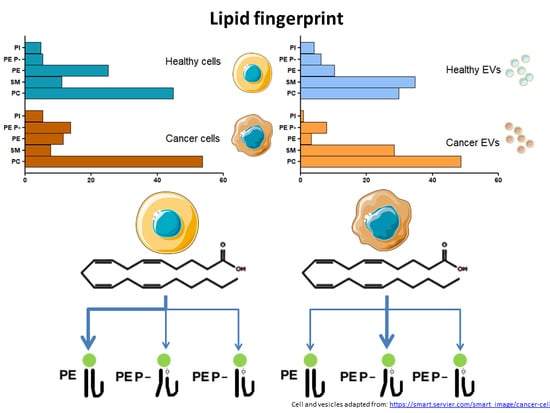
Common and Differential Traits of the Membrane Lipidome of Colon Cancer Cell Lines and Their Secreted Vesicles: Impact on Studies Using Cell Lines
Abstract
1. Introduction
2. Results
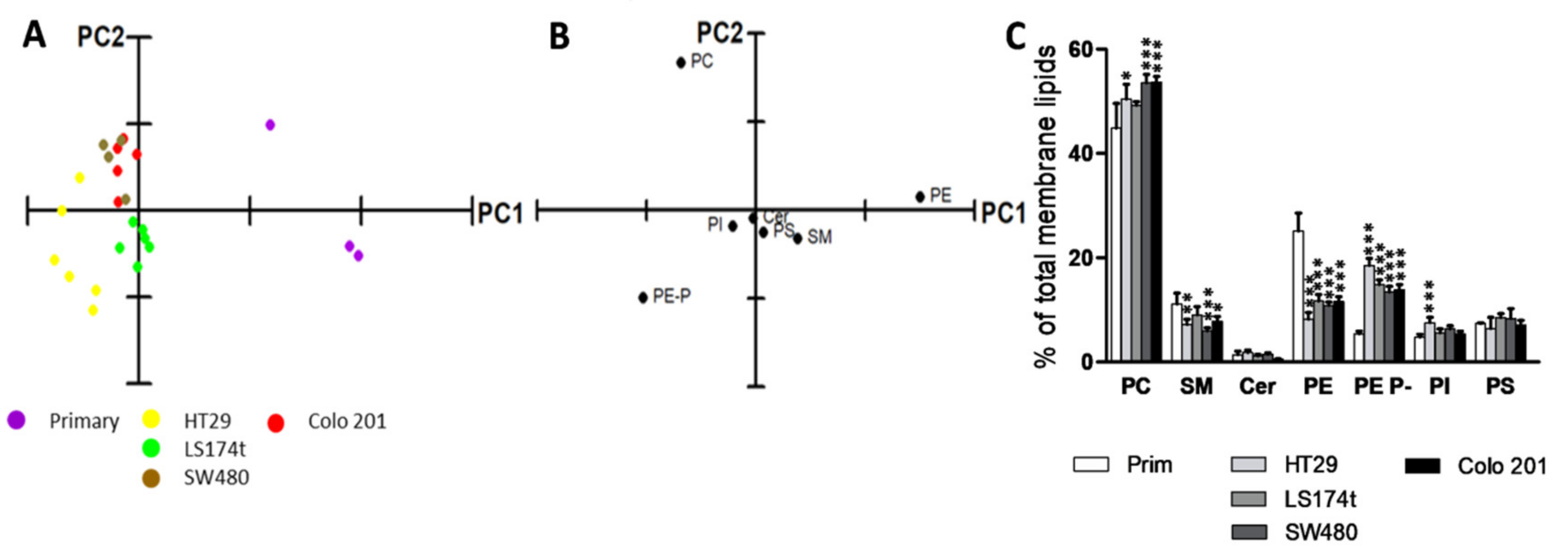
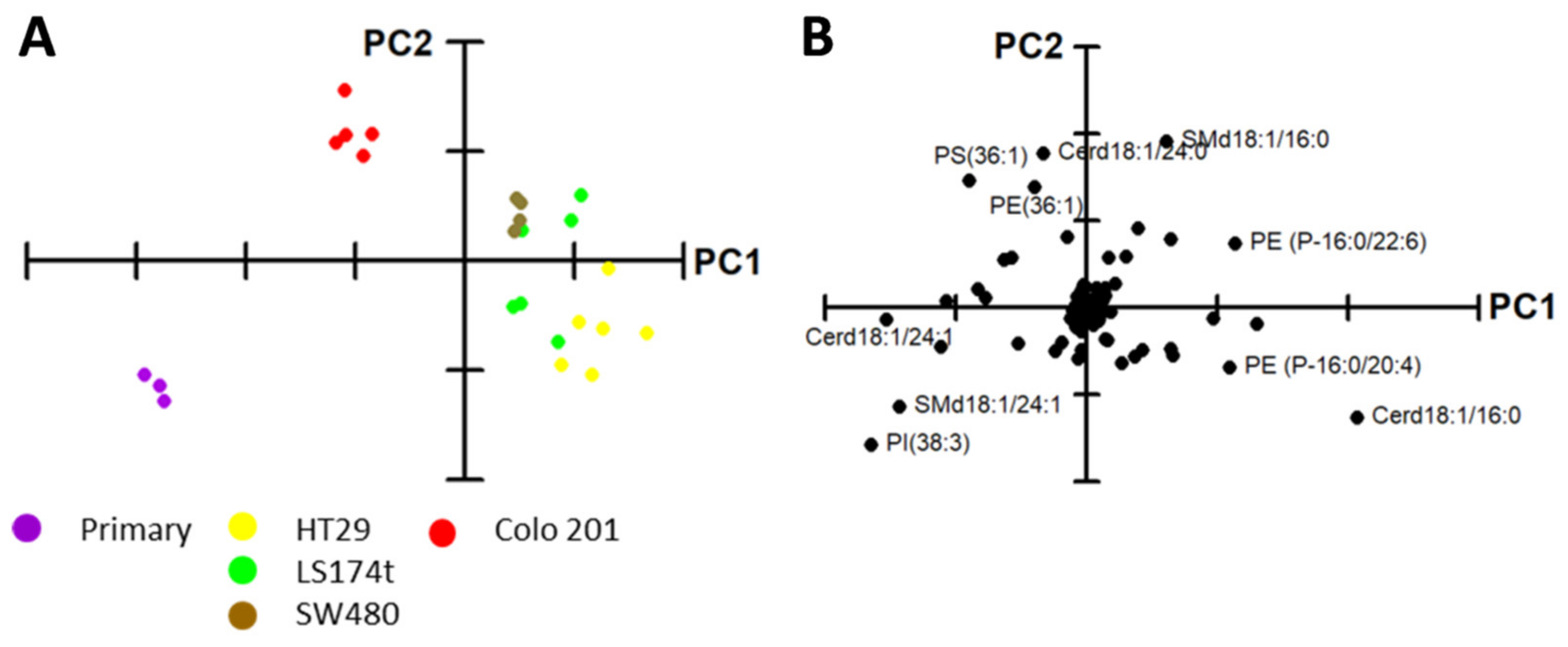
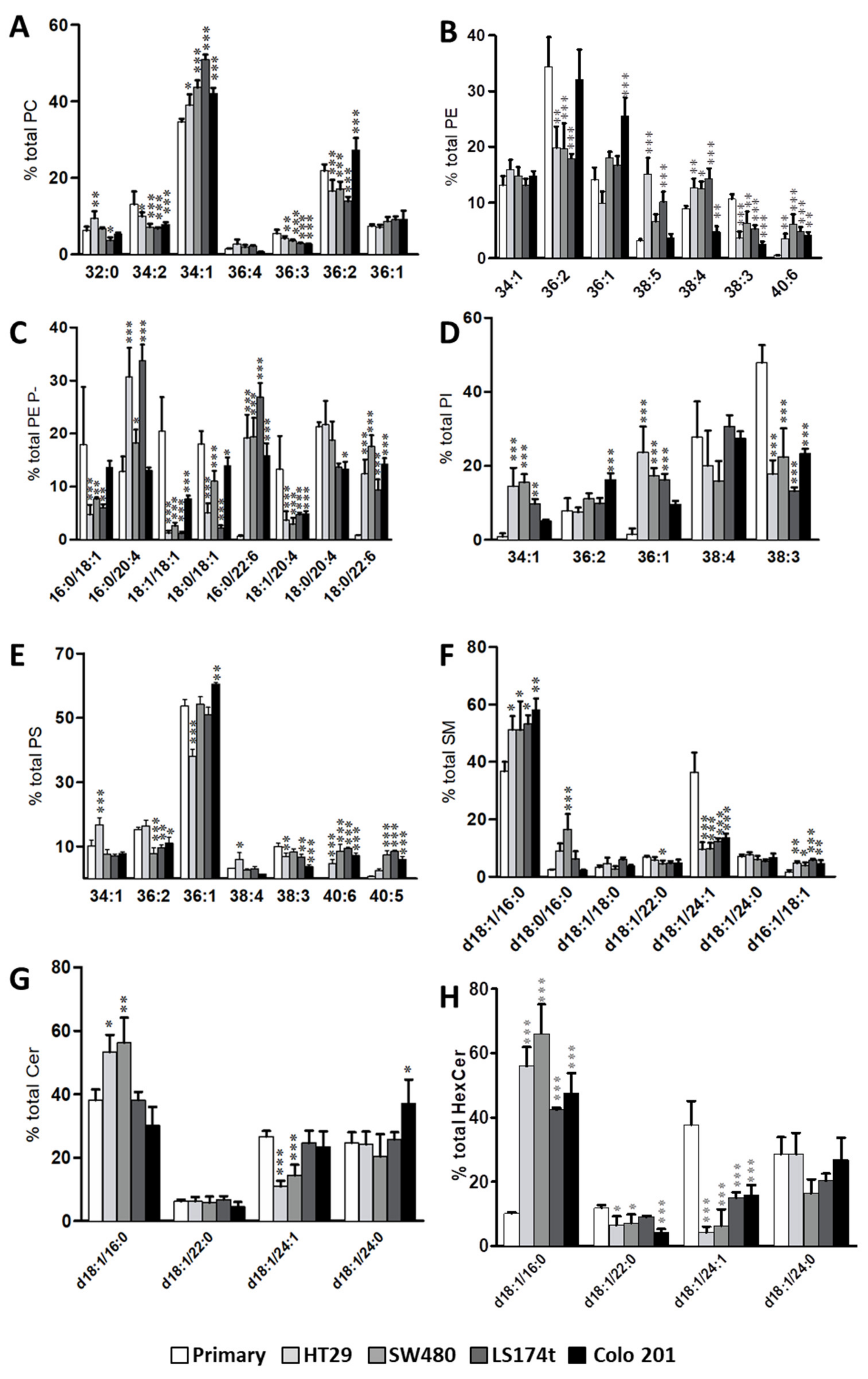
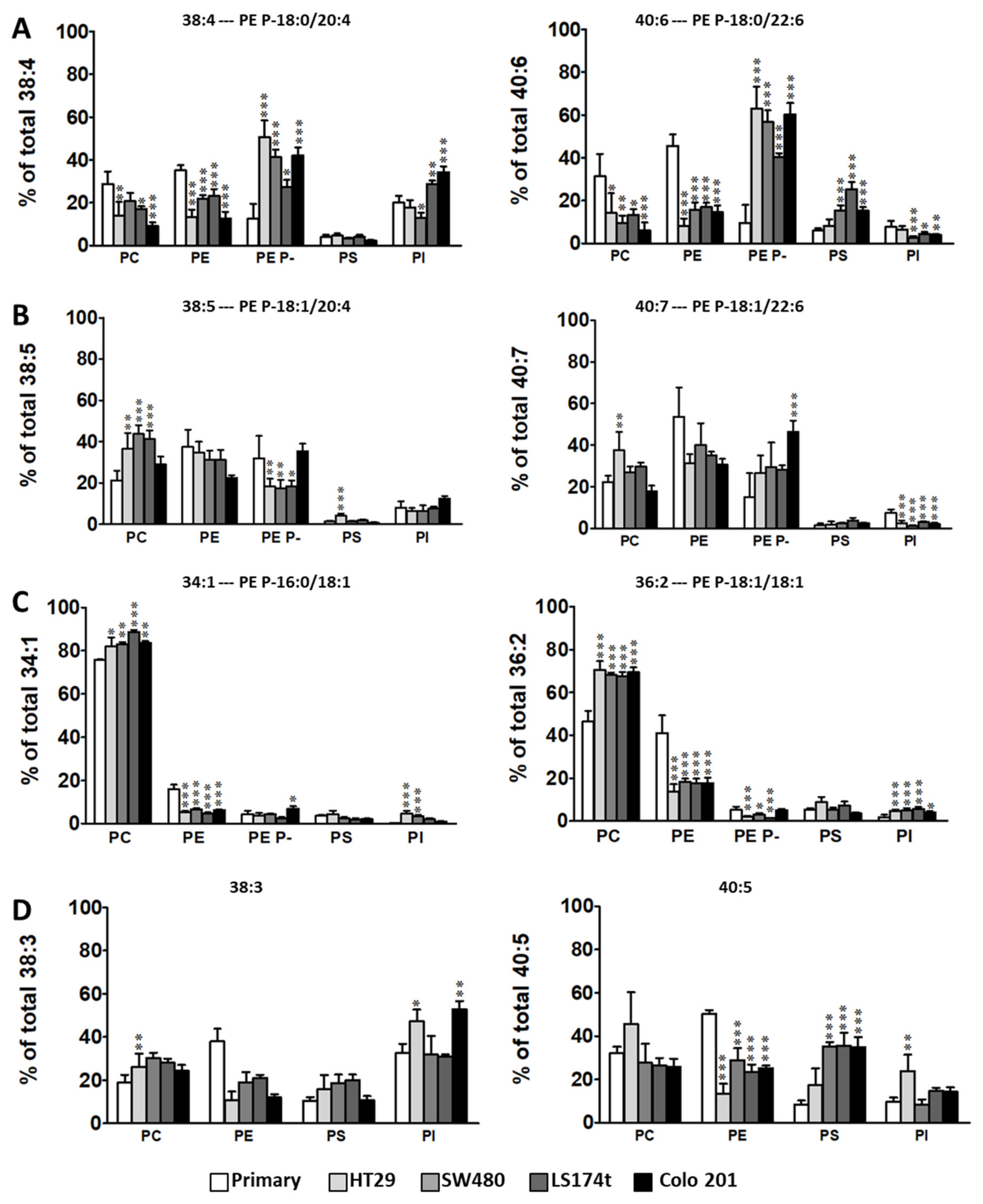
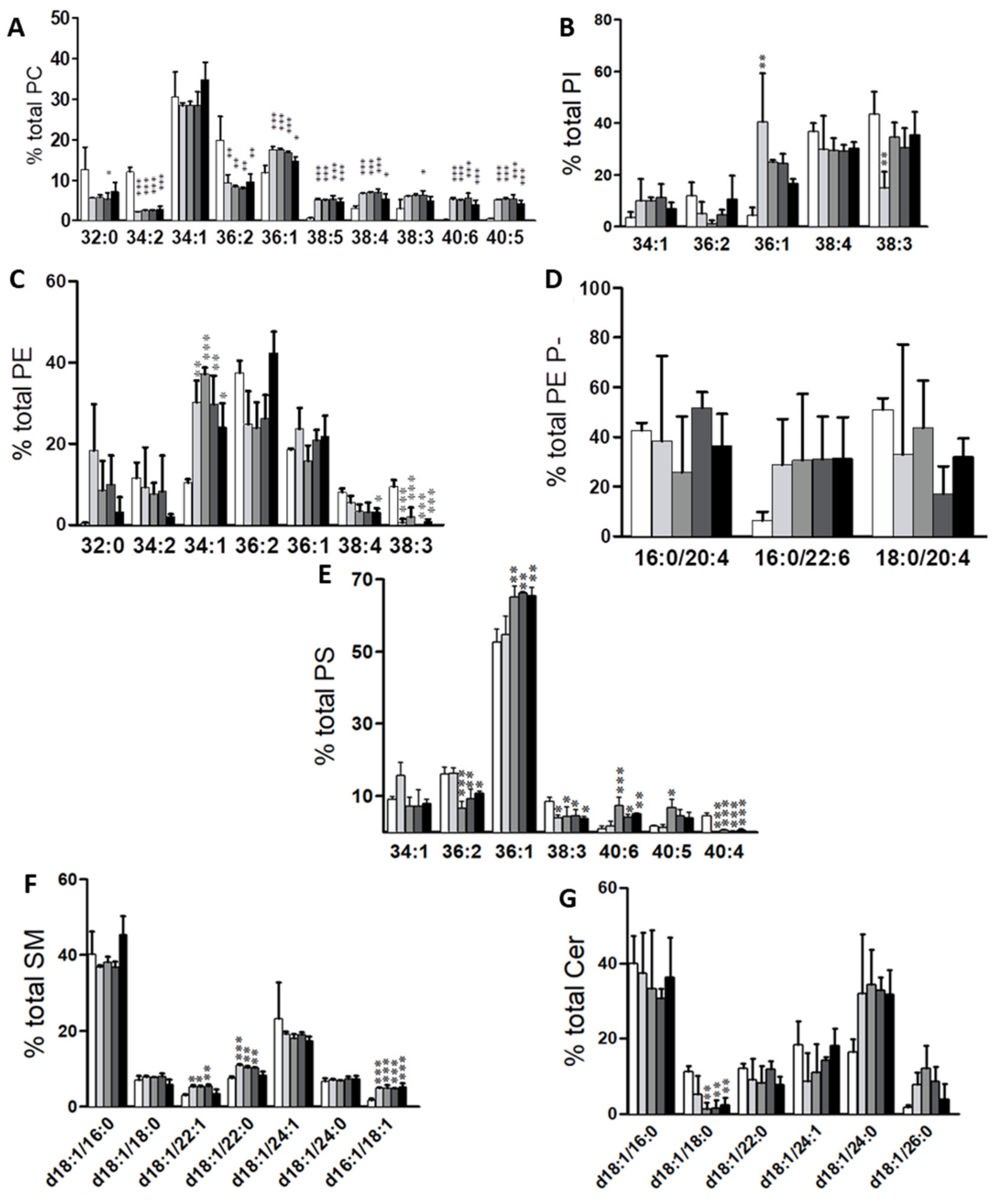
2.1. Identification of Potential Lipid Markers for Cellular Malignization in Commercial Cell Line
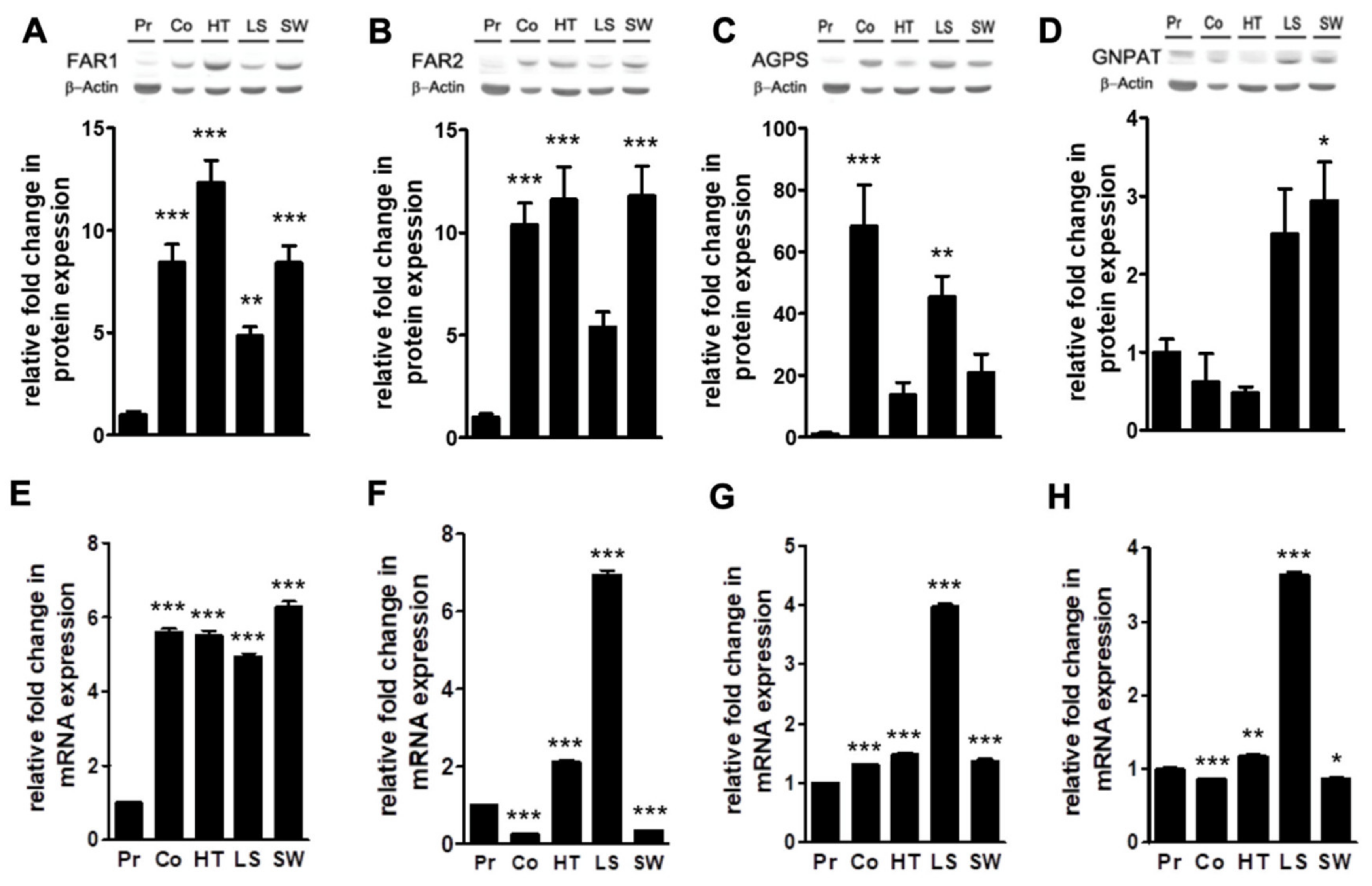
2.2. Levels of Plasmalogen are Coordinated with Enzyme Changes
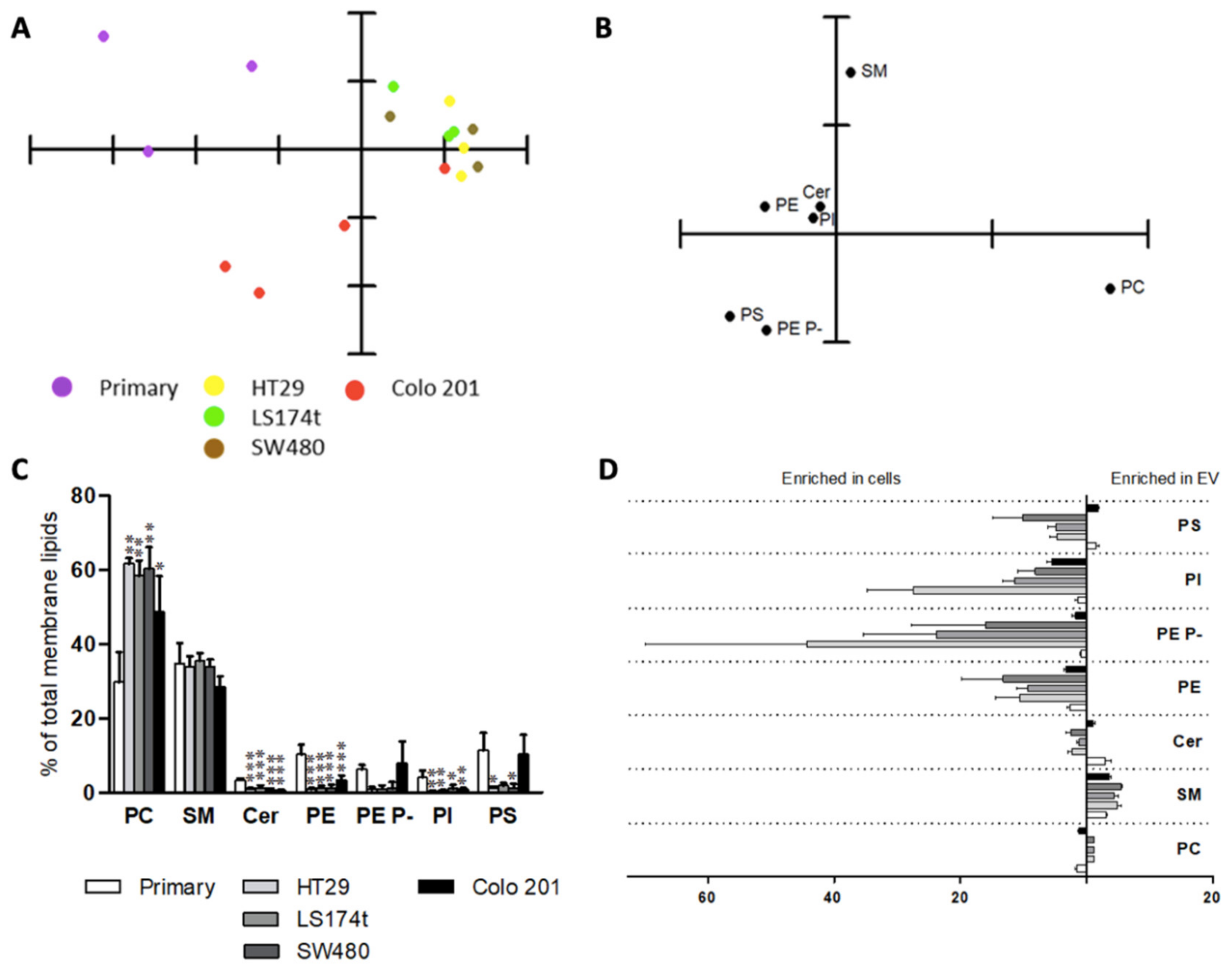
2.3. Extracellular Vesicle Lipids as Biomarkers of Malignization in a Cell Culture Model
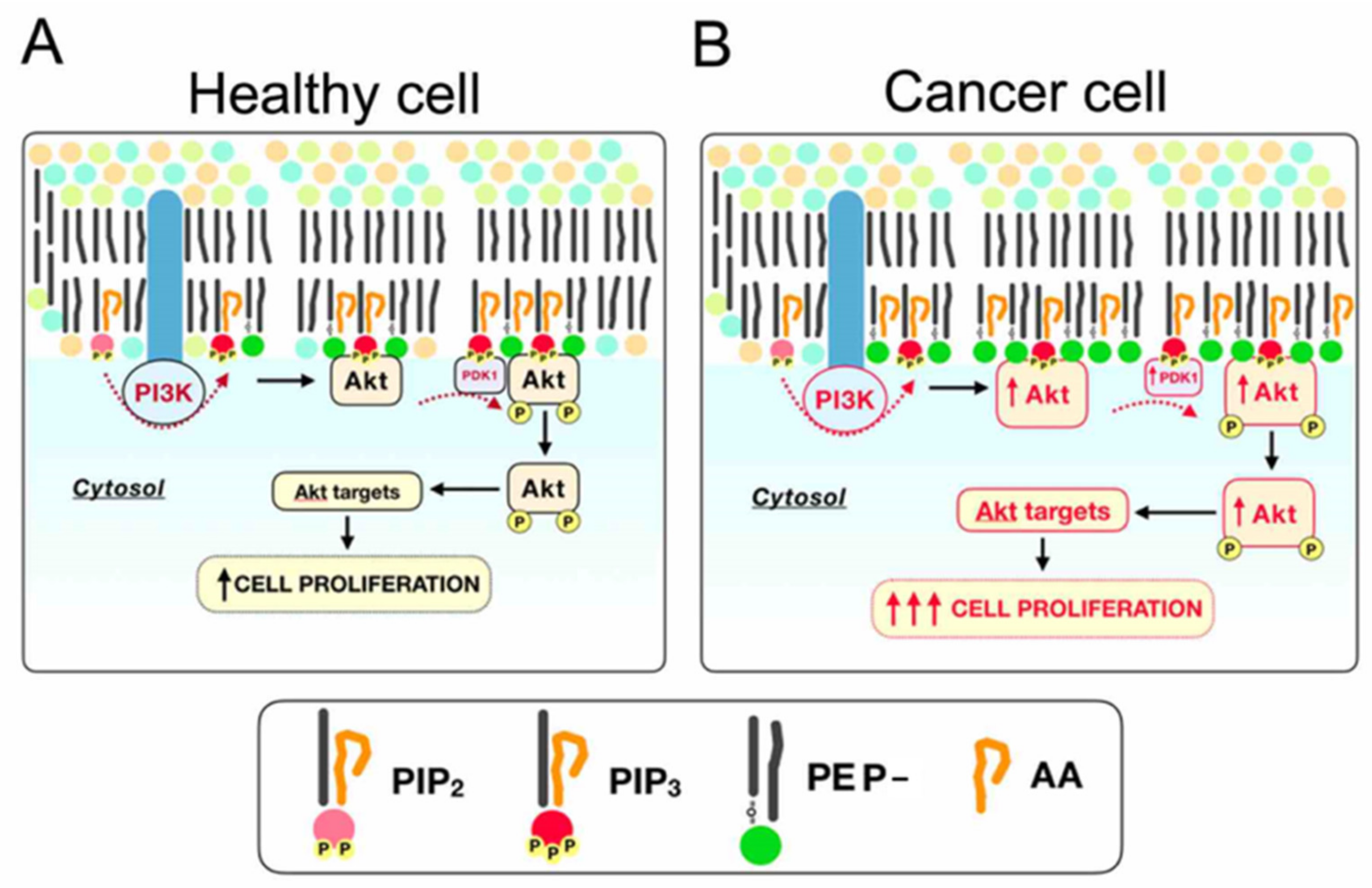
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture Protocol
4.2. Extracellular Vesicle Isolation
4.3. Lipid Extraction and LC-MS
4.3.1. Lipid Standard Solutions
4.3.2. Lipid Extractions
4.4. MS Analysis
4.5. Protein Expression Determined by Western Blot
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- Araghi, M.; Soerjomataram, I.; Bardot, A.; Ferlay, J.; Cabasag, C.J.; Morrison, D.S.; De, P.; Tervonen, H.; Walsh, P.M.; Bucher, O.; et al. Changes in colorectal cancer incidence in seven high-income countries: A population-based study. Lancet Gastroenterol. Hepatol. 2019, 4, 511–518. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Mapstone, M.; Cheema, A.K.; Fiandaca, M.S.; Zhong, X.; Mhyre, T.R.; Macarthur, L.H.; Hall, W.J.; Fisher, S.G.; Peterson, D.R.; Haley, J.M.; et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat. Med. 2014, 20, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Cui, X.; Zhang, N.; Li, M.; Bai, Y.; Han, X.; Shi, Y.; Liu, H. Comprehensive lipid profiling of plasma in patients with benign breast tumor and breast cancer reveals novel biomarkers. Anal. Bioanal. Chem. 2015, 407, 5065–5077. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Mao, J.; Ai, J.; Deng, Y.; Roth, M.R.; Pound, C.; Henegar, J.; Welti, R.; Bigler, S.A. Identification of Plasma Lipid Biomarkers for Prostate Cancer by Lipidomics and Bioinformatics. PLoS ONE 2012, 7, e48889. [Google Scholar] [CrossRef]
- Guijas, C.; Meana, C.; Astudillo, A.M.; Balboa, M.A.; Balsinde, J. Foamy Monocytes Are Enriched in cis-7-Hexadecenoic Fatty Acid (16:1n-9), a Possible Biomarker for Early Detection of Cardiovascular Disease. Cell Chem. Biol. 2016, 23, 689–699. [Google Scholar] [CrossRef]
