Whole Genome Analysis of Ovarian Granulosa Cell Tumors Reveals Tumor Heterogeneity and a High-Grade TP53-Specific Subgroup
Abstract
:1. Introduction
2. Results
2.1. Description of WGS Cohort
2.2. Common Copy Number Alterations in Chromosome 12, 14 and 22
2.3. Mutational Signatures in AGCTs Are Related to Ageing and Platinum Treatment
2.4. Variants in Known Cancer Genes Were Detected in FOXL2, TERT, KMT2D, PIK3CA and TP53
2.5. Subgroup of Patients with High-Grade AGCT Characterized by TP53 Mutation
2.6. FOXL2-Wildtype AGCTs Resemble FOXL2-Mutant AGCTs
2.7. Investigation of Shared Variants between AGCT Patients Confirms a Limited Number of Recurrent Mutations
2.7.1. Overlapping Single Nucleotide Variants
2.7.2. Overlapping Gene Loss and Structural Variants
2.8. Novel Candidate Gene Analysis
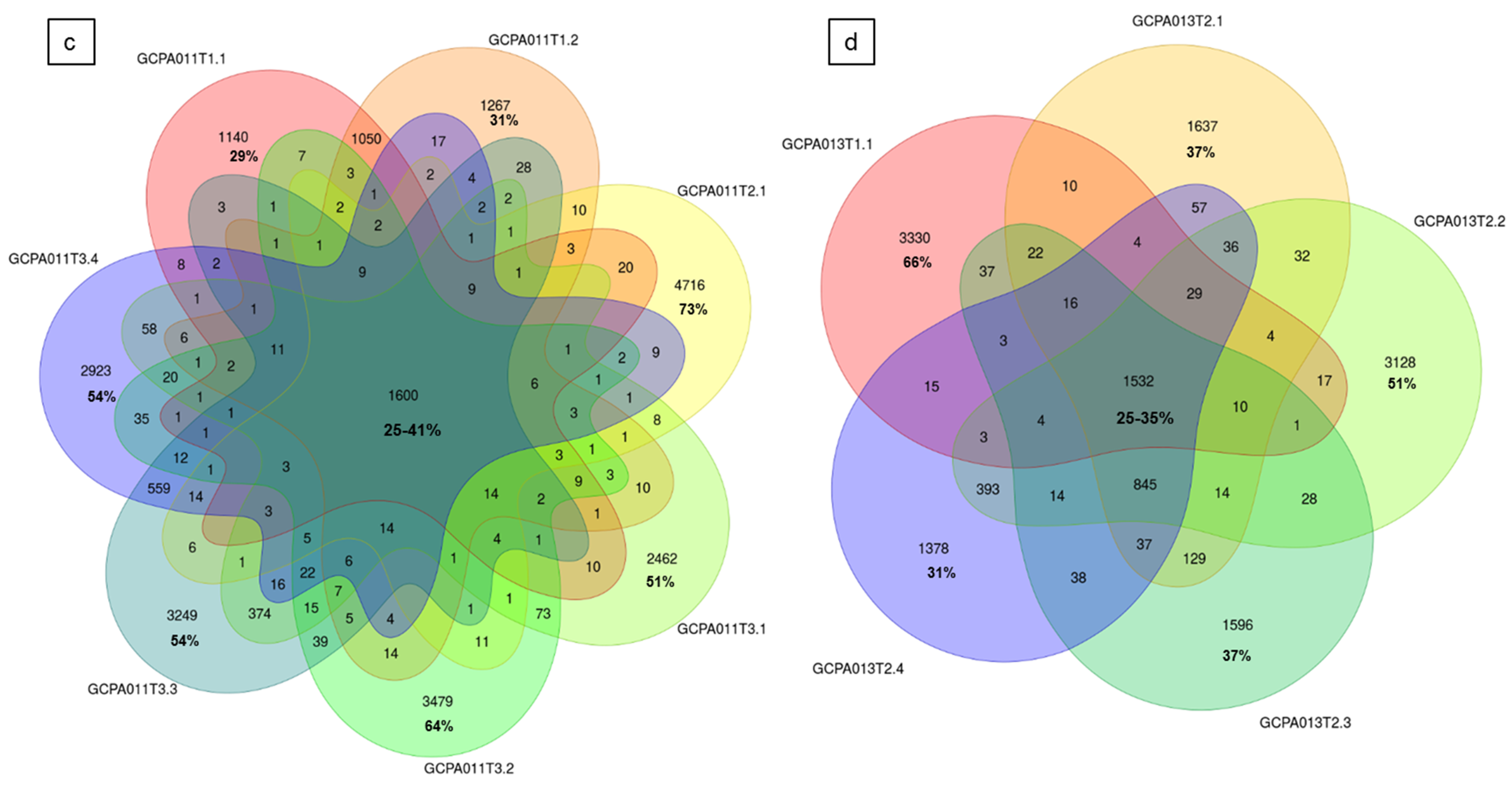
2.9. Intra-Patient Comparisons Reveal Tumor Heterogeneity
3. Discussion
4. Materials and Methods
4.1. Patient Cohort and Study Inclusion Criteria
4.2. Tissue Acquisition
4.3. DNA Isolation, Quantification and Qualification
4.4. Whole-Genome Sequencing and Variant Calling
4.5. WGS Data Analysis
4.6. CNV Analysis
4.7. Exonic Variant Analysis
4.8. Whole Genome Variant Analysis
4.9. Mutational Signature Analysis
4.10. Tumor Heterogeneity Assessment
4.11. Homologous Recombination
4.12. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bryk, S.; Pukkala, E.; Martinsen, J.-I.; Unkila-Kallio, L.; Tryggvadottir, L.; Sparen, P.; Kjaerheim, K.; Weiderpass, E.; Riska, A. Incidence and occupational variation of ovarian granulosa cell tumours in Finland, Iceland, Norway and Sweden during 1953–2012: A longitudinal cohort study. BJOG 2017, 124, 143–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meurs, H.S.; Bleeker, M.C.G.; Van der Velden, J.; Overbeek, L.I.H.; Kenter, G.G.; Buist, M.R. The incidence of endometrial hyperplasia and cancer in 1031 patients with a granulosa cell tumor of the ovary: Long-term follow-up in a population-based cohort study. Int. J. Gynecol. Cancer 2013, 23, 1417–1422. [Google Scholar] [CrossRef] [PubMed]
- Ohel, G.; Kaneti, H.; Schenker, J.G. Granulosa cell tumors in Israel: A study of 172 cases. Gynecol. Oncol. 1983, 15, 278–286. [Google Scholar] [CrossRef]
- Schumer, S.T.; Cannistra, S.A. Granulosa cell tumor of the ovary. J. Clin. Oncol. 2003, 21, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, S.; Fuller, P.J. Management of granulosa cell tumour of the ovary. Curr. Opin. Oncol. 2008, 20, 560–564. [Google Scholar] [CrossRef] [PubMed]
