Role of Anillin in Tumour: From a Prognostic Biomarker to a Novel Target
Abstract
:1. Introduction
2. Role of ANLN in Normal Cell
2.1. Findings from Drosophila and C. elegans
2.2. Binding Partners
2.2.1. Binding Partners Related to Cytokinesis
2.2.2. Other Binding Partners
2.3. Role of ANLN during Cytokinensis
2.4. Mechanism of ANLN-Controlled Cytoskeletal Dynamics

2.4.1. ANLN in Nucleus
2.4.2. ANLN in Cytosol
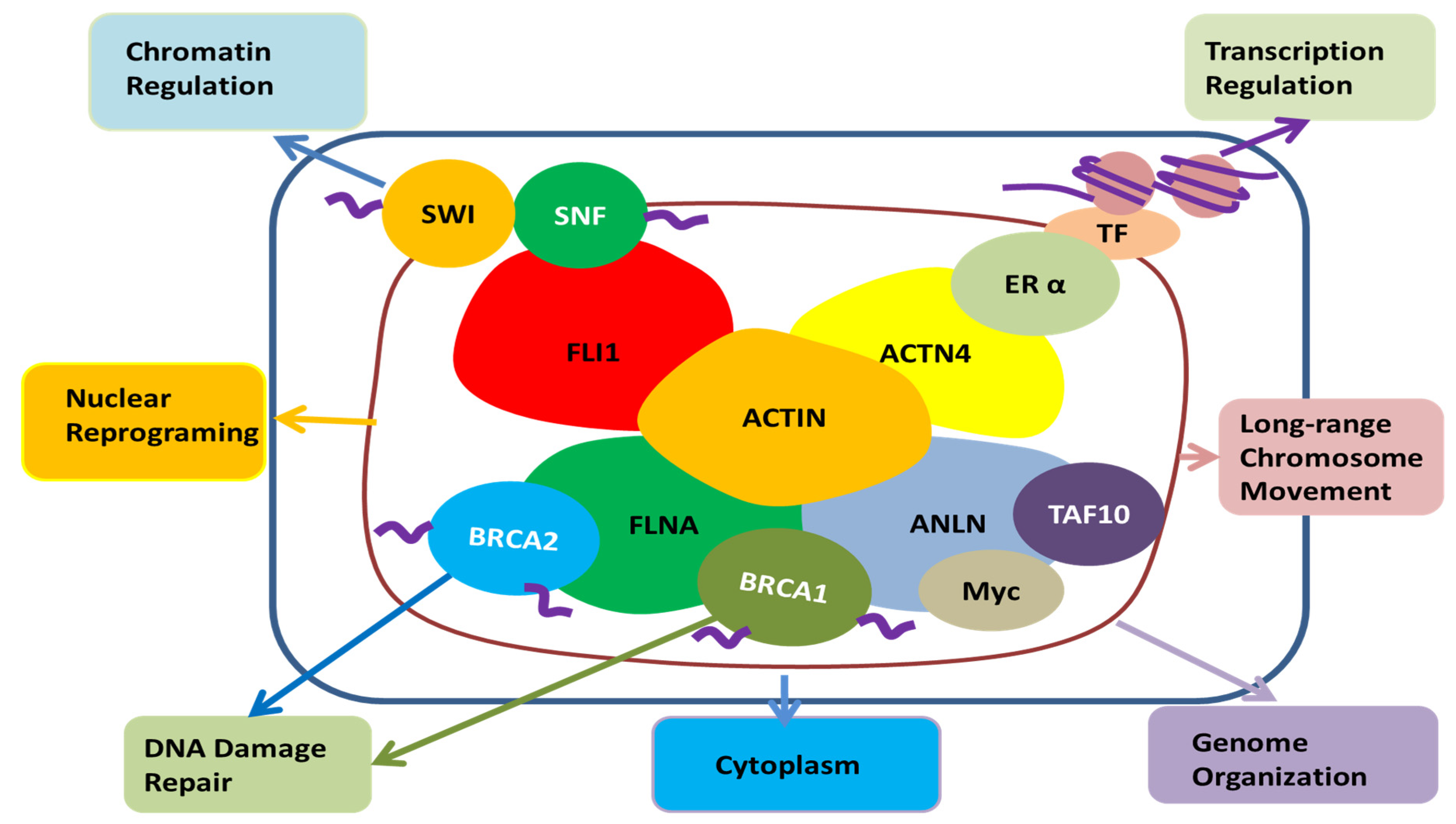
3. Role of ANLN in Tumour Cells
3.1. What Is the Role of ANLN on Proliferation and Cell Death of Cancer Cells?
3.2. What Is the Role of ANLN on Invasion and Metastasis of Cancer Cells?
3.3. What Is the Role of ANLN in Replicative Immortality and DNA Repair?
3.4. Possible Role of ANLN on Other Hallmarks of Cancer
4. Biomarker and Potential Therapeutic Options
4.1. ANLN as a Biomarker
| Tissue | Samples Overexpressed/Total Sample Tested | Percentages of Samples Overexpressed (%) |
|---|---|---|
| Adrenal gland | 4/79 | 5.06 |
| Breast | 125/1104 | 11.32 |
| Central nervous system | 30/697 | 4.3 |
| Cervix | 19/307 | 6.19 |
| Endometrium | 45/602 | 7.48 |
| Hematopoietic and lymphoid | 10/211 | 4.52 |
| Kidney | 33/600 | 5.5 |
| Large intestine | 29/610 | 4.75 |
| Liver | 25/373 | 6.7 |
| Lung | 148/1019 | 14.52 |
| Oesophagus | 18/125 | 14.4 |
| Ovary | 12/266 | 4.51 |
| Pancreas | 15/179 | 8.38 |
| Prostate | 46/498 | 9.24 |
| Skin | 43/473 | 9.09 |
| Soft tissue | 23/263 | 8.75 |
| Stomach | 32/285 | 11.23 |
| Thyroid | 28/513 | 5.46 |
| Upper respiratory tract | 54/522 | 10.34 |
| Urinary tract | 34/408 | 8.33 |
4.2. Potential Therapeutic Options
5. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Piekny, A.J.; Maddox, A.S. The myriad roles of anillin during cytokinesis. Semin. Cell Dev. Biol. 2010, 21, 881–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Hall, P.A.; Todd, C.B.; Hyland, P.L.; McDade, S.S.; Grabsch, H.; Dattani, M.; Hillan, K.J.; Russell, S.E. The septin-binding protein anillin is overexpressed in diverse human tumors. Clin. Cancer Res. 2005, 11, 6780–6786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olakowski, M.; Tyszkiewicz, T.; Jarzab, M.; Krol, R.; Oczko-Wojciechowska, M.; Kowalska, M.; Kowal, M.; Gala, G.M.; Kajor, M.; Lange, D.; et al. NBL1 and anillin (ANLN) genes over-expression in pancreatic carcinoma. Folia Histochem. Cytobiol. 2009, 47, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Dai, H.; Gong, Y.; Zhang, C.; Shu, J.; Luo, Y.; Jiang, Y.; Liu, W.; Bie, P. ANLN-induced EZH2 upregulation promotes pancreatic cancer progression by mediating miR-218-5p/LASP1 signaling axis. J. Exp. Clin. Cancer Res. 2019, 38, 347. [Google Scholar] [CrossRef] [Green Version]
- Idichi, T.; Seki, N.; Kurahara, H.; Yonemori, K.; Osako, Y.; Arai, T.; Okato, A.; Kita, Y.; Arigami, T.; Mataki, Y.; et al. Regulation of actin-binding protein ANLN by antitumor miR-217 inhibits cancer cell aggressiveness in pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 53180–53193. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Shen, W.; Cui, L.; Chen, W.; Hu, X.; Fu, J. Overexpression of anillin (ANLN) is correlated with colorectal cancer progression and poor prognosis. Cancer Biomark. 2016, 16, 459–465. [Google Scholar] [CrossRef]
- Magnusson, K.; Gremel, G.; Ryden, L.; Ponten, V.; Uhlen, M.; Dimberg, A.; Jirstrom, K.; Ponten, F. ANLN is a prognostic biomarker independent of Ki-67 and essential for cell cycle progression in primary breast cancer. BMC Cancer 2016, 16, 904. [Google Scholar] [CrossRef]
- Long, X.; Zhou, W.; Wang, Y.; Liu, S. Prognostic significance of ANLN in lung adenocarcinoma. Oncol. Lett. 2018, 16, 1835–1840. [Google Scholar] [CrossRef]
- Gbadegesin, R.A.; Hall, G.; Adeyemo, A.; Hanke, N.; Tossidou, I.; Burchette, J.; Wu, G.; Homstad, A.; Sparks, M.A.; Gomez, J.; et al. Mutations in the gene that encodes the F-actin binding protein anillin cause FSGS. J. Am. Soc. Nephrol. 2014, 25, 1991–2002. [Google Scholar] [CrossRef]
- Zhang, L.; Maddox, A.S. Anillin. Curr. Biol. 2010, 20, R135–R136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, C.M.; Alberts, B.M. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 1995, 131, 165–178. [Google Scholar] [CrossRef]
- Hickson, G.R.; O’Farrell, P.H. Rho-dependent control of anillin behavior during cytokinesis. J. Cell Biol. 2008, 180, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Echard, A.; Hickson, G.R.; Foley, E.; O’Farrell, P.H. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 2004, 14, 1685–1693. [Google Scholar] [CrossRef] [Green Version]
- Maddox, A.S.; Lewellyn, L.; Desai, A.; Oegema, K. Anillin and the septins promote asymmetric ingression of the cytokinetic furrow. Dev. Cell 2007, 12, 827–835. [Google Scholar] [CrossRef] [Green Version]
- Piekny, A.J.; Glotzer, M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr. Biol. 2008, 18, 30–36. [Google Scholar] [CrossRef] [Green Version]
- Hickson, G.R.; O’Farrell, P.H. Anillin: A pivotal organizer of the cytokinetic machinery. Biochem. Soc. Trans. 2008, 36, 439–441. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.G.; Field, C.M.; Alberts, B.M. Actin-binding proteins from Drosophila embryos: A complex network of interacting proteins detected by F-actin affinity chromatography. J. Cell Biol. 1989, 109, 2963–2975. [Google Scholar] [CrossRef] [Green Version]