- Bestard-Escalas, J.; Garate, J.; Maimó-Barceló, A.; Fernández, R.; Lopez, D.H.; Lage, S.; Reigada, R.; Khorrami, S.; Ginard, D.; Reyes, J.; et al. Lipid fingerprint image accurately conveys human colon cell pathophysiologic state: A solid candidate as biomarker. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1942–1950. [Google Scholar] [CrossRef]
- Wallner, S.; Orso, E.; Grandl, M.; Konovalova, T.; Liebisch, G.; Schmitz, G. Phosphatidylcholine and phosphatidylethanolamine plasmalogens in lipid loaded human macrophages. PLoS ONE 2018, 13, 1–21. [Google Scholar] [CrossRef]
- Hofmanová, J.; Slavík, J.; Ovesná, P.; Tylichová, Z.; Dušek, L.; Straková, N.; Vaculová, A.H.; Ciganek, M.; Kala, Z.; Jíra, M.; et al. Phospholipid profiling enables to discriminate tumor- And non-tumor-derived human colon epithelial cells: Phospholipidome similarities and differences in colon cancer cell lines and in patient-derived cell samples. PLoS ONE 2020, 15, e0228010. [Google Scholar] [CrossRef]
- Zemski Berry, K.A.; Gordon, W.C.; Murphy, R.C.; Bazan, N.G. Spatial organization of lipids in the human retina and optic nerve by MALDI imaging mass spectrometry. J. Lipid Res. 2014, 55, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Carriel, V.; Lage, S.; Garate, J.; Díez-García, J.; Ochoa, B.; Castro, B.; Alaminos, M.; Fernández, J.A. Deciphering the Lipid Architecture of the Rat Sciatic Nerve Using Imaging Mass Spectrometry. ACS Chem. Neurosci. 2016, 7, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Eberlin, L.S.; Norton, I.; Dill, A.L.; Golby, A.J.; Ligon, K.L.; Santagata, S.; Graham Cooks, R.; Agar, N.Y.R. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012, 72, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Maimó-Barceló, A.; Garate, J.; Bestard-Escalas, J.; Fernández, R.; Berthold, L.; Lopez, D.H.; Fernández, J.A.; Barceló-Coblijn, G. Confirmation of sub-cellular resolution using oversampling imaging mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 7935–7941. [Google Scholar] [CrossRef]
- Lopez, D.H.; Bestard-Escalas, J.; Garate, J.; Maimó-Barceló, A.; Fernández, R.; Reigada, R.; Khorrami, S.; Ginard, D.; Okazaki, T.; Fernández, J.A.; et al. Tissue-selective alteration of ethanolamine plasmalogen metabolism in dedifferentiated colon mucosa. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 928–938. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Subra, C.; Grand, D.; Laulagnier, K.; Stella, A.; Lambeau, G.; Paillasse, M.; De Medina, P.; Monsarrat, B.; Perret, B.; Silvente-Poirot, S.; et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010, 51, 2105–2120. [Google Scholar] [CrossRef]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.M.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.W.; Geuze, H.J.; Stoorvogel, W. Proteomic and biochemical analyses of human B cell-derived exosomes: Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M.; Schwille, P.; et al. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Laulagnier, K.; Grand, D.; Dujardin, A.; Hamdi, S.; Vincent-Schneider, H.; Lankar, D.; Salles, J.-P.; Bonnerot, C.; Perret, B.; Record, M. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 2004, 572, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Carayon, K.; Chaoui, K.; Ronzier, E.; Lazar, I.; Bertrand-Michel, J.; Roques, V.; Balor, S.; Terce, F.; Lopez, A.; Salomé, L.; et al. Proteolipidic Composition of Exosomes Changes during Reticulocyte Maturation. J. Biol. Chem. 2011, 286, 34426–34439. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, H. Role of LBPA and Alix in Multivesicular Liposome Formation and Endosome Organization. Science 2004, 303, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Poliakov, A.; Liu, C.; Liu, Y.; Deng, Z.; Wang, J.; Cheng, Z.; Shah, S.V.; Wang, G.-J.; Zhang, L.; et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer 2009, 124, 2621–2633. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2013, 1831, 1302–1309. [Google Scholar] [CrossRef]
- Lydic, T.A.; Townsend, S.; Adda, C.G.; Collins, C.; Mathivanan, S.; Reid, G.E. Rapid and comprehensive ‘shotgun’ lipidome profiling of colorectal cancer cell derived exosomes. Methods 2015, 87, 83–95. [Google Scholar] [CrossRef]
- Cansell, M.; Gouygou, J.-P.; Jozefonvicz, J.; Letourneur, D. Lipid composition of cultured endothelial cells in relation to their growth. Lipids 1997, 32, 39–44. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzás, E.I.; Lötvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2, 20677. [Google Scholar] [CrossRef]
- Martin, M.L.; Liebisch, G.; Lehneis, S.; Schmitz, G.; Alonso-Sande, M.; Bestard-Escalas, J.; Lopez, D.H.; García-Verdugo, J.M.; Soriano-Navarro, M.; Busquets, X.; et al. Sustained activation of sphingomyelin synthase by 2-hydroxyoleic acid induces sphingolipidosis in tumor cells. J. Lipid Res. 2013, 54, 1457–1465. [Google Scholar] [CrossRef]