- McConechy, M.K.; Farkkila, A.; Horlings, H.M.; Talhouk, A.; Unkila-Kallio, L.; Van Meurs, H.S.; Winnie, Y.; Rozenberg, N.; Andersson, N.; Zaby, K.; et al. Molecularly defined adult granulosa cell tumor of the ovary: The clinical phenotype. J. Natl. Cancer Inst. 2016, 108, djw134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meurs, H.S.; Schuit, E.; Horlings, H.M.; Van der Velden, J.; Van Driel, W.J.; Mol, B.W.J.; Kenter, G.G.; Buist, M.R. Development and internal validation of a prognostic model to predict recurrence free survival in patients with adult granulosa cell tumors of the ovary. Gynecol. Oncol. 2014, 134, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Farkkila, A.; Haltia, U.-M.; Tapper, J.; McConechy, M.K.; Huntsman, D.G.; Heikinheimo, M. Pathogenesis and treatment of adult-type granulosa cell tumor of the ovary. Ann. Med. 2017, 49, 435–447. [Google Scholar] [CrossRef]
- Shah, S.P.; Kobel, M.; Senz, J.; Morin, R.D.; Clarke, B.A.; Wiegand, K.C.; Leung, G.; Zayed, A.; Mehl, E.; Kalloger, S.E.; et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009, 360, 2719–2729. [Google Scholar] [CrossRef]
- Schmidt, D.; Ovitt, C.E.; Anlag, K.; Fehsenfeld, S.; Gredsted, L.; Treier, A.-C.; Treier, M. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 2004, 131, 933–942. [Google Scholar] [CrossRef] [Green Version]
- Georges, A.; Auguste, A.; Bessiere, L.; Vanet, A.; Todeschini, A.-L.; Veitia, R.A. FOXL2: A central transcription factor of the ovary. J. Mol. Endocrinol. 2014, 52, R17–R33. [Google Scholar] [CrossRef] [PubMed]
- Caburet, S.; Anttonen, M.; Todeschini, A.L.; Unkila-Kallio, L.; Mestivier, D.; Butzow, R.; Veitia, R.A. Combined comparative genomic hybridization and transcriptomic analyses of ovarian granulosa cell tumors point to novel candidate driver genes. BMC Cancer 2015, 15. [Google Scholar] [CrossRef]
- Hillman, R.T.; Celestino, J.; Terranova, C.; Beird, H.C.; Gumbs, C.; Little, L.; Nguyen, T.; Thornton, R.; Tippen, S.; Zhang, J.; et al. KMT2D/MLL2 inactivation is associated with recurrence in adult-type granulosa cell tumors of the ovary. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Alexiadis, M.; Rowley, S.M.; Chu, S.; Leung, D.T.H.; Stewart, C.J.R.; Amarasinghe, K.C.; Campbell, I.G.; Fuller, P.J. Mutational landscape of ovarian adult granulosa cell tumors from whole exome and targeted TERT promoter sequencing. Mol. Cancer Res. 2019, 17, 177–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayr, D.; Kaltz-Wittmer, C.; Arbogast, S.; Amann, G.; Aust, D.E.; Diebold, J. Characteristic pattern of genetic aberrations in ovarian granulosa cell tumors. Mod. Pathol. 2002, 15, 951–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Berghe, I.; Dal Cin, P.; De Groef, K.; Michielssen, P.; Van den Berghe, H. Monosomy 22 and trisomy 14 may be early events in the tumorigenesis of adult granulosa cell tumor. Cancer Genet. Cytogenet. 1999, 112, 46–48. [Google Scholar] [CrossRef]
