Functional Characterization of POFUT1 Variants Associated with Colorectal Cancer
Abstract
:1. Introduction
2. Results
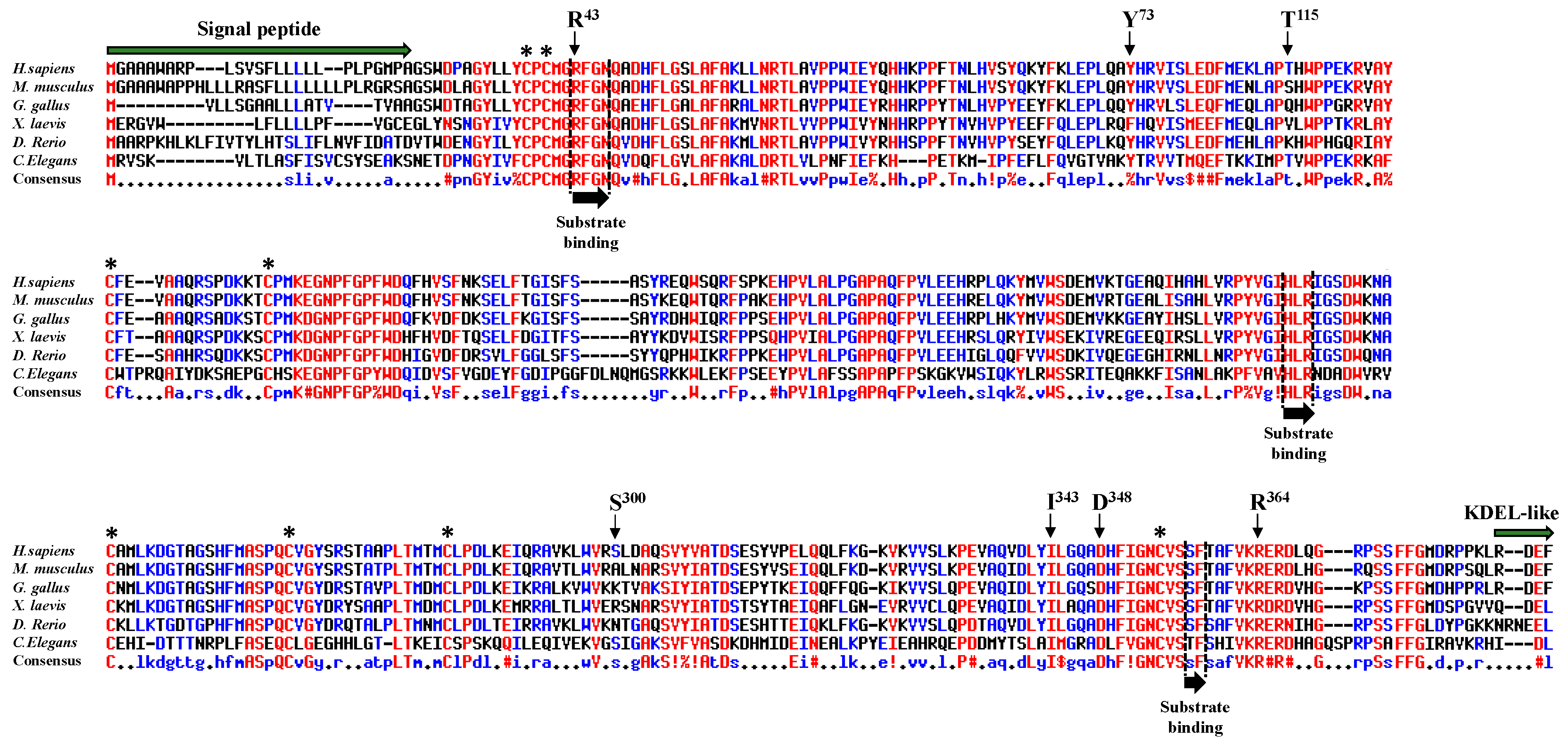
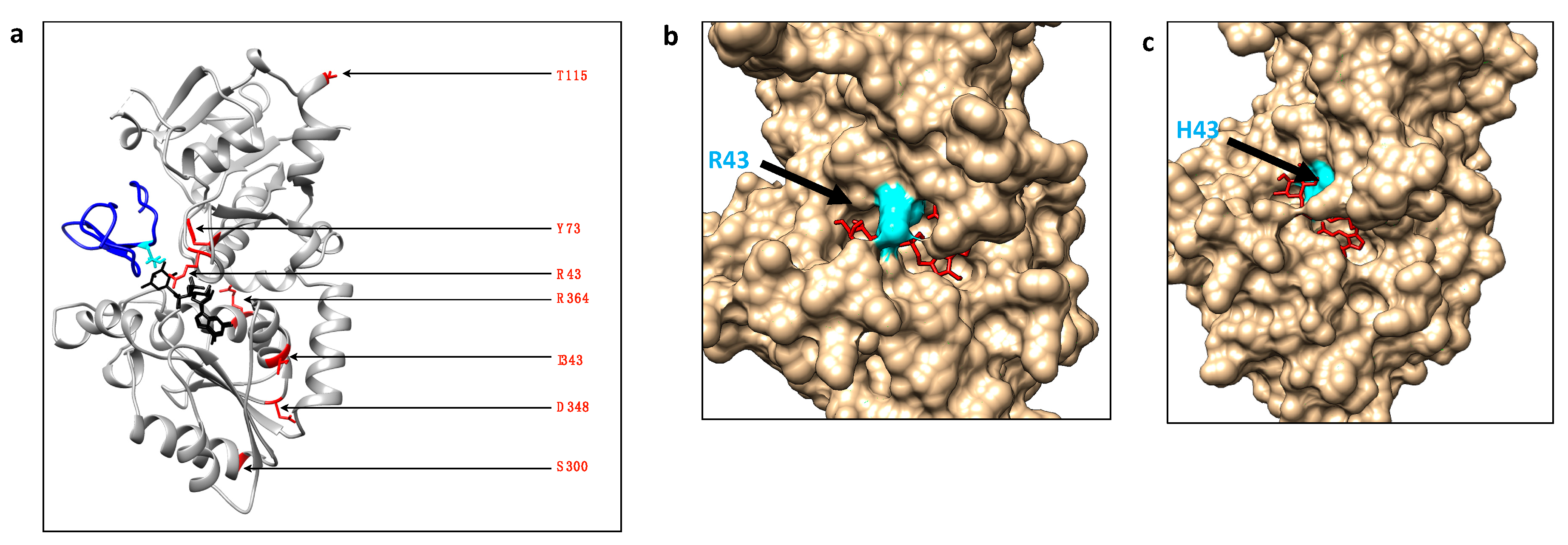
2.1. Frequency and Location of Missense Mutations Affecting POFUT1 in CRC
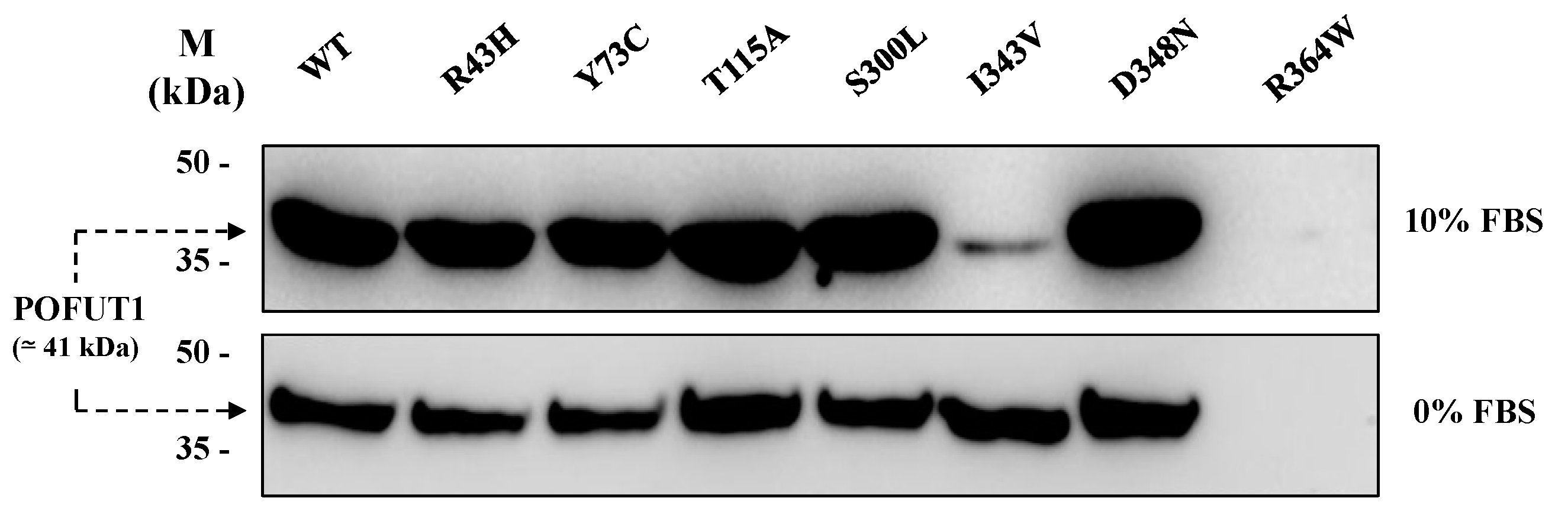
2.2. Production of Soluble Forms for Human WT and Mutated POFUT1 Variants
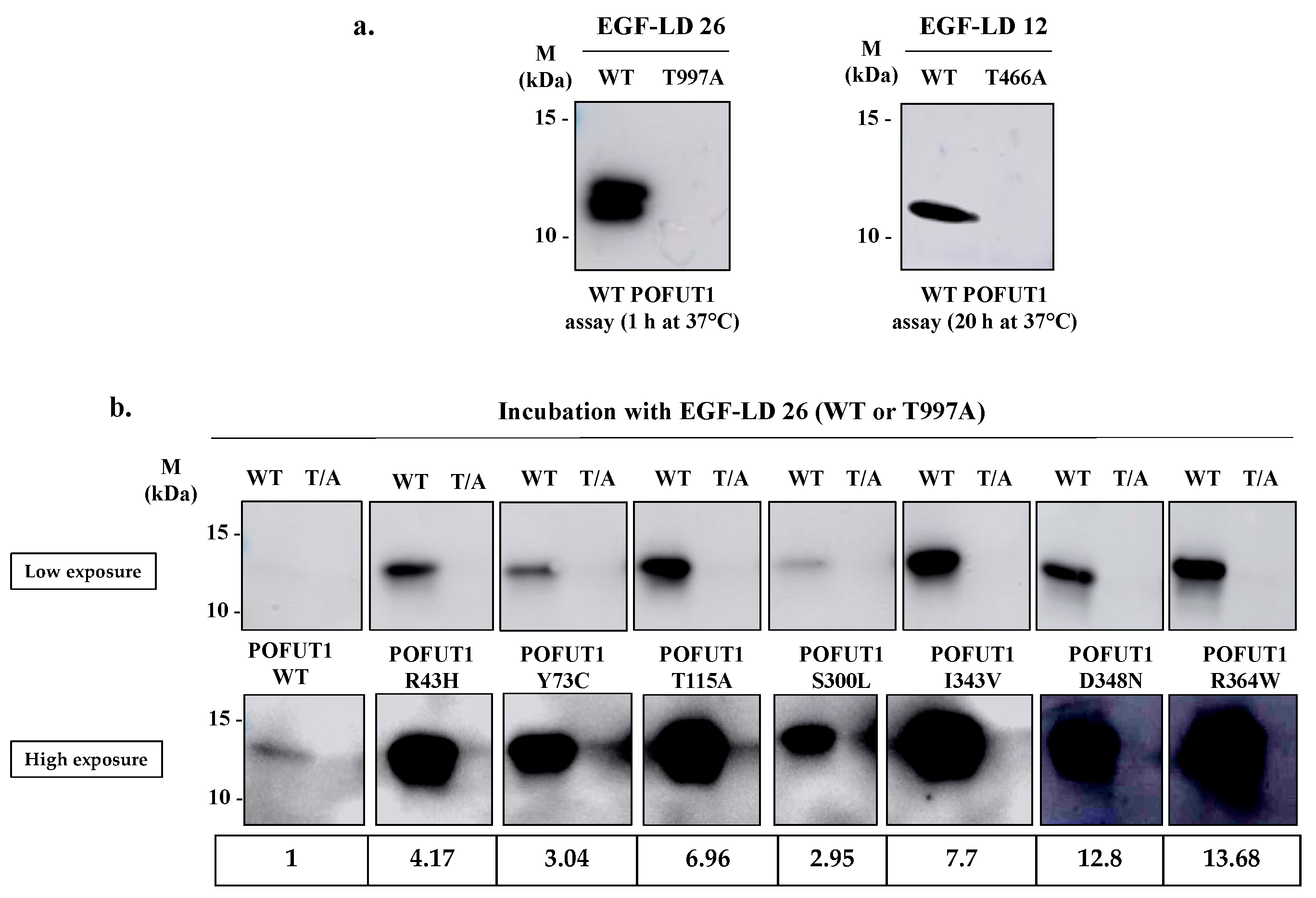
2.3. Evidencing of the O-Fucosyltransferase Activity for POFUT1 Variants
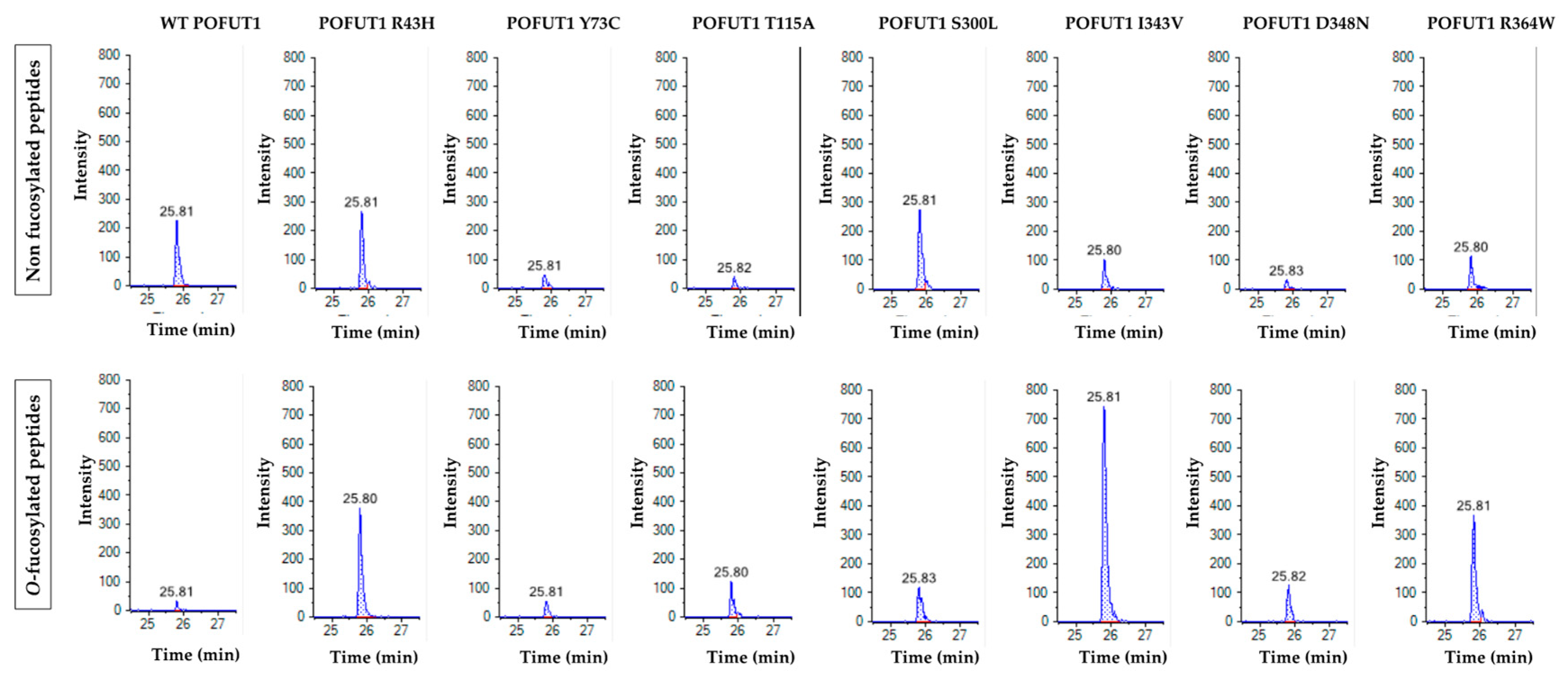
2.4. Quantitative MRM-MS Analysis of the O-Fucosyltransferase Activity for Each POFUT1 Variant
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructs to Produce Recombinant EGF-LDs and POFUT1 Variants
4.2. Cell Culture and Transfection
4.3. Cell Lines
4.4. Protein Production and Purification
4.5. SDS-PAGE and Blotting Techniques
4.6. Glycosyltransferase Reaction
4.7. Click Chemistry Reactions
4.8. Targeted Mass Spectrometry
4.9. Alignments and Automatic Molecular Modelling
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nguyen, H.T.; Duong, H.-Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol Lett 2018, 16, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, B.; Postma, C.; Mongera, S.; Hopmans, E.; Diskin, S.; van de Wiel, M.A.; van Criekinge, W.; Thas, O.; Matthai, A.; Cuesta, M.A.; et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut 2009, 58, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Mackinnon, R.N.; Selan, C.; Wall, M.; Baker, E.; Nandurkar, H.; Campbell, L.J. The