- Goldbach, P.; Wong, R.; Beise, N.; Sarpal, R.; Trimble, W.S.; Brill, J.A. Stabilization of the actomyosin ring enables spermatocyte cytokinesis in Drosophila. Mol. Biol. Cell 2010, 21, 1482–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giansanti, M.G.; Bonaccorsi, S.; Gatti, M. The role of anillin in meiotic cytokinesis of Drosophila males. J. Cell Sci. 1999, 112, 2323–2334. [Google Scholar] [PubMed]
- Straight, A.F.; Cheung, A.; Limouze, J.; Chen, I.; Westwood, N.J.; Sellers, J.R.; Mitchison, T.J. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 2003, 299, 1743–1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oegema, K.; Savoian, M.S.; Mitchison, T.J.; Field, C.M. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J. Cell Biol. 2000, 150, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Field, C.M.; Coughlin, M.L.; Straight, A.F.; Mitchison, T.J. Self-and actin-templated assembly of mammalian septins. Dev. Cell 2002, 3, 791–802. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.L. The contractile ring. Curr. Biol. 2011, 21, R976–R978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddox, A.S.; Habermann, B.; Desai, A.; Oegema, K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development 2005, 132, 2837–2848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Furihata, M.; Tsunoda, T.; Ashida, S.; Takata, R.; Obara, W.; Yoshioka, H.; Daigo, Y.; Nasu, Y.; Kumon, H.; et al. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007, 67, 5117–5125. [Google Scholar] [CrossRef] [Green Version]
- Straight, A.F.; Field, C.M.; Mitchison, T.J. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell 2005, 16, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Field, C.M.; Coughlin, M.; Doberstein, S.; Marty, T.; Sullivan, W. Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development 2005, 132, 2849–2860. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, C.; Daigo, Y.; Ishikawa, N.; Kato, T.; Hayama, S.; Ito, T.; Tsuchiya, E.; Nakamura, Y. ANLN plays a critical role in human lung carcinogenesis through the activation of RHOA and by involvement in the phosphoinositide 3-kinase/AKT pathway. Cancer Res. 2005, 65, 11314–11325. [Google Scholar] [CrossRef] [Green Version]
- Gregory, S.L.; Ebrahimi, S.; Milverton, J.; Jones, W.M.; Bejsovec, A.; Saint, R. Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Curr. Biol. 2008, 18, 25–29. [Google Scholar] [CrossRef]
- D’Avino, P.P.; Takeda, T.; Capalbo, L.; Zhang, W.; Lilley, K.S.; Laue, E.D.; Glover, D.M. Interaction between anillin and RacGAP50C connects the actomyosin contractile ring with spindle microtubules at the cell division site. J. Cell Sci. 2008, 121, 1151–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frenette, P.E.H.A.; Loloyan, M.; Kinalb, M.; Pakarianc, P.; Piekny, A. An Anillin-Ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PLoS ONE 2012, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monzo, P.G.; Gauthier, N.C.; Keslair, F.; Loubat, A.; Field, C.M.; Le Marchand-Brustel, Y.; Cormont, M. Clues to CD2-associated protein involvement in cytokinesis. Mol. Biol. Cell 2005, 16, 2891–2902. [Google Scholar] [CrossRef] [Green Version]
- Xia, L.; Su, X.; Shen, J.; Meng, Q.; Yan, J.; Zhang, C.; Chen, Y.; Wang, H.; Xu, M. ANLN functions as a key candidate gene in cervical cancer as determined by integrated bioinformatic analysis. Cancer Manag. Res. 2018, 10, 663–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sisson, J.C.; Field, C.; Ventura, R.; Royou, A.; Sullivan, W. Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J. Cell Biol. 2000, 151, 905–918. [Google Scholar] [CrossRef] [Green Version]
- Goldschmidt-Clermont, P.J.; Furman, M.I.; Wachsstock, D.; Safer, D.; Nachmias, V.T.; Pollard, T.D. The control of actin nucleotide exchange by thymosin beta 4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol. Biol. Cell 1992, 3, 1015–1024. [Google Scholar] [CrossRef] [Green Version]
- Witke, W.; Podtelejnikov, A.V.; Di Nardo, A.; Sutherland, J.D.; Gurniak, C.B.; Dotti, C.; Mann, M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998, 17, 967–976. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, R.P.; Veltman, D.; Machesky, L.M. Actin-bundling proteins in cancer progression at a glance. J. Cell Sci. 2012, 125, 1073–1079. [Google Scholar] [CrossRef] [Green Version]
- Sohrmann, M.; Fankhauser, C.; Brodbeck, C.; Simanis, V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996, 10, 2707–2719. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Okawa, K.; Miki, T.; Sakamoto, S.; Morinaga, T.; Segawa, K.; Arakawa, T.; Kinoshita, M.; Ishizaki, T.; Narumiya, S. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol. Biol. Cell 2010, 21, 3193–3204. [Google Scholar] [CrossRef] [Green Version]
- Haglund, K.; Nezis, I.P.; Lemus, D.; Grabbe, C.; Wesche, J.; Liestøl, K.; Dikic, I.; Palmer, R.; Stenmark, H. Cindr interacts with anillin to control cytokinesis in Drosophila melanogaster. Curr. Biol. 2010, 20, 944–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.-M.; Fang, G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J. Biol. Chem. 2005, 280, 33516–33524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, M.; Glotzer, M. Control of Cortical Contractility During Cytokinesis; Portland Press Ltd.: London, UK, 2008. [Google Scholar]