- Lin, Y.-S.; Eng, H.-L.; Jan, Y.-J.; Lee, H.-S.; Ho, W.L.; Liou, C.-P.; Lee, W.-Y.; Tzeng, C.-C. Molecular cytogenetics of ovarian granulosa cell tumors by comparative genomic hybridization. Gynecol. Oncol. 2005, 97, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Kraus, F.; Dremaux, J.; Altakfi, W.; Goux, M.; Pontois, L. FOXL2 homozygous genotype and chromosome instability are associated with recurrence in adult granulosa cell tumors of the ovary. Oncotarget 2020, 11, 419–428. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.K.; Bashashati, A.; Anglesio, M.S.; Cochrane, D.R.; Grewal, D.S.; Ha, G.; McPherson, A.; Horlings, H.M.; Senz, J.; Prentice, L.M.; et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat. Genet. 2017, 49, 856–864. [Google Scholar] [CrossRef]
- Pilsworth, J.A.; Cochrane, D.R.; Xia, Z.; Aubert, G.; Färkkilä, A.E.M.; Horlings, H.M.; Yanagida, S.; Yang, W.; Lim, J.L.P.; Wang, Y.K.; et al. TERT promoter mutation in adult granulosa cell tumor of the ovary. Mod. Pathol. 2018, 31, 1107–1115. [Google Scholar] [CrossRef]
- Teer, J.K.; Yoder, S.; Gjyshi, A.; Nicosia, S.V.; Zhang, C.; Monteiro, A.N.A. Mutational heterogeneity in non-serous ovarian cancers. Sci. Rep. 2017, 7, 9728. [Google Scholar] [CrossRef] [PubMed]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23. [Google Scholar] [CrossRef]
- Kopper, O.; De Witte, C.J.; Lohmussaar, K.; Valle-Inclan, J.E.; Hami, N.; Kester, L.; Balgobind, A.V.; Korving, J.; Proost, N.; Begthel, H.; et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019, 25, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Pich, O.; Muiños, F.; Lolkema, M.P.; Steeghs, N.; Gonzalez-perez, A. The mutational footprints of cancer therapies. Nat. Genet. 2019, 51. [Google Scholar] [CrossRef] [PubMed]
- Taruscio, D.; Carcangiu, M.L.; Ward, D.C. Detection of trisomy 12 on ovarian sex cord stromal tumors by fluorescence in situ hybridization. Diagn. Mol. Pathol. 1993, 2, 94–98. [Google Scholar] [CrossRef]
- Speleman, F.; Dermaut, B.; De Potter, C.R.; Van Gele, M.; Van Roy, N.; De Paepe, A.; Laureys, G. Monosomy 22 in a mixed germ cell-sex cord-stromal tumor of the ovary. Genes. Chromosomes Cancer 1997, 19, 192–194. [Google Scholar] [CrossRef]
- Dal Cin, P.; Qi, H.; Pauwels, P.; Backx, C.; Van den Berghe, H. Monosomy 22 in a fibrothecoma. Cancer Genet. Cytogenet. 1997, 99, 129–131. [Google Scholar] [CrossRef]
- Manegold, E.; Tietze, L.; Gunther, K.; Fleischer, A.; Amo-Takyi, B.K.; Schroder, W.; Handt, S. Trisomy 8 as sole karyotypic aberration in an ovarian metastasizing Sertoli-Leydig cell tumor. Hum. Pathol. 2001, 32, 559–562. [Google Scholar] [CrossRef]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 2018, 18, 696–705. [Google Scholar] [CrossRef]
