Discovery of Novel Recurrent Mutations and Clinically Meaningful Subgroups in Nodal Marginal Zone Lymphoma
Abstract
:1. Introduction
2. Results
2.1. Clinicopathological Characteristics of the NMZL Cohorts
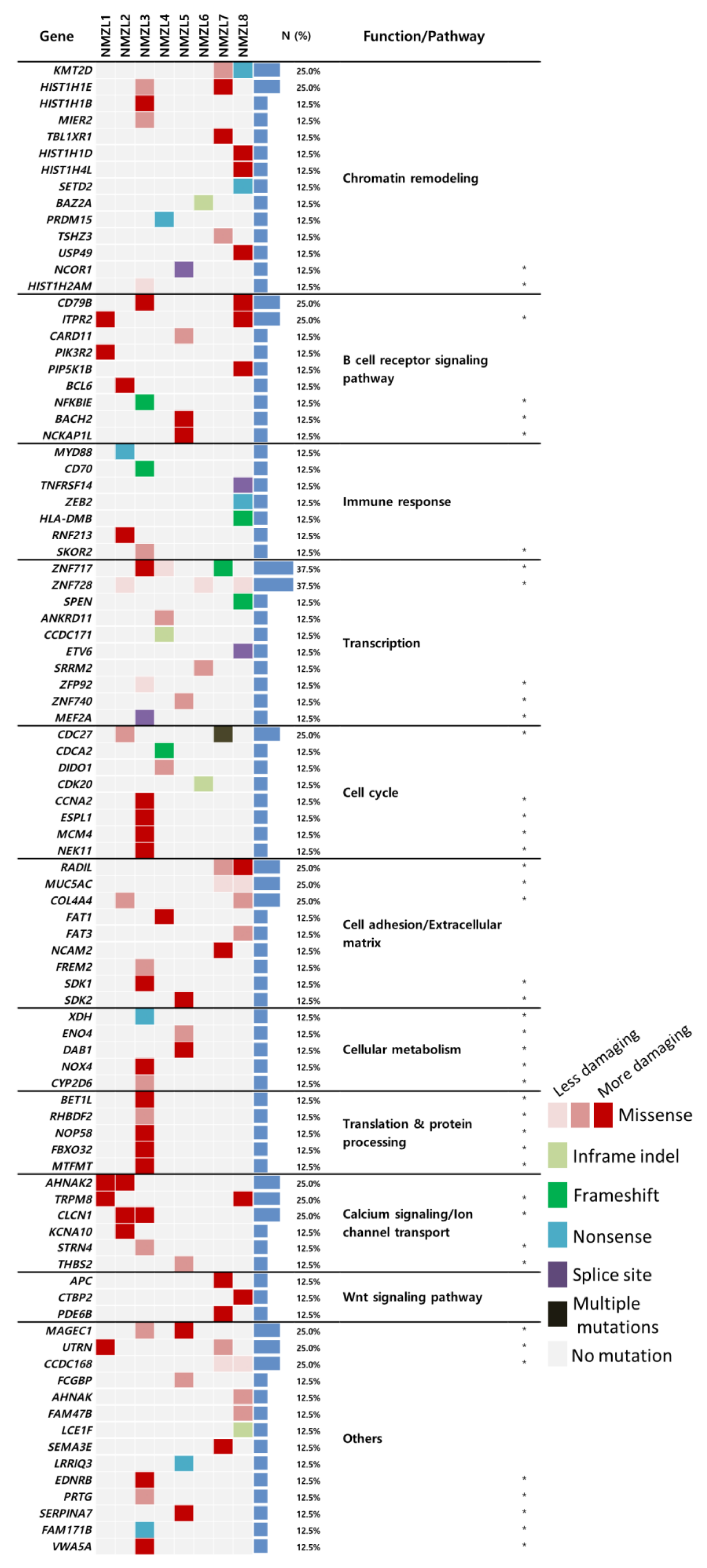
2.2. Mutational Landscape of NMZL and Identification of Candidate Genes
2.3. Transcriptomic Features of NMZLs in Comparison with Non-Neoplastic LNs
2.4. Major Upstream Regulators and Their Roles in the Pathogenesis of NMZL
2.5. Characterization of NMZL Subgroups according to GEPs
2.6. NMZL Subgroups and Their Clinicopathological Implications
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. WES and Processing of Variants
4.3. RNA-seq and Gene Expression Analysis
4.4. Direct Sequencing and Immunohistochemistry (IHC)
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Teras, L.R.; DeSantis, C.E.; Cerhan, J.R.; Morton, L.M.; Jemal, A.; Flowers, C.R. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA A Cancer J. Clin. 2016, 66, 443–459. [Google Scholar] [CrossRef]
- Kim, J.-M.; Ko, Y.-H.; Lee, S.-S.; Huh, J.; Kang, C.S.; Kim, C.W.; Kang, Y.K.; Go, J.H.; Kim, M.K.; Kim, W.-S.; et al. WHO Classification of Malignant Lymphomas in Korea: Report of the Third Nationwide Study. Korean J. Pathol. 2011, 45, 254–257. [Google Scholar] [CrossRef]
- Camacho, F.I.; Algara, P.; Mollejo, M.; García, J.F.; Montalbán, C.; Martinez, N.; Sánchez-Beato, M.; Piris, M.A. Nodal marginal zone lymphoma: A heterogeneous tumor: A comprehensive analysis of a series of 27 cases. Am. J. Surg. Pathol. 2003, 27, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Arcaini, L.; Paulli, M.; Burcheri, S.; Rossi, A.; Spina, M.; Passamonti, F.; Lucioni, M.; Motta, T.; Canzonieri, V.; Montanari, M.; et al. Primary nodal marginal zone B-cell lymphoma: Clinical features and prognostic assessment of a rare disease. Br. J. Haematol. 2007, 136, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Traverse-Glehen, A.; Felman, P.; Callet-Bauchu, E.; Gazzo, S.; Baseggio, L.; Bryon, P.A.; Thieblemont, C.; Coiffier, B.; Salles, G.; Berger, F. A clinicopathological study of nodal marginal zone B-cell lymphoma. A report on 21 cases. Histopathology 2006, 48, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Spina, V.; Khiabanian, H.; Messina, M.; Monti, S.; Cascione, L.; Bruscaggin, A.; Spaccarotella, E.; Holmes, A.B.; Arcaini, L.; Lucioni, M.; et al. The genetics of nodal marginal zone lymphoma. Blood 2016, 128, 1362–1373. [Google Scholar] [CrossRef] [PubMed]
- Pillonel, V.; Juskevicius, D.; Ng, C.K.Y.; Bodmer, A.; Zettl, A.; Jucker, D.; Dirnhofer, S.; Tzankov, A. High-throughput sequencing of nodal marginal zone lymphomas identifies recurrent BRAF mutations. Leukemia 2018, 32, 2412–2426. [Google Scholar] [CrossRef] [Green Version]