paradox of 20q11.21 amplification in a subset of cases of myeloid malignancy with chromosome 20 deletion. Genes Chromosomes Cancer 2010, 49, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lin, C.; Li, N.; Du, Y.; Yang, C.; Bai, Y.; Feng, Z.; Su, C.; Wu, R.; Song, S.; et al. PLAGL2 and POFUT1 are regulated by an evolutionarily conserved bidirectional promoter and are collaboratively involved in colorectal cancer by maintaining stemness. EBioMedicine 2019, 45, 124–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germot, A.; Maftah, A. POFUT1 and PLAGL2 gene pair linked by a bidirectional promoter: The two in one of tumour progression in colorectal cancer? EBioMedicine 2019, 46, 25–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chabanais, J.; Labrousse, F.; Chaunavel, A.; Germot, A.; Maftah, A. POFUT1 as a Promising Novel Biomarker of Colorectal Cancer. Cancers (Basel) 2018, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Li, D.; Li, N.; Su, C.; Yang, C.; Lin, C.; Chen, M.; Wu, R.; Li, X.; Hu, G. POFUT1 promotes colorectal cancer development through the activation of Notch1 signaling. Cell Death & Disease 2018, 9. [Google Scholar] [CrossRef]
- Albesa-Jové, D.; Guerin, M.E. The conformational plasticity of glycosyltransferases. Curr. Opin. Struct. Biol. 2016, 40, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Lira-Navarrete, E.; Valero-González, J.; Martínez-Júlvez, M.; Tejero, T.; Merino, P.; Panjikar, S.; Hurtado-Guerrero, R. Structural Insights into the Mechanism of Protein O-Fucosylation. PLoS One 2011, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Han, K.; Pak, J.E.; Satkunarajah, M.; Zhou, D.; Rini, J.M. Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat. Chem. Biol. 2017, 13, 757–763. [Google Scholar] [CrossRef]
- Loriol, C.; Audfray, A.; Dupuy, F.; Germot, A.; Maftah, A. The two N-glycans present on bovine Pofut1 are differently involved in its solubility and activity. FEBS J. 2007, 274, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Moloney, D.J.; Panin, V.M.; Johnston, S.H.; Chen, J.; Shao, L.; Wilson, R.; Wang, Y.; Stanley, P.; Irvine, K.D.; Haltiwanger, R.S.; et al. Fringe is a glycosyltransferase that modifies Notch. Nature 2000, 406, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Panin, V.M.; Shao, L.; Lei, L.; Moloney, D.J.; Irvine, K.D.; Haltiwanger, R.S. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J. Biol. Chem. 2002, 277, 29945–29952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arboleda-Velasquez, J.F.; Rampal, R.; Fung, E.; Darland, D.C.; Liu, M.; Martinez, M.C.; Donahue, C.P.; Navarro-Gonzalez, M.F.; Libby, P.; D’Amore, P.A.; et al. CADASIL mutations impair Notch3 glycosylation by Fringe. Hum. Mol. Genet. 2005, 14, 1631–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakuda, S.; Haltiwanger, R.S. Deciphering the Fringe-Mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands. Dev. Cell 2017, 40, 193–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamura, Y.; Saga, Y. Pofut1 is required for the proper localization of the Notch receptor during mouse development. Mechanisms of Development 2008, 125, 663–673. [Google Scholar] [CrossRef]