- Marquardt, J.; Chen, X.; Bi, E. Architecture, remodeling, and functions of the septin cytoskeleton. Cytoskeleton 2019, 76, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, K.; Zieger, B. The mammalian septin interactome. Front. Cell Dev. Biol. 2017, 5, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weirich, C.S.; Erzberger, J.P.; Barral, Y. The septin family of GTPases: Architecture and dynamics. Nat. Rev. Mol. Cell Biol. 2008, 9, 478–489. [Google Scholar] [CrossRef]
- Renshaw, M.J.; Liu, J.; Lavoie, B.D.; Wilde, A. Anillin-dependent organization of septin filaments promotes intercellular bridge elongation and Chmp4B targeting to the abscission site. Open Biol. 2014, 4, 130190. [Google Scholar] [CrossRef] [Green Version]
- Mostowy, S.; Cossart, P. Septins: The fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012, 13, 183–194. [Google Scholar] [CrossRef]
- Shen, D.-W.; Pouliot, L.M.; Gillet, J.-P.; Ma, W.; Johnson, A.C.; Hall, M.D.; Gottesman, M.M. The transcription factor GCF2 is an upstream repressor of the small GTPAse RhoA, regulating membrane protein trafficking, sensitivity to doxorubicin, and resistance to cisplatin. Mol. Pharm. 2012, 9, 1822–1833. [Google Scholar] [CrossRef] [Green Version]
- Valderrama, F.; Cordeiro, J.V.; Schleich, S.; Frischknecht, F.; Way, M. Vaccinia virus-induced cell motility requires F11L-mediated inhibition of RhoA signaling. Science 2006, 311, 377–381. [Google Scholar] [CrossRef]
- Hirai, A.; Nakamura, S.; Noguchi, Y.; Yasuda, T.; Kitagawa, M.; Tatsuno, I.; Oeda, T.; Tahara, K.; Terano, T.; Narumiya, S. Geranylgeranylated rho small GTPase (s) are essential for the degradation of p27Kip1 and facilitate the progression from G1 to S phase in growth-stimulated rat FRTL-5 cells. J. Biol. Chem. 1997, 272, 13–16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Zhou, X.; Lang, R.A.; Guo, F. RhoA of the Rho family small GTPases is essential for B lymphocyte development. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalpana, G.; Figy, C.; Yeung, M.; Yeung, K.C. Reduced RhoA expression enhances breast cancer metastasis with a concomitant increase in CCR5 and CXCR4 chemokines signaling. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Guan, R.; Lee, I.-J.; Liu, Y.; Chen, M.; Wang, J.; Wu, J.-Q.; Chen, Z. Mechanistic insights into the anchorage of the contractile ring by anillin and Mid1. Dev. Cell 2015, 33, 413–426. [Google Scholar] [CrossRef] [Green Version]
- Somers, W.G.; Saint, R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell 2003, 4, 29–39. [Google Scholar] [CrossRef]
- Wu, H.; Guo, J.; Zhou, Y.-T.; Gao, X.-D. The anillin-related region of Bud4 is the major functional determinant for Bud4’s function in septin organization during bud growth and axial bud site selection in budding yeast. Eukaryot. Cell 2015, 14, 241–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Johnson, J.M.; Lera, R.F.; Brahma, S.; Burkard, M.E. Anillin phosphorylation controls timely membrane association and successful cytokinesis. PLoS Genet. 2017, 13, e1006511. [Google Scholar] [CrossRef]
- Dai, X.; Chen, X.; Hakizimana, O.; Mei, Y. Genetic interactions between ANLN and KDR are prognostic for breast cancer survival. Oncol Rep. 2019, 42, 2255–2266. [Google Scholar] [CrossRef]
- Dema, A.; Macaluso, F.; Sgrò, F.; Berto, G.E.; Bianchi, F.T.; Chiotto, A.A.; Pallavicini, G.; Di Cunto, F.; Gai, M. Citron kinase-dependent F-actin maintenance at midbody secondary ingression sites mediates abscission. Co. Biol. 2018, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, J.Y.; Gao, J.; Sun, X.; Cao, M.D.; Shi, L.; Xia, T.S.; Zhou, W.B.; Wang, S.; Ding, Q.; Wei, J.F. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene 2019, 38, 6123–6141. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, M.; Rao, X.; Zhang, W.; Li, X.; Wang, L.; Huang, G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Oncol. Targets 2019, 12, 3421–3428. [Google Scholar] [CrossRef] [Green Version]
- Lan, T.; Li, H.; Zhang, D.; Xu, L.; Liu, H.; Hao, X.; Yan, X.; Liao, H.; Chen, X.; Xie, K.; et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 2019, 18, 186. [Google Scholar] [CrossRef]
- Hoffman, B.; Liebermann, D.A. The proto-oncogene c-myc and apoptosis. Oncogene 1998, 17, 3351–3357. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Hou, Y.; Ashktorab, H.; Gao, L.; Xu, Y.; Wu, K.; Zhai, J.; Zhang, L. The impact of C-MYC gene expression on gastric cancer cell. Mol. Cell Biochem. 2010, 344, 125–135. [Google Scholar] [CrossRef] [Green Version]
- Heidelberger, J.B.; Voigt, A.; Borisova, M.E.; Petrosino, G.; Ruf, S.; Wagner, S.A.; Beli, P. Proteomic profiling of VCP substrates links VCP to K6-linked ubiquitylation and c-Myc function. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Kalkat, M.; Resetca, D.; Lourenco, C.; Chan, P.K.; Wei, Y.; Shiah, Y.J.; Vitkin, N.; Tong, Y.; Sunnerhagen, M.; Done, S.J.; et al. MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol. Cell 2018, 72, 836–848.e7. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Mei, Y.; Chen, X.; Cai, D. ANLN and KDR are jointly prognostic of breast cancer survival and can be modulated for triple negative breast cancer control. Front. Genet. 2019, 10, 790. [Google Scholar] [CrossRef]