- Da Cruz Paula, A.; da Silva, E.M.; Segura, S.E.; Pareja, F.; Bi, R.; Selenica, P.; Kim, S.H.; Ferrando, L.; Vahdatinia, M.; Soslow, R.A.; et al. Genomic profiling of primary and recurrent adult granulosa cell tumors of the ovary. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 2020. [Google Scholar] [CrossRef]
- Fashedemi, Y.; Coutts, M.; Wise, O.; Bonhomme, B.; Baker, G.; Kelly, P.J.; Soubeyran, I.; Catherwood, M.A.; Croce, S.; Glenn McCluggage, W. Adult granulosa cell tumor with high-grade transformation: Report of a series with FOXL2 mutation analysis. Am. J. Surg. Pathol. 2019, 43, 1229–1238. [Google Scholar] [CrossRef]
- Watanabe, K.; Abiko, K.; Minamiguchi, S.; Maeda, H.; Murakami, R.; Kitamura, S.; Horie, A.; Mandai, M. Aggressive adult granulosa cell tumor of the ovary without a FOXL2 mutation: A case report. J. Obstet. Gynaecol. Res. 2019, 45, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Leary, A.; Gatalica, Z. Comprehensive molecular profiling of adult ovarian granulosa cell tumors (GCT) identifies candidate actionable targets. In International Journal of Gynecological Cancer; BMJ Publishing Group: London, UK, 2019. [Google Scholar]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- AACR Project GENIE Consortium. AACR Project GENIE: Powering precision medicine through an international consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heravi-Moussavi, A.; Anglesio, M.S.; Cheng, S.-W.G.; Senz, J.; Yang, W.; Prentice, L.; Fejes, A.P.; Chow, C.; Tone, A.; Kalloger, S.E.; et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N. Engl. J. Med. 2012, 366, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Vedanayagam, J.; Chatila, W.K.; Aksoy, B.A.; Majumdar, S.; Skanderup, A.J.; Demir, E.; Schultz, N.; Sander, C.; Lai, E.C. Cancer-associated mutations in DICER1 RNase IIIa and IIIb domains exert similar effects on miRNA biogenesis. Nat. Commun. 2019, 10, 3682. [Google Scholar] [CrossRef] [PubMed]
- Cluzet, V.; Devillers, M.M.; Petit, F.; Chauvin, S.; François, C.M.; Giton, F.; Genestie, C.; Joëlle, C.; Guigon, C.C.J. Aberrant granulosa cell-fate related to inactivated p53/Rb signaling contributes to granulosa cell tumors and to FOXL2 downregulation in the mouse ovary. Oncogene 2020, 39, 1875–1890. [Google Scholar] [CrossRef] [PubMed]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020, 578. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Lupski, J.R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 2015, 24, R102–R110. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-Y.; Ebbert, K.; Cordeiro, M.H.; Romero, M.M.; Whelan, K.A.; Suarez, A.A.; Woodruff, T.K.; Kurita, T. Constitutive activation of PI3K in oocyte induces ovarian granulosa cell tumors. Cancer Res. 2016, 76, 3851–3861. [Google Scholar] [CrossRef] [Green Version]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Priestley, P.; Baber, J.; Lolkema, M.P.; Steeghs, N.; De Bruijn, E.; Shale, C.; Duyvesteyn, K.; Haidari, S.; Van Hoeck, A.; Onstenk, W.; et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 2019, 575, 210–216. [Google Scholar] [CrossRef] [Green Version]
- Joshi, K.; Robert de Massy, M.; Ismail, M.; Reading, J.L.; Uddin, I.; Woolston, A.; Hatipoglu, E.; Oakes, T.; Rosenthal, R.; Peacock, T.; et al. Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer. Nat. Med. 2019, 25, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Staaf, J.; Glodzik, D.; Bosch, A.; Vallon-Christersson, J.; Reutersward, C.; Hakkinen, J.; Degasperi, A.; Amarante, T.D.; Saal, L.H.; Hegardt, C.; et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat. Med. 2019, 25, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.J.; Getz, G.; Stuart, J.M.; Korbel, J.O.; Stein, L.D. Net—ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Pan-cancer analysis of whole genomes. bioRxiv 2017, 3, 162784. [Google Scholar] [CrossRef] [Green Version]
- Vinagre, J.; Almeida, A.; Populo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Uribe, P.; Adrianzen-Ruesta, M.P.; Deng, Z.; Echevarria-Vargas, I.; Mender, I.; Saheb, S.; Liu, Q.; Altieri, D.C.; Murphy, M.E.; Shay, J.W.; et al. Exploiting TERT dependency as a therapeutic strategy for NRAS-mutant melanoma. Oncogene 2018, 37, 4058–4072. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Saunders, C.T.; Wong, W.S.W.; Swamy, S.; Becq, J.; Murray, L.J.; Cheetham, R.K. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012, 28, 1811–1817. [Google Scholar] [CrossRef] [Green Version]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, D.L.; Schroder, J.; Penington, J.S.; Do, H.; Molania, R.; Dobrovic, A.; Speed, T.P.; Papenfuss, A.T. GRIDSS: Sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017, 27, 2050–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angus, L.; Smid, M.; Wilting, S.M.; Van Riet, J.; Van Hoeck, A.; Nguyen, L.; Nik-Zainal, S.; Steenbruggen, T.G.; Tjan-Heijnen, V.C.G.; Labots, M.; et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat. Genet. 2019, 51, 1450–1458. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Shao, Y.; Tian, L.; Flasch, D.A.; Mulder, H.L.; Edmonson, M.N.; Liu, Y.; Chen, X.; Newman, S.; Nakitandwe, J.; et al. Analysis of error profiles in deep next-generation sequencing data. Genome Biol. 2019, 20, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blokzijl, F.; Janssen, R.; Van Boxtel, R.; Cuppen, E. MutationalPatterns: Comprehensive genome-wide analysis of mutational processes. Genome Med. 2018, 10, 33. [Google Scholar] [CrossRef]