- Shi, S.; Stahl, M.; Lu, L.; Stanley, P. Canonical Notch Signaling Is Dispensable for Early Cell Fate Specifications in Mammals. Mol. Cell Biol. 2005, 25, 9503–9508. [Google Scholar] [CrossRef] [Green Version]
- Oka, C.; Nakano, T.; Wakeham, A.; Okazaki, S.; de la Pompa, J.L.; Mori, C.; Sakai, T.; Kawaichi, M.; Shiota, K.; Mak, T.W.; et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development 1995, 121, 3291–3301. [Google Scholar]
- Ma, L.; Dong, P.; Liu, L.; Gao, Q.; Duan, M.; Zhang, S.; Chen, S.; Xue, R.; Wang, X. Overexpression of protein O-fucosyltransferase 1 accelerates hepatocellular carcinoma progression via the Notch signaling pathway. Biochem. Biophys. Res. Commun. 2016, 473, 503–510. [Google Scholar] [CrossRef]
- Dong, S.; Wang, Z.; Huang, B.; Zhang, J.; Ge, Y.; Fan, Q.; Wang, Z. Bioinformatics insight into glycosyltransferase gene expression in gastric cancer: POFUT1 is a potential biomarker. Biochem. Biophys. Res. Commun. 2017, 483, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.; Ogawara, K.; Kimura, R.; Shimizu, F.; Baba, T.; Minakawa, Y.; Higo, M.; Kasamatsu, A.; Endo-Sakamoto, Y.; Shiiba, M.; et al. Protein O-fucosyltransferase 1: A potential diagnostic marker and therapeutic target for human oral cancer. Int. J. Oncol. 