- Jurica, M.S.; Licklider, L.J.; Gygi, S.R.; Grigorieff, N.; Moore, M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 2002, 8, 426–439. [Google Scholar] [CrossRef] [Green Version]
- Grote, M.; Wolf, E.; Will, C.L.; Lemm, I.; Agafonov, D.E.; Schomburg, A.; Fischle, W.; Urlaub, H.; Luhrmann, R. Molecular architecture of the human Prp19/CDC5L complex. Mol. Cell Biol. 2010, 30, 2105–2119. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yan, C.; Hang, J.; Finci, L.I.; Lei, J.; Shi, Y. An atomic structure of the human spliceosome. Cell 2017, 169, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Bertram, K.; Agafonov, D.E.; Liu, W.T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Urlaub, H.; Kastner, B.; Stark, H.; Luhrmann, R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 2017, 542, 318–323. [Google Scholar] [CrossRef] [Green Version]
- Haselbach, D.; Komarov, I.; Agafonov, D.E.; Hartmuth, K.; Graf, B.; Dybkov, O.; Urlaub, H.; Kastner, B.; Luhrmann, R.; Stark, H. Structure and conformational dynamics of the human spliceosomal B(act) complex. Cell 2018, 172, 454–464. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yan, C.; Zhan, X.; Li, L.; Lei, J.; Shi, Y. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018, 28, 307–322. [Google Scholar] [CrossRef]
- Zhan, X.; Yan, C.; Zhang, X.; Lei, J.; Shi, Y. Structure of a human catalytic step I spliceosome. Science 2018, 359, 537–545. [Google Scholar] [CrossRef] [Green Version]
- Fica, S.M.; Oubridge, C.; Wilkinson, M.E.; Newman, A.J.; Nagai, K. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science 2019, 363, 710–714. [Google Scholar] [CrossRef]
- Mu, R.; Wang, Y.B.; Wu, M.; Yang, Y.; Song, W.; Li, T.; Zhang, W.N.; Tan, B.; Li, A.L.; Wang, N.; et al. Depletion of pre-mRNA splicing factor Cdc5L inhibits mitotic progression and triggers mitotic catastrophe. Cell Death Dis. 2014, 5, e1151. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Mao, W.; Wang, L.; Liu, M.; Zhang, W.; Wu, Y.; Zhang, J.; Mao, S.; Geng, J.; Yao, X. Depletion of CDC5L inhibits bladder cancer tumorigenesis. J. Cancer 2020, 11, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Lleres, D.; Denegri, M.; Biggiogera, M.; Ajuh, P.; Lamond, A.I. Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep. 2010, 11, 445–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ajuh, P.; Kuster, B.; Panov, K.; Zomerdijk, J.C.; Mann, M.; Lamond, A.I. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 2000, 19, 6569–6581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulos, P.; Gutierrez, L.; Demmers, J.; Scheer, E.; Pourfarzad, F.; Papageorgiou, D.N.; Karkoulia, E.; Strouboulis, J.; van de Werken, H.J.; van der Linden, R.; et al. TAF10 Interacts with the GATA1 transcription factor and controls mouse erythropoiesis. Mol. Cell Biol. 2015, 35, 2103–2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardot, P.; Vincent, S.D.; Fournier, M.; Hubaud, A.; Joint, M.; Tora, L.; Pourquie, O. The TAF10-containing TFIID and SAGA transcriptional complexes are dispensable for early somitogenesis in the mouse embryo. Development 2017, 144, 3808–3818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pahi, Z.; Borsos, B.N.; Vedelek, B.; Shidlovskii, Y.V.; Georgieva, S.G.; Boros, I.M.; Pankotai, T. TAF10 and TAF10b partially redundant roles during Drosophila melanogaster morphogenesis. Transcription 2017, 8, 297–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatarakis, A.; Margaritis, T.; Martinez-Jimenez, C.P.; Kouskouti, A.; Mohan, W.S.; Haroniti, A.; Kafetzopoulos, D.; Tora, L.; Talianidis, I. Dominant and redundant functions of TFIID involved in the regulation of hepatic genes. Mol. Cell 2008, 31, 531–543. [Google Scholar] [CrossRef]
- Jeronimo, C.; Forget, D.; Bouchard, A.; Li, Q.; Chua, G.; Poitras, C.; Therien, C.; Bergeron, D.; Bourassa, S.; Greenblatt, J.; et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 2007, 27, 262–274. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Miki, Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004, 95, 866–871. [Google Scholar] [CrossRef]
- Wang, Y.; Cortez, D.; Yazdi, P.; Neff, N.; Elledge, S.J.; Qin, J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000, 14, 927–939. [Google Scholar]
- Narod, S.A. BRCA mutations in the management of breast cancer: The state of the art. Nat. Rev. Clin. Oncol. 2010, 7, 702–707. [Google Scholar] [CrossRef]
- Gupta, R.; Somyajit, K.; Narita, T.; Maskey, E.; Stanlie, A.; Kremer, M.; Typas, D.; Lammers, M.; Mailand, N.; Nussenzweig, A.; et al. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 2018, 173, 972–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Oostende-Triplet, C.; Jaramillo-Garcia, M.; Haji-Bik, H.; Beaudet, D.; Piekny, A. Anillin interacts with microtubules and is part of the astral pathway that defines cortical domains. J. Cell Sci. 2014, 127, 3699–3710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somma, M.P.; Fasulo, B.; Cenci, G.; Cundari, E.; Gatti, M. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol Cell 2002, 13, 2448–2460. [Google Scholar] [CrossRef] [Green Version]
- Engel, F.B.; Schebesta, M.; Keating, M.T. Anillin localization defect in cardiomyocyte binucleation. J. Mol. Cell Cardiol. 2006, 41, 601–612. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, P.P. How to scaffold the contractile ring for a safe cytokinesis-lessons from anillin-related proteins. J. Cell Sci. 2009, 122, 1071–1079. [Google Scholar] [CrossRef] [Green Version]