- Lemonnier, L.A.; Dillehay, D.L.; Vespremi, M.J.; Abrams, J.; Brody, E.; Schmelz, E.M. Sphingomyelin in the suppression of colon tumors: Prevention versus intervention. Arch. Biochem. Biophys. 2003, 419, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Husband, D.; Imray, C.; Rowley, S.; Lawson, N. Arachidonic acid and docosahexaenoic acid are increased in human colorectal cancer. Gut 1991, 32, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Pienimaeki-Roemer, A.; Kuhlmann, K.; Böttcher, A.; Konovalova, T.; Black, A.; Orsó, E.; Liebisch, G.; Ahrens, M.; Eisenacher, M.; Meyer, H.E.; et al. Lipidomic and proteomic characterization of platelet extracellular vesicle subfractions from senescent platelets. Transfusion 2015, 55, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Beskow, L.M. Lessons from HeLa Cells: The Ethics and Policy of Biospecimens. Annu. Rev. Genomics Hum. Genet. 2016, 17, 395–417. [Google Scholar] [CrossRef]
- Masters, J.R. HeLa cells 50 years on: The good, the bad and the ugly. Nat. Rev. Cancer 2002, 2, 315–319. [Google Scholar] [CrossRef]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef]
- Planutis, A.K.; Holcombe, R.F.; Planoutene, M.V.; Planoutis, K.S. SW480 colorectal cancer cells that naturally express Lgr5 are more sensitive to the most common chemotherapeutic agents than Lgr5-negative SW480 cells. Anticancer Drugs 2015, 26, 942–947. [Google Scholar] [CrossRef]
- Zhu, L.; Ma, W.; Zhang, M.; Lee, M.M.-L.; Wong, W.-Y.; Chan, B.D.; Yang, Q.; Wong, W.-T.; Tai, W.C.-S.; Lee, C.-S. Scalable synthesis enabling multilevel bio-evaluations of natural products for discovery of lead compounds. Nat. Commun. 2018, 9, 1283. [Google Scholar] [CrossRef]
- Klijn, C.; Durinck, S.; Stawiski, E.W.; Haverty, P.M.; Jiang, Z.; Liu, H.; Degenhardt, J.; Mayba, O.; Gnad, F.; Liu, J.; et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015, 33, 306–312. [Google Scholar] [CrossRef]
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic analysis. Anal. Bioanal. Chem. 2020, 90, 4249–4257. [Google Scholar] [CrossRef] [PubMed]
- Barcelo-Coblijn, G.; Martin, M.L.; de Almeida, R.F.M.; Noguera-Salva, M.A.; Marcilla-Etxenike, A.; Guardiola-Serrano, F.; Luth, A.; Kleuser, B.; Halver, J.E.; Escriba, P.V. Sphingomyelin and sphingomyelin synthase (SMS) in the malignant transformation of glioma cells and in 2-hydroxyoleic acid therapy. Proc. Natl. Acad. Sci. USA 2011, 108, 19569–19574. [Google Scholar] [CrossRef] [PubMed]