2013, 43, 1864–1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, G.; Tian, L.; Yu, Y.; Li, F.; Wang, X.; Li, C.; Deng, S.; Yu, X.; Cai, X.; Zuo, Z.; et al. Overexpression of Pofut1 and activated Notch1 may be associated with poor prognosis in breast cancer. Biochem. Biophys. Res. Commun. 2017, 491, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Rampal, R.; Arboleda-Velasquez, J.F.; Nita-Lazar, A.; Kosik, K.S.; Haltiwanger, R.S. Highly conserved O-fucose sites have distinct effects on Notch1 function. J. Biol. Chem. 2005, 280, 32133–32140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennarubia, F.; Pinault, E.; Maftah, A.; Legardinier, S. In vitro acellular method to reveal O -fucosylation on EGF-like domains. Glycobiology 2019, 29, 192–198. [Google Scholar] [CrossRef]
- Dingerdissen, H.M.; Torcivia-Rodriguez, J.; Hu, Y.; Chang, T.-C.; Mazumder, R.; Kahsay, R. BioMuta and BioXpress: Mutation and expression knowledgebases for cancer biomarker discovery. Nucleic Acids Res 2018, 46, D1128–D1136. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.-J.; Shamsaddini, A.; Pan, Y.; Smith, K.; Crichton, D.J.; Simonyan, V.; Mazumder, R. A framework for organizing cancer-related variations from existing databases, publications and NGS data using a High-performance Integrated Virtual Environment (HIVE). Database (Oxford) 2014, 2014, bau022. [Google Scholar] [CrossRef]