- Mishima, M.; Kaitna, S.; Glotzer, M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2002, 2, 41–54. [Google Scholar] [CrossRef] [Green Version]
- D’Avino, P.P.; Savoian, M.S.; Capalbo, L.; Glover, D.M. RacGAP50C is sufficient to signal cleavage furrow formation during cytokinesis. J. Cell Sci. 2006, 119, 4402–4408. [Google Scholar] [CrossRef] [Green Version]
- Manukyan, A.; Ludwig, K.; Sanchez-Manchinelly, S.; Parsons, S.J.; Stukenberg, P.T. A complex of p190RhoGAP-A and anillin modulates RhoA-GTP and the cytokinetic furrow in human cells. J. Cell Sci. 2015, 128, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Silverman-Gavrila, R.V.; Hales, K.G.; Wilde, A. Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol. Biol. Cell 2008, 19, 3735–3744. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Jiang, X.; Zhang, J.; Xie, S.; Hua, Y.; Wang, R.; Yang, Y. Identification of significant gene and pathways involved in HBV-related hepatocellular carcinoma by bioinformatics analysis. Peer J. 2019, 7, e7408. [Google Scholar] [CrossRef] [Green Version]
- Plessner, M.; Grosse, R. Dynamizing nuclear actin filaments. Curr. Opin. Cell Biol. 2019, 56, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Plessner, M.; Melak, M.; Chinchilla, P.; Baarlink, C.; Grosse, R. Nuclear F-actin formation and reorganization upon cell spreading. J. Biol Chem. 2015, 290, 11209–11216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baarlink, C.; Wang, H.; Grosse, R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 2013, 340, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mao, Y. Diaphanous formin mDia2 regulates CENP-A levels at centromeres. J. Cell Biol. 2016, 213, 415–424. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Wang, Z.; Shen, N.; Pi, W.; Jiang, W.; Huang, J.; Hu, Y.; Li, X.; Sun, L. Knockdown of ANLN by lentivirus inhibits cell growth and migration in human breast cancer. Mol. Cell. Biochem. 2015, 398, 11–19. [Google Scholar] [CrossRef]
- Kyheröinen, S.; Vartiainen, M.K. Nuclear actin dynamics in gene expression and genome organization. Semin. Cell Dev. Biol. 2019, 102, 105–112. [Google Scholar] [CrossRef]
- Toret, C.P.; D’Ambrosio, M.V.; Vale, R.D.; Simon, M.A.; Nelson, W.J. A genome-wide screen identifies conserved protein hubs required for cadherin-mediated cell-cell adhesion. J. Cell Biol. 2014, 204, 265–279. [Google Scholar] [CrossRef] [Green Version]
- Reyes, C.C.; Jin, M.; Breznau, E.B.; Espino, R.; Delgado-Gonzalo, R.; Goryachev, A.B.; Miller, A.L. Anillin regulates cell-cell junction integrity by organizing junctional accumulation of Rho-GTP and actomyosin. Curr. Biol. 2014, 24, 1263–1270. [Google Scholar] [CrossRef] [Green Version]
- Rehain, K.; Maddox, A.S. Neuron migration: Anillin protects leading edge actin. Curr. Biol. 2015, 25, R423–R425. [Google Scholar] [CrossRef] [Green Version]
- Chuang, H.Y.; Ou, Y.H. Overexpression of anillin in colorectal cancer promotes the cell proliferation, cell mobility and cell invasion. In Proceedings of the AACR Annual Meeting 2014, San Diego, CA, USA, 5–9 April 2014. [Google Scholar]
- Tian, D.; Diao, M.; Jiang, Y.; Sun, L.; Zhang, Y.; Chen, Z.; Huang, S.; Ou, G. Anillin regulates neuronal migration and neurite growth by linking RhoG to the actin cytoskeleton. Curr. Biol. 2015, 25, 1135–1145. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Naydenov, N.G.; Dozmorov, M.G.; Koblinski, J.E.; Ivanov, A.I. Anillin regulates breast cancer cell migration, growth, and metastasis by non-canonical mechanisms involving control of cell stemness and differentiation. Breast Cancer Res. 2020, 22, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Chadha, G.K.; Feygin, A.; Ivanov, A.I. F-actin binding protein, anillin, regulates integrity of intercellular junctions in human epithelial cells. Cell Mol. Life Sci. 2015, 72, 3185–3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Nguyen, L.H.; Zhou, K.; Tu, H.C.; Sehgal, A.; Nassour, I.; Li, L.; Gopal, P.; Goodman, J.; Singal, A.G.; et al. Knockdown of anillin actin binding protein blocks cytokinesis in hepatocytes and reduces liver tumor development in mice without affecting regeneration. Gastroenterology 2018, 154, 1421–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, Y.F.; Huang, Y.L.; Wang, J.L.; Deng, M.H.; Xia, T.L.; Zeng, M.S.; Chen, M.S.; Wang, H.B.; Huang, Y.H. Anillin is required for tumor growth and regulated by miR-15a/miR-16-1 in HBV-related hepatocellular carcinoma. Aging 2018, 10, 1884–1901. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Mo, Y.; Midorikawa, K.; Zhang, Z.; Huang, G.; Ma, N.; Zhao, W.; Hiraku, Y.; Oikawa, S.; Murata, M. The potent tumor suppressor miR-497 inhibits cancer phenotypes in nasopharyngeal carcinoma by targeting ANLN and HSPA4L. Oncotarget 2015, 6, 35893–35907. [Google Scholar] [CrossRef] [Green Version]