- Nguyen, L.; Martens, J.; Van Hoeck, A.; Cuppen, E. Pan-cancer landscape of homologous recombination deficiency. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]






| Patient ID | Sample ID | Primary/ Recurrence | Sample Location | Tumor Purity | Reference DNA | Age at Diagnosis | Initial Tumor Stage | Disease Status |
|---|---|---|---|---|---|---|---|---|
| GCPA002 | Recurrence | abdominal wall | 90% | Saliva | 35 | IC | NED | |
| GCPA005 | Recurrence | mesentery | 80% | Blood | 66 | IC | DOD | |
| GCPA006 | Recurrence | pelvic wall, left | 90% | Blood | 37 | IC | NED | |
| GCPA008 | T1.1 | Recurrence | small pelvis | 90% | Blood | 53 | IA | NED |
| T2.1 | Recurrence | lung | 90% | Blood | ||||
| GCPA011 | T1.1 | Recurrence | diaphragm right side | 90% | Blood | 53 | IC | AWD |
| T1.2 | Recurrence | diaphragm right side | 90% | Blood | ||||
| T2.1 | Recurrence | small bowel meso | 70% | Blood | ||||
| T3.1 | Recurrence | liver | 80% | Blood | ||||
| T3.2 | Recurrence | pelvis right side | 60% | Blood | ||||
| T3.3 | Recurrence | ligamentum triangulare left | 70% | Blood | ||||
| T3.4 | Recurrence | greater curvature stomach | 60% | Blood | ||||
| GCPA013 | T1.1 | Recurrence | paracolic right | 90% | Blood | 50 | IC | AWD |
| T2.1 | Recurrence | bladder peritoneum | 80% | Blood | ||||
| T2.2 | Recurrence | iliac left | 60% | Blood | ||||
| T2.3 | Recurrence | promontory | 70% | Blood | ||||
| T2.4 | Recurrence | iliac right | 90% | Blood | ||||
| GCPA016 | Recurrence | abdominal wall | 60% | Blood | 69 | IIB | AWD | |
| GCPA017 | Recurrence | below liver | 80% | Blood | 36 | IC | AWD | |
| GCPA019 | Recurrence | lateral of psoas muscle | 80% | Blood | 57 | IA | AWD | |
| GCPA021 | Recurrence | bladder peritoneum | 70% | Blood | 61 | IC | NED | |
| GCPA022 | T1.1 | Primary | ovary (left side within tumor) | 70% | Blood | 57 | IA | NED |
| T1.2 | Primary | ovary (right side within tumor) | 80% | Blood | ||||
| GCPA023 | T1.1 | Recurrence | peritoneal cavity | 80% | Blood | 48 | IA-C * | AWD |
| T2.1 | Recurrence | mesentery rectosigmoid | 80% | Blood | ||||
| GCPA024 | Recurrence | rectosigmoid | 60% | Blood | 30 | IA-C * | AWD | |
| GCPA030 | Recurrence | bladder peritoneum | 90% | Blood | 43 | IA-C * | AWD | |
| GCPA031 | Primary | ovary left | 80% | Blood | 49 | IA | NED | |
| GCPA032 | Recurrence | obturatorius loge right | 40% | Blood | 61 | unknown | NED | |