- Dória, M.L.; Cotrim, C.Z.; Simões, C.; Macedo, B.; Domingues, P.; Domingues, M.R.; Helguero, L.A. Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J. Cell. Physiol. 2013, 228, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Marien, E.; Meister, M.; Muley, T.; Fieuws, S.; Bordel, S.; Derua, R.; Spraggins, J.; Van de Plas, R.; Dehairs, J.; Wouters, J.; et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int. J. Cancer 2015, 137, 1539–1548. [Google Scholar] [CrossRef]
- Benjamin, D.I.; Cozzo, A.; Ji, X.; Roberts, L.S.; Louie, S.M.; Mulvihill, M.M.; Luo, K.; Nomura, D.K. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc. Natl. Acad. Sci. USA 2013, 110, 14912–14917. [Google Scholar] [CrossRef]
- Smith, R.E.; Lespi, P.; Di Luca, M.; Bustos, C.; Marra, F.A.; de Alaniz, M.J.T.; Marra, C.A. A Reliable Biomarker Derived from Plasmalogens to Evaluate Malignancy and Metastatic Capacity of Human Cancers. Lipids 2008, 43, 79–89. [Google Scholar] [CrossRef]
- Fernández, R.; Garate, J.; Lage, S.; Terés, S.; Higuera, M.; Bestard-Escalas, J.; López, D.H.; Guardiola-Serrano, F.; Escribá, P.V.; Barceló-Coblijn, G.; et al. Identification of Biomarkers of Necrosis in Xenografts Using Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2016, 27, 244–254. [Google Scholar] [CrossRef]
- Olivera, A.; Buckley, N.E.; Spiegel, S. Sphingomyelinase and cell-permeable ceramide analogs stimulate cellular proliferation in quiescent Swiss 3T3 fibroblasts. J. Biol. Chem. 1992, 267, 26121–26127. [Google Scholar]
- Abe, M.; Makino, A.; Hullin-Matsuda, F.; Kamijo, K.; Ohno-Iwashita, Y.; Hanada, K.; Mizuno, H.; Miyawaki, A.; Kobayashi, T. A Role for Sphingomyelin-Rich Lipid Domains in the Accumulation of Phosphatidylinositol-4,5-Bisphosphate to the Cleavage Furrow during Cytokinesis. Mol. Cell. Biol. 2012, 32, 1396–1407. [Google Scholar] [CrossRef]
- Chughtai, K.; Jiang, L.; Greenwood, T.R.; Glunde, K.; Heeren, R.M.A. Mass spectrometry images acylcarnitines, phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor models. J. Lipid Res. 2013, 54, 333–344. [Google Scholar] [CrossRef]
- Patterson, N.H.; Alabdulkarim, B.; Lazaris, A.; Thomas, A.; Marcinkiewicz, M.M.; Gao, Z.H.; Vermeulen, P.B.; Chaurand, P.; Metrakos, P. Assessment of pathological response to therapy using lipid mass spectrometry imaging. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Biermann, J.; Just, W.W.; Wanders, R.J.A.; van den Bosch, H. Alkyl-dihydroxyacetone phosphate synthase and dihydroxyacetone phosphate acyltransferase form a protein complex in peroxisomes. Eur. J. Biochem. 