- Zeng, S.; Yu, X.; Ma, C.; Song, R.; Zhang, Z.; Zi, X.; Chen, X.; Wang, Y.; Yu, Y.; Zhao, J.; et al. Transcriptome sequencing identifies ANLN as a promising prognostic biomarker in bladder urothelial carcinoma. Sci. Rep. 2017, 7, 3151. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, Y.; Long, J.; Guo, K.; Ge, C.; Du, R. microRNA-218 suppresses the proliferation, invasion and promotes apoptosis of pancreatic cancer cells by targeting HMGB1. Chin. J. Cancer Res. 2015, 27, 247–257. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, H.; Yuan, S.; Zhou, B.; Zhao, W.; Pan, Y.; Qi, D. Overexpression of ANLN in lung adenocarcinoma is associated with metastasis. Thorac. Cancer 2019, 10, 1702–1709. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, R.S.; Karemore, G.; Gudjonsson, T.; Rask, M.B.; Neumann, B.; Heriche, J.K.; Pepperkok, R.; Ellenberg, J.; Gerlich, D.W.; Lukas, J.; et al. Profiling DNA damage response following mitotic perturbations. Nat. Commun. 2016, 7, 13887. [Google Scholar] [CrossRef]
- Pandi, N.S.; Manimuthu, M.; Harunipriya, P.; Murugesan, M.; Asha, G.V.; Rajendran, S. In silico analysis of expression pattern of a Wnt/beta-catenin responsive gene ANLN in gastric cancer. Gene 2014, 545, 23–29. [Google Scholar] [CrossRef]
- Zhou, Q.; Fukushima, P.; DeGraff, W.; Mitchell, J.B.; Stetler Stevenson, M.; Ashkenazi, A.; Steeg, P.S. Radiation and the Apo2L/TRAIL apoptotic pathway preferentially inhibit the colonization of premalignant human breast cells overexpressing cyclin D1. Cancer Res. 2000, 60, 2611–2615. [Google Scholar] [PubMed]
- Yang, G.; Xiong, G.; Cao, Z.; Zheng, S.; You, L.; Zhang, T.; Zhao, Y. miR-497 expression, function and clinical application in cancer. Oncotarget 2016, 7, 55900–55911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blagosklonny, M.V. Cell immortality and hallmarks of cancer. Cell Cycle 2003, 2, 296–299. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xie, L.Y.; Allan, S.; Beach, D.; Hannon, G.J. Myc activates telomerase. Genes Dev. 1998, 12, 1769–1774. [Google Scholar] [CrossRef] [Green Version]
- Biermann, J.; Nemes, S.; Parris, T.Z.; Engqvist, H.; Werner Ronnerman, E.; Kovacs, A.; Karlsson, P.; Helou, K. A 17-marker panel for global genomic instability in breast cancer. Genomics 2020, 112, 1151–1161. [Google Scholar] [CrossRef]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y. Cancer energy metabolism: Shutting power off cancer factory. Biomol Ther. 2018, 26, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Zhou, K.; Luo, X.; Li, L.; Tu, H.C.; Sehgal, A.; Nguyen, L.H.; Zhang, Y.; Gopal, P.; Tarlow, B.D.; et al. The polyploid state plays a tumor-suppressive role in the liver. Dev. Cell 2018, 47, 390. [Google Scholar] [CrossRef] [Green Version]
- Desdouets, C.; Avila, M.A. Inhibiting cytokinesis in the liver: A new way to reduce tumor development. Gastroenterology 2018, 154, 1229–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swiderska, E.; Strycharz, J.; Wróblewski, A.; Szemraj, J.; Drzewoski, J.; Sliwińska, A. Role of PI3K/AKT pathway in insulin-mediated glucose uptake. In Blood Glucose Levels; IntechOpen: London, UK, 2018. [Google Scholar]
- Lien, E.C.; Lyssiotis, C.A.; Cantley, L.C. Metabolic reprogramming by the PI3K-Akt-mTOR pathway in cancer. Recent Res. Cancer Res. 2016, 207, 39–72. [Google Scholar] [CrossRef]
- Junttila, M.R.; de Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Bhome, R.; Bullock, M.D.; Al Saihati, H.A.; Goh, R.W.; Primrose, J.N.; Sayan, A.E.; Mirnezami, A.H. A top-down view of the tumor microenvironment: Structure, cells and signaling. Front. Cell Dev. Biol. 2015, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423. [Google Scholar] [CrossRef]
- Kowal, J.; Kornete, M.; Joyce, J.A. Re-education of macrophages as a therapeutic strategy in cancer. Immunotherapy 2019, 11, 677–689. [Google Scholar] [CrossRef]
- Folkman, J. Tumor angiogenesis: Therapeutic implications. New Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar]
- Naoyo, N.; Hirohisa, Y.; Takashi, N.; Toshiharu, K.; Masamichi, K. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar]
- Guo, S.; Colbert, L.S.; Fuller, M.; Zhang, Y.; Gonzalez-Perez, R.R. Vascular endothelial growth factor receptor-2 in breast cancer. Biochim. Biophys. Acta Rev. Cancer 2010, 1806, 108–121. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Kitadai, Y.; Bucana, C.D.; Cleary, K.R.; Ellis, L.M. Expression of vascular endothelial growth factor and its receptor, KDR, correlates with vascularity, metastasis, and proliferation of human colon cancer. Cancer Res. 1995, 55, 3964–3968. [Google Scholar]
- Karar, J.; Maity, A. PI3K/AKT/mTOR pathway in angiogenesis. Front. Mol. Neurosci. 2011, 4, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copie-Bergman, C.; Gaulard, P.; Lavergne-Slove, A.; Brousse, N.; Fléjou, J.; Dordonne, K.; De Mascarel, A.; Wotherspoon, A. Proposal for a new histological grading system for post-treatment evaluation of gastric MALT lymphoma. Gut 2003, 52, 1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greten, F.R.; Grivennikov, S.I. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Holopainen, S.; Hytönen, M.K.; Syrjä, P.; Arumilli, M.; Järvinen, A.-K.; Rajamäki, M.; Lohi, H. ANLN truncation causes a familial fatal acute respiratory distress syndrome in Dalmatian dogs. PLoS Genet. 2017, 13, e1006625. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, D.S.; Glavan, B.J.; Holden, T.D.; Fong, C.; Black, R.A.; Rona, G.; Tejera, P.; Christiani, D.C.; Wurfel, M.M. Inflammation and immune-related candidate gene associations with acute lung injury susceptibility and severity: A validation study. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Arcaroli, J.J.; Liu, N.; Yi, N.; Abraham, E. Association between IL-32 genotypes and outcome in infection-associated acute lung injury. Crit. Care 2011, 15, R138. [Google Scholar] [CrossRef] [Green Version]