| GCPA044 | Primary | ovary | 50% | Blood | 61 | IC | NED | |
| GCPA046 | Recurrence | spleen | 90% | Blood | 54 | IA-C * | AWD | |
| GCPA048 | Primary | left ovary | 80% | Blood | 35 | IC | NED | |
| GCPA049 | Recurrence | omentum | 90% | Blood | 52 | unknown | NED | |
| GCPA050 | Recurrence | pouch of Douglas | 60% | Saliva | 32 | IA | NED | |
| GCPA051 | Recurrence | pouch of Douglas | 90% | Blood | 39 | IA-C * | NED | |
| GCPA052 | Recurrence | suprarenal infrahepatic | 90% | Blood | 37 | IC | NED | |
| GCPA053 | Recurrence | ileocecal | 80% | Saliva | 39 | IA | NED | |
| GCPA054 | Primary | ovary | 90% | Blood | 75 | IC | NED | |
| GCPA055 | Primary | ovary | 40% | Blood | 65 | IA | NED | |
| GCPA057 | Primary | ovary | 80% | Saliva | 65 | unknown | NED | |
| GCPA058 | Recurrence | liver | 80% | Saliva | 53 | IA | AWD | |
| GCPA059 | Primary | ovary | 90% | Saliva | 66 | IA | AWD | |
| GCPA060 | Recurrence | ileum | 90% | Saliva | 47 | IA | NED | |
| GCPA061 | Primary | ovary | 80% | Saliva | 71 | IA | NED | |
| GCPA065 | Primary | ovary | 90% | Blood | 61 | IA | NED | |
| GCPA066 | Primary | ovary | 70% | Saliva | 29 | IA | NED |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roze, J.; Monroe, G.; Kutzera, J.; Groeneweg, J.; Stelloo, E.; Paijens, S.; Nijman, H.; van Meurs, H.; van Lonkhuijzen, L.; Piek, J.; et al. Whole Genome Analysis of Ovarian Granulosa Cell Tumors Reveals Tumor Heterogeneity and a High-Grade TP53-Specific Subgroup. Cancers 2020, 12, 1308. https://doi.org/10.3390/cancers12051308
Roze J, Monroe G, Kutzera J, Groeneweg J, Stelloo E, Paijens S, Nijman H, van Meurs H, van Lonkhuijzen L, Piek J, et al. Whole Genome Analysis of Ovarian Granulosa Cell Tumors Reveals Tumor Heterogeneity and a High-Grade TP53-Specific Subgroup. Cancers. 2020; 12(5):1308. https://doi.org/10.3390/cancers12051308
Chicago/Turabian StyleRoze, Joline, Glen Monroe, Joachim Kutzera, Jolijn Groeneweg, Ellen Stelloo, Sterre Paijens, Hans Nijman, Hannah van Meurs, Luc van Lonkhuijzen, Jurgen Piek, and et al. 2020. "Whole Genome Analysis of Ovarian Granulosa Cell Tumors Reveals Tumor Heterogeneity and a High-Grade TP53-Specific Subgroup" Cancers 12, no. 5: 1308. https://doi.org/10.3390/cancers12051308
APA StyleRoze, J., Monroe, G., Kutzera, J., Groeneweg, J., Stelloo, E., Paijens, S., Nijman, H., van Meurs, H., van Lonkhuijzen, L., Piek, J., Lok, C., Jonges, G., Witteveen, P., Verheijen, R., van Haaften, G., & Zweemer, R. (2020). Whole Genome Analysis of Ovarian Granulosa Cell Tumors Reveals Tumor Heterogeneity and a High-Grade TP53-Specific Subgroup. Cancers, 12(5), 1308. https://doi.org/10.3390/cancers12051308