1999, 261, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Abe, Y.; Ali, F.; Youssef, M.; Honsho, M.; Fujiki, Y.; Katafuchi, T. Reduction of Ether-Type Glycerophospholipids, Plasmalogens, by NF-κB Signal Leading to Microglial Activation. J. Neurosci. 2017, 37, 4074–4092. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.E.; Hajra, A.K. Mechanism and specificity of formation of long chain alcohols by developing rat brain. J. Biol. Chem. 1981, 256, 9542–9550. [Google Scholar]
- Cheng, J.B.; Russell, D.W. Mammalian Wax Biosynthesis. J. Biol. Chem. 2004, 279, 37789–37797. [Google Scholar] [CrossRef]
- Rizzo, W.B. Fatty aldehyde and fatty alcohol metabolism: Review and importance for epidermal structure and function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1841, 377–389. [Google Scholar] [CrossRef]
- Honsho, M.; Asaoku, S.; Fujiki, Y. Posttranslational Regulation of Fatty Acyl-CoA Reductase 1, Far1, Controls Ether Glycerophospholipid Synthesis. J. Biol. Chem. 2010, 285, 8537–8542. [Google Scholar] [CrossRef]
- Honsho, M.; Fujiki, Y. Plasmalogen homeostasis—Regulation of plasmalogen biosynthesis and its physiological consequence in mammals. FEBS Lett. 2017, 591, 2720–2729. [Google Scholar] [CrossRef]
- Iwamoto, K.; Hayakawa, T.; Murate, M.; Makino, A.; Ito, K.; Fujisawa, T.; Kobayashi, T. Curvature-dependent recognition of ethanolamine phospholipids by duramycin and cinnamycin. Biophys. J. 2007, 93, 1608–1619. [Google Scholar] [CrossRef]
- Marconescu, A.; Thorpe, P.E. Coincident exposure of phosphatidylethanolamine and anionic phospholipids on the surface of irradiated cells. Biochim. Biophys. Acta Biomembr. 2008, 1778, 2217–2224. [Google Scholar] [CrossRef]
- Stafford, J.H.; Thorpe, P.E. Increased Exposure of Phosphatidylethanolamine on the Surface of Tumor Vascular Endothelium. Neoplasia 2011, 13, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Emoto, K.; Toyama-Sorimachi, N.; Karasuyama, H.; Inoue, K.; Umeda, M. Exposure of Phosphatidylethanolamine on the Surface of Apoptotic Cells. Exp. Cell Res. 1997, 232, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Honsho, M.; Abe, Y.; Fujiki, Y. Plasmalogen biosynthesis is spatiotemporally regulated by sensing plasmalogens in the inner leaflet of plasma membranes. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gaposchkin, D.P.; Zoeller, R.A. Plasmalogen status influences docosahexaenoic acid levels in a macrophage cell line. Insights using ether lipid-deficient variants. J. Lipid Res. 1999, 40, 495–503. [Google Scholar] [PubMed]