- Brenner, D.R.; Scherer, D.; Muir, K.; Schildkraut, J.; Boffetta, P.; Spitz, M.R.; Le Marchand, L.; Chan, A.T.; Goode, E.L.; Ulrich, C.M. A review of the application of inflammatory biomarkers in epidemiologic cancer research. Cancer Epidemiol. Prev. Biomark. 2014, 23, 1729–1751. [Google Scholar] [CrossRef] [Green Version]
- Zelová, H.; Hošek, J. TNF-α signalling and inflammation: Interactions between old acquaintances. Inflamm. Res. 2013, 62, 641–651. [Google Scholar] [CrossRef]
- Dame, T.M.; Orenzoff, B.L.; Palmer, L.E.; Furie, M.B. IFN-γ alters the response of Borrelia burgdorferi-activated endothelium to favor chronic inflammation. J. Immunol. 2007, 178, 1172–1179. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Seliger, B. Strategies of tumor immune evasion. Bio Drugs 2005, 19, 347–354. [Google Scholar] [CrossRef]
- Meissner, M.; Reichert, T.E.; Kunkel, M.; Gooding, W.; Whiteside, T.L.; Ferrone, S.; Seliger, B. Defects in the human leukocyte antigen class I antigen processing machinery in head and neck squamous cell carcinoma: Association with clinical outcome. Clin. Cancer Res. 2005, 11, 2552–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shevach, E.M. Fatal attraction: Tumors beckon regulatory T cells. Nat. Med. 2004, 10, 900–901. [Google Scholar] [CrossRef] [PubMed]
- Georgouli, M.; Herraiz, C.; Crosas-Molist, E.; Fanshawe, B.; Maiques, O.; Perdrix, A.; Pandya, P.; Rodriguez-Hernandez, I.; Ilieva, K.M.; Cantelli, G.; et al. Regional activation of myosin II in cancer cells drives tumor progression via a secretory cross-talk with the immune microenvironment. Cell 2019, 176, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Venkataramani, V.; Tanev, D.I.; Strahle, C.; Studier-Fischer, A.; Fankhauser, L.; Kessler, T.; Korber, C.; Kardorff, M.; Ratliff, M.; Xie, R.; et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 2019, 573, 532–538. [Google Scholar] [CrossRef]
- Lee, C.H.; Cho, J.; Lee, K. Tumour regression via integrative regulation of neurological, inflammatory, and hypoxic tumour microenvironment. Biomol. Ther. 2020, 28, 119–130. [Google Scholar] [CrossRef]
- Arese, M.; Bussolino, F.; Pergolizzi, M.; Bizzozero, L.; Pascal, D. Tumor progression: The neuronal input. Ann. Transl. Med. 2018, 6, 89. [Google Scholar] [CrossRef] [Green Version]
- Scanlon, C.S.; Banerjee, R.; Inglehart, R.C.; Liu, M.; Russo, N.; Hariharan, A.; van Tubergen, E.A.; Corson, S.L.; Asangani, I.A.; Mistretta, C.M.; et al. Galanin modulates the neural niche to favour perineural invasion in head and neck cancer. Nat. Commun. 2015, 6, 6885. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.W.; Nagaraja, A.S.; Lutgendorf, S.K.; Green, P.A.; Sood, A.K. Sympathetic nervous system regulation of the tumour microenvironment. Nat. Rev. Cancer 2015, 15, 563–572. [Google Scholar] [CrossRef] [Green Version]
- Makale, M.T.; McDonald, C.R.; Hattangadi-Gluth, J.A.; Kesari, S. Mechanisms of radiotherapy-associated cognitive disability in patients with brain tumours. Nat. Rev. Neurol. 2017, 13, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Erwig, M.S.; Patzig, J.; Steyer, A.M.; Dibaj, P.; Heilmann, M.; Heilmann, I.; Jung, R.B.; Kusch, K.; Mobius, W.; Jahn, O.; et al. Anillin facilitates septin assembly to prevent pathological outfoldings of central nervous system myelin. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Catenaccio, A.; Llavero Hurtado, M.; Diaz, P.; Lamont, D.J.; Wishart, T.M.; Court, F.A. Molecular analysis of axonal-intrinsic and glial-associated co-regulation of axon degeneration. Cell Death Dis. 2017, 8, e3166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patzig, J.; Erwig, M.S.; Tenzer, S.; Kusch, K.; Dibaj, P.; Mobius, W.; Goebbels, S.; Schaeren-Wiemers, N.; Nave, K.A.; Werner, H.B. Septin/anillin filaments scaffold central nervous system myelin to accelerate nerve conduction. Elife 2016, 5. [Google Scholar] [CrossRef]
- Qiu, R.; Runxiang, Q.; Geng, A.; Liu, J.; Xu, C.W.; Menon, M.B.; Gaestel, M.; Lu, Q. SEPT7 interacts with KIF20A and regulates the proliferative state of neural progenitor cells during cortical development. Cereb. Cortex 2019. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Nitschke, K.; Heinkele, J.; Weis, C.A.; Worst, T.S.; Eckstein, M.; Porubsky, S.; Erben, P. ANLN and TLE2 in muscle invasive bladder cancer: A functional and clinical evaluation based on in silico and in vitro data. Cancers 2019, 11, 1840. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, S.; Seki, N.; Sugimoto, T.; Horiguchi, S.; Tanzawa, H.; Hanazawa, T.; Okamoto, Y. Identification of molecular targets in head and neck squamous cell carcinomas based on genome-wide gene expression profiling. Oncol. Rep. 2007, 18, 1489–1497. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
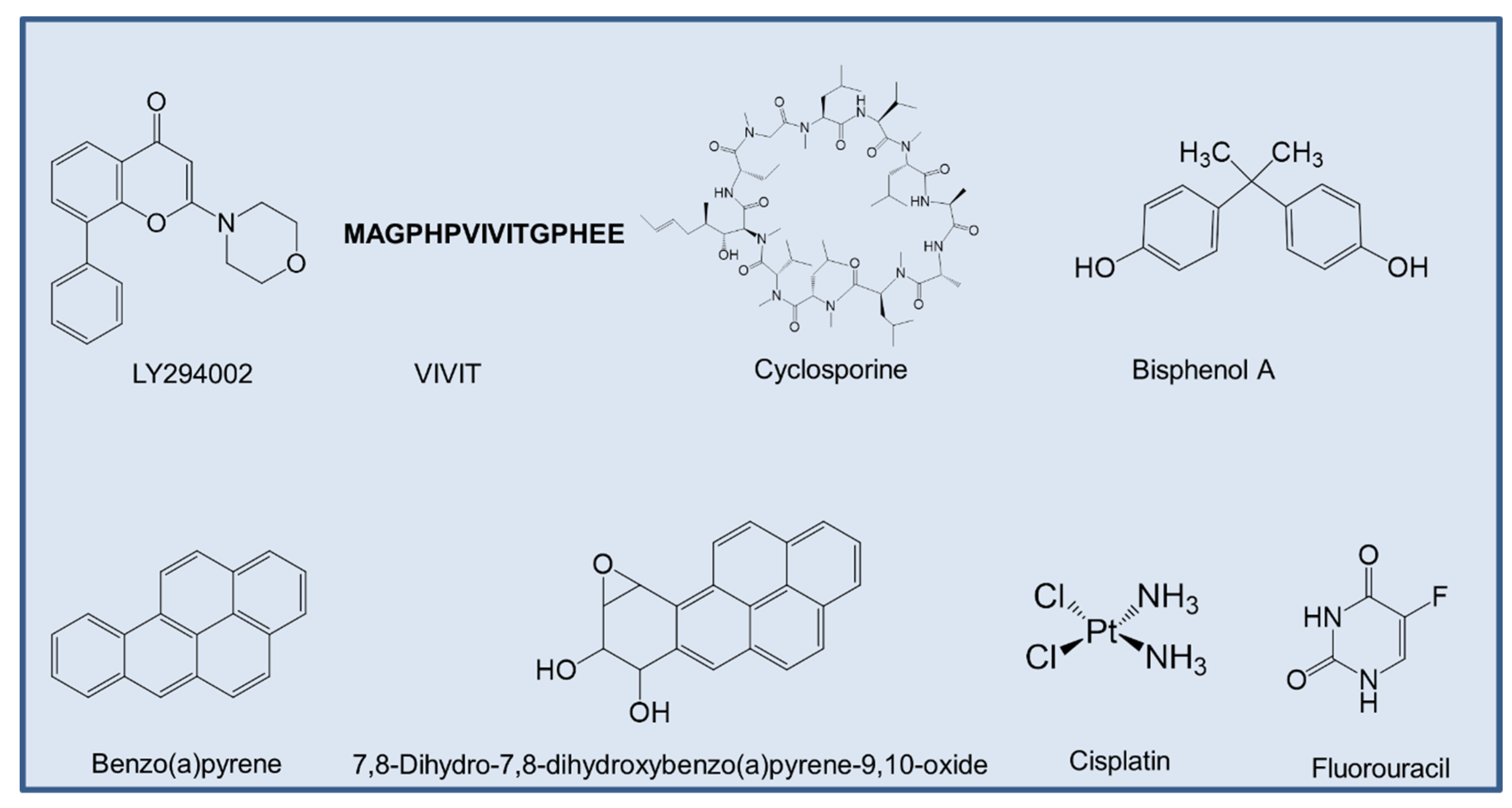
- Vlahos, C.J.; Matter, W.F.; Hui, K.Y.; Brown, R.F. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 1994, 269, 5241–5248. [Google Scholar]
- Yu, H.; van Berkel, T.J.; Biessen, E.A. Therapeutic potential of VIVIT, a selective peptide inhibitor of nuclear factor of activated T cells, in cardiovascular disorders. Cardiovasc. Drug Rev. 2007, 25, 175–187. [Google Scholar] [CrossRef]