- Pan, Y.; Karagiannis, K.; Zhang, H.; Dingerdissen, H.; Shamsaddini, A.; Wan, Q.; Simonyan, V.; Mazumder, R. Human germline and pan-cancer variomes and their distinct functional profiles. Nucleic Acids Res. 2014, 42, 11570–11588. [Google Scholar] [CrossRef] [Green Version]
- Lira-Navarrete, E.; Hurtado-Guerrero, R. A perspective on structural and mechanistic aspects of protein O -fucosylation. Acta Crystallographica Section F Structural Biology Communications 2018, 74, 443–450. [Google Scholar] [CrossRef]
- McMillan, B.J.; Zimmerman, B.; Egan, E.D.; Lofgren, M.; Xu, X.; Hesser, A.; Blacklow, S.C. Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations. Glycobiology 2017, 27, 777–786. [Google Scholar] [CrossRef] [Green Version]
- Sherwood, C.A.; Eastham, A.; Lee, L.W.; Risler, J.; Mirzaei, H.; Falkner, J.A.; Martin, D.B. Rapid optimization of MRM-MS instrument parameters by subtle alteration of precursor and product m/z targets. J. Proteome Res. 2009, 8, 3746–3751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komor, M.A.; de Wit, M.; van den Berg, J.; Martens de Kemp, S.R.; Delis-van Diemen, P.M.; Bolijn, A.S.; Tijssen, M.; Schelfhorst, T.; Piersma, S.R.; Chiasserini, D.; et al. Molecular characterization of colorectal adenomas reveals POFUT1 as a candidate driver of tumor progression. Int. J. Cancer 2019. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Peng, Z.; Zhang, Y.; Yang, J. COACH-D: Improved protein-ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 2018, 46, W438–W442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Roy, A.; Zhang, Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Kumar, V.; Nordstrøm, L.U.; Feng, L.; Takeuchi, H.; Hao, H.; Luca, V.C.; Garcia, K.C.; Stanley, P.; Wu, P.; et