- D’Souza, K.; Epand, R.M. Enrichment of phosphatidylinositols with specific acyl chains. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.I.S.; Sprecher, H. Distribution of arachidonic acid in choline- and ethanolamine-containing phosphoglycerides in subfractionated human neutrophils. J. Biol. Chem. 1989, 264, 17718–17726. [Google Scholar] [PubMed]
- Hossain, M.S.; Ifuku, M.; Take, S.; Kawamura, J.; Miake, K.; Katafuchi, T. Plasmalogens Rescue Neuronal Cell Death through an Activation of AKT and ERK Survival Signaling. PLoS ONE 2013, 8, e83508. [Google Scholar] [CrossRef]
- Dsouza, K.; Epand, R.M. The phosphatidylinositol synthase-catalyzed formation of phosphatidylinositol does not exhibit acyl chain specificity. Biochemistry 2015, 54, 1151–1153. [Google Scholar] [CrossRef]
- Shulga, Y.V.; Anderson, R.A.; Topham, M.K.; Epand, R.M. Phosphatidylinositol-4-phosphate 5-Kinase Isoforms Exhibit Acyl Chain Selectivity for Both Substrate and Lipid Activator. J. Biol. Chem. 2012, 287, 35953–35963. [Google Scholar] [CrossRef]
- Traynor-Kaplan, A.; Kruse, M.; Dickson, E.J.; Dai, G.; Vivas, O.; Yu, H.; Whittington, D.; Hille, B. Fatty-acyl chain profiles of cellular phosphoinositides. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 513–522. [Google Scholar] [CrossRef]
- Da Silva, T.F.; Eira, J.; Lopes, A.T.; Malheiro, A.R.; Sousa, V.; Luoma, A.; Avila, R.L.; Wanders, R.J.A.; Just, W.W.; Kirschner, D.A.; et al. Peripheral nervous system plasmalogens regulate Schwann cell differentiation and myelination. J. Clin. Investig. 2014, 124, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Gulhati, P.; Rampy, B.A.; Han, Y.; Rychahou, P.G.; Doan, H.Q.; Weiss, H.L.; Evers, B.M. Novel Expression Patterns of PI3K/Akt/mTOR Signaling Pathway Components in Colorectal Cancer. J. Am. Coll. Surg. 2010, 210, 767. [Google Scholar] [CrossRef] [PubMed]
- Ogiso, H.; Taniguchi, M.; Araya, S.; Aoki, S.; Wardhani, L.; Yamashita, Y.; Ueda, Y.; Okazaki, T. Comparative Analysis of Biological Sphingolipids with Glycerophospholipids and Diacylglycerol by LC-MS/MS. Metabolites 2014, 4, 98–114. [Google Scholar] [CrossRef]