- Hall, G.; Lane, B.M.; Khan, K.; Pediaditakis, I.; Xiao, J.; Wu, G.; Wang, L.; Kovalik, M.E.; Chryst-Stangl, M.; Davis, E.E.; et al. The human FSGS-causing ANLN R431C mutation induces dysregulated PI3K/AKT/mTOR/Rac1 signaling in podocytes. J. Am. Soc. Nephrol. 2018, 29, 2110–2122. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The comparative toxicogenomics database: Update 2019. Nucleic. Acids Res. 2019, 47, D948–D954. [Google Scholar] [CrossRef]
- Canafax, D.M.; Ascher, N.L. Cyclosporine immunosuppression. Clin. Pharm. 1983, 2, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; He, B.; Lai, L.; Chen, Q.; Liu, Y.; Guo, Q.; Wang, Q. Cyclosporine A inhibits breast cancer cell growth by downregulating the expression of pyruvate kinase subtype M2. Int. J. Mol. Med. 2012, 30, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharm. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Huret, J.-L.; Ahmad, M.; Arsaban, M.; Bernheim, A.; Cigna, J.; Desangles, F.; Guignard, J.-C.; Jacquemot-Perbal, M.-C.; Labarussias, M.; Leberre, V. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic. Acids Res. 2012, 41, D920–D924. [Google Scholar] [CrossRef]
- Ohue, Y.; Nishikawa, H. Regulatory T (treg) cells in cancer: Can treg cells be a new therapeutic target? Cancer Sci. 2019, 110, 2080. [Google Scholar] [CrossRef]



| Partners of ANLN | System | Binding Regions of ANLN | Binding Regions of Partners | Ref | Role of ANLN to Partners | Characteristics in the Association between ANLN and the Partners |
|---|---|---|---|---|---|---|
| Actin | Drosophila | Amino acids 258–340 | Full length | [12] |
| |
| Human | Amino acids 231–454 | Full length | [22] | |||
| X. Leavis | Amino acids 255–418 | Full length | [23] | |||
| Septins (a group of GTP-binding proteins) | Human | Amino acids 929–1125 | Full length | [22] |
|
|
| Human | Amino acid 748–1116 | Full length | [23] | |||
| Drosophila | Amino acids 815–1201 | Full length | [26] | |||
| Myosin | X. Leavis | Amino acids 142–254 | Full length | [27] |
|
|
| RhoA | Human | Amino acids 608–943 | Full length | [16] |
|
|
| RacGap | Drosophila | Amino acids 517–1212 | Amino acids 83–309 | [30] |
|
|
| Drosophila | Amino acids 929–129 | Amino acids 136–371 | [31] | |||
| Ect2 | Human | Amino acids 926–980 | Full length | [1] |
|
|
| Human | Amino acids 421–621 | Amino acids 608–940 | [32] | |||
| CD2A | Human | Amino acids 1–155 | Amino acids 1–175 | [33] |
| |
| Drosophila | Amino acids 1–328 | Full length | [34] | |||
| Drosophila | Amino acids 930–1239 | Full length | [34] | |||
| Microtubules | Drosophila | Full length | Full length | [35] |
|
|
| Hallmarks of Cancer | Cancer Type | Cancer Cell Line | Effect | References |
|---|---|---|---|---|
| Proliferation | Liver cancer | H2.35, QGY-7703, BEL-7404, Hep3B, MHCC-97L, Huh7, HepG2, PLC/PRC/5, BEL-7405, HepG2.215, SMMC-7721 and Sk-Hep-1 | Upregulate | [115,116] |
| Lung cancer | A549, LC319, PC-3, PC-9, PC-14, A427, NCI-H1373 | Upregulate | [29] | |
| Nasopharyngeal carcinoma | HK1, CNE1, HONE1 | Upregulate | [117] | |
| Bladder urothelial carcinoma | J82, 5637 | Upregulate | [118] | |
| Pancreatic cancer | BxPC-3, SW1990 | Upregulate | [5] | |
| Breast cancer | MDA-MB-231 | Upregulate | [106] | |
| Apoptosis | Breast cancer | MDA-MB-231 | Downregulate | [106] |
| Liver cancer | H2.35, QGY-7703, BEL-7404, Hep3B, MHC C-97L, Huh7, HepG2, PLC/PRC/5, BEL-7405, HepaG2.215, SMMC-7721 and Sk-Hep-1 | Downregulate | [116] | |
| Pancreatic cancer | AsPC-1, BxPC-3, and PANC-1 | Downregulate | [119] | |
| Nasopharyngeal carcinoma | HK1, CNE1, HONE1 | Downregulate | [117] | |
| Bladder urothelial carcinoma | J82, 5637 | No effect | [118] | |
| Invasion and metastasis | Pancreatic cancer | BxPC-3, SW1990 | Upregulate | [5] |
| Breast cancer | MDA-MB-231 | Upregulate | [106] | |
| Lung cancer | NIH3T3, COS-7, A549, PC9 | Upregulate | [29,120] | |
| Bladder urothelial carcinoma | J82, 5637 cells | Upregulate | [118] | |
| Nasopharyngeal carcinoma | HK1, CNE1, HONE1 | Upregulate | [117] | |
| Growth suppressor | Not experimentally determined | |||
| Cell immortality | Not experimentally determined | |||
| Genome instability and mutation | Bone cancer | U2OS | Downregulate | [121] |
| Cancer metabolism | Not experimentally determined | |||
| Angiogenesis | Not experimentally determined | |||
| Inflammation | Not experimentally determined | |||
| Immune evasion | Not experimentally determined | |||
| Nerve connection | Not experimentally determined | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuan, N.M.; Lee, C.H. Role of Anillin in Tumour: From a Prognostic Biomarker to a Novel Target. Cancers 2020, 12, 1600. https://doi.org/10.3390/cancers12061600
Tuan NM, Lee CH. Role of Anillin in Tumour: From a Prognostic Biomarker to a Novel Target. Cancers. 2020; 12(6):1600. https://doi.org/10.3390/cancers12061600
Chicago/Turabian StyleTuan, Nguyen Minh, and Chang Hoon Lee. 2020. "Role of Anillin in Tumour: From a Prognostic Biomarker to a Novel Target" Cancers 12, no. 6: 1600. https://doi.org/10.3390/cancers12061600
APA StyleTuan, N. M., & Lee, C. H. (2020). Role of Anillin in Tumour: From a Prognostic Biomarker to a Novel Target. Cancers, 12(6), 1600. https://doi.org/10.3390/cancers12061600