al. Inhibition of Delta-induced Notch signaling using fucose analogs. Nat Chem Biol 2018, 14, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Der Vartanian, A.; Audfray, A.; Al Jaam, B.; Janot, M.; Legardinier, S.; Maftah, A.; Germot, A. Protein O-Fucosyltransferase 1 Expression Impacts Myogenic C2C12 Cell Commitment via the Notch Signaling Pathway. Mol Cell Biol 2015, 35, 391–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deschuyter, M.; Pennarubia, F.; Pinault, E.; Legardinier, S.; Maftah, A. Functional Characterization of POFUT1 Variants Associated with Colorectal Cancer. Cancers 2020, 12, 1430. https://doi.org/10.3390/cancers12061430
Deschuyter M, Pennarubia F, Pinault E, Legardinier S, Maftah A. Functional Characterization of POFUT1 Variants Associated with Colorectal Cancer. Cancers. 2020; 12(6):1430. https://doi.org/10.3390/cancers12061430
Chicago/Turabian StyleDeschuyter, Marlène, Florian Pennarubia, Emilie Pinault, Sébastien Legardinier, and Abderrahman Maftah. 2020. "Functional Characterization of POFUT1 Variants Associated with Colorectal Cancer" Cancers 12, no. 6: 1430. https://doi.org/10.3390/cancers12061430
APA StyleDeschuyter, M., Pennarubia, F., Pinault, E., Legardinier, S., & Maftah, A. (2020). Functional Characterization of POFUT1 Variants Associated with Colorectal Cancer. Cancers, 12(6), 1430. https://doi.org/10.3390/cancers12061430