- Atilla-Gokcumen, G.E.; Muro, E.; Relat-Goberna, J.; Sasse, S.; Bedigian, A.; Coughlin, M.L.; Garcia-Manyes, S.; Eggert, U.S. Dividing Cells Regulate Their Lipid Composition and Localization. Cell 2014, 156, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Emoto, K.; Kobayashi, T.; Yamaji, A.; Aizawa, H.; Yahara, I.; Inoue, K.; Umeda, M. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc. Natl. Acad. Sci. USA 1996, 93, 12867–12872. [Google Scholar] [CrossRef] [PubMed]
- Valli, A.; Rodriguez, M.; Moutsianas, L.; Fischer, R.; Fedele, V.; Huang, H.L.; Van Stiphout, R.; Jones, D.; Mccarthy, M.; Vinaxia, M.; et al. Hypoxia induces a lipogenic cancer cell phenotype via HIF1α-dependent and -independent pathways. Oncotarget 2015, 6, 1920–1941. [Google Scholar] [CrossRef]
- Jones, D.T.; Valli, A.; Haider, S.; Zhang, Q.; Smethurst, E.A.; Schug, Z.T.; Peck, B.; Aboagye, E.O.; Critchlow, S.E.; Schulze, A.; et al. 3D Growth of Cancer Cells Elicits Sensitivity to Kinase Inhibitors but Not Lipid Metabolism Modifiers. Mol. Cancer Ther. 2019, 18, 376–388. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bestard-Escalas, J.; Maimó-Barceló, A.; Lopez, D.H.; Reigada, R.; Guardiola-Serrano, F.; Ramos-Vivas, J.; Hornemann, T.; Okazaki, T.; Barceló-Coblijn, G. Common and Differential Traits of the Membrane Lipidome of Colon Cancer Cell Lines and Their Secreted Vesicles: Impact on Studies Using Cell Lines. Cancers 2020, 12, 1293. https://doi.org/10.3390/cancers12051293
Bestard-Escalas J, Maimó-Barceló A, Lopez DH, Reigada R, Guardiola-Serrano F, Ramos-Vivas J, Hornemann T, Okazaki T, Barceló-Coblijn G. Common and Differential Traits of the Membrane Lipidome of Colon Cancer Cell Lines and Their Secreted Vesicles: Impact on Studies Using Cell Lines. Cancers. 2020; 12(5):1293. https://doi.org/10.3390/cancers12051293
Chicago/Turabian StyleBestard-Escalas, Joan, Albert Maimó-Barceló, Daniel H. Lopez, Rebeca Reigada, Francisca Guardiola-Serrano, José Ramos-Vivas, Thorsten Hornemann, Toshiro Okazaki, and Gwendolyn Barceló-Coblijn. 2020. "Common and Differential Traits of the Membrane Lipidome of Colon Cancer Cell Lines and Their Secreted Vesicles: Impact on Studies Using Cell Lines" Cancers 12, no. 5: 1293. https://doi.org/10.3390/cancers12051293
APA StyleBestard-Escalas, J., Maimó-Barceló, A., Lopez, D. H., Reigada, R., Guardiola-Serrano, F., Ramos-Vivas, J., Hornemann, T., Okazaki, T., & Barceló-Coblijn, G. (2020). Common and Differential Traits of the Membrane Lipidome of Colon Cancer Cell Lines and Their Secreted Vesicles: Impact on Studies Using Cell Lines. Cancers, 12(5), 1293. https://doi.org/10.3390/cancers12051293