- Rinaldi, A.; Mian, M.; Chigrinova, E.; Arcaini, L.; Bhagat, G.; Novak, U.; Rancoita, P.M.V.; De Campos, C.P.; Forconi, F.; Gascoyne, R.D.; et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood 2011, 117, 1595–1604. [Google Scholar] [CrossRef]
- Arribas, A.J.; Campos-Martin, Y.; Gomez-Abad, C.; Algara, P.; Sanchez-Beato, M.; Rodriguez-Pinilla, M.S.; Montes-Moreno, S.; Martínez, N.; Alves-Ferreira, J.; Piris, M.A.; et al. Nodal marginal zone lymphoma: Gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood 2012, 119, e9–e21. [Google Scholar] [CrossRef]
- Braggio, E.; Dogan, A.; Keats, J.J.; Chng, W.J.; Huang, G.; Matthews, J.M.; Maurer, M.J.; Law, M.E.; Bosler, D.S.; Barrett, M.; et al. Genomic analysis of marginal zone and lymphoplasmacytic lymphomas identified common and disease-specific abnormalities. Mod. Pathol. 2012, 25, 651–660. [Google Scholar] [CrossRef]
- Mansouri, L.; Sutton, L.-A.; Ljungström, V.; Bondza, S.; Arngården, L.; Bhoi, S.; Larsson, J.; Cortese, D.; Kalushkova, A.; Plevova, K.; et al. Functional loss of IκBε leads to NF-κB deregulation in aggressive chronic lymphocytic leukemia. J. Exp. Med. 2015, 212, 833–843. [Google Scholar] [CrossRef]
- Yoon, S.; Kim, S.-Y.; Nam, D. Improving Gene-Set Enrichment Analysis of RNA-Seq Data with Small Replicates. PLoS ONE 2016, 11, e0165919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaffer, A.L.; Wright, G.; Yang, L.; Powell, J.; Ngo, V.; Lamy, L.; Lam, L.T.; Davis, R.E.; Staudt, L.M. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol. Rev. 2006, 210, 67–85. [Google Scholar] [CrossRef]
- Weller, S. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood 2004, 104, 3647–3654. [Google Scholar] [CrossRef]
- Yang, Y.; Shaffer, A.L.; Emre, N.C.T.; Ceribelli, M.; Zhang, M.; Wright, G.; Xiao, W.; Powell, J.; Platig, J.; Kohlhammer, H.; et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 2012, 21, 723–737. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dierlamm, J.; Rosenberg, C.; Stul, M.; Pittaluga, S.; Wlodarska, I.; Michaux, L.; Dehaen, M.; Verhoef, G.; Thomas, J.; de Kelver, W.; et al. Characteristic pattern of chromosomal gains and losses in marginal zone B cell lymphoma detected by comparative genomic hybridization. Leukemia 1997, 11, 747–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whiteside, S.T. I kappa B epsilon, a novel member of the Ikappa B family, controls RelA and cRel NF-kappa B activity. EMBO J. 1997, 16, 1413–1426. [Google Scholar] [CrossRef]
- Vela, V.; Juskevicius, D.; Gerlach, M.M.; Meyer, P.; Graber, A.; Cathomas, G.; Dirnhofer, S.; Tzankov, A. High throughput sequencing reveals high specificity of TNFAIP3 mutations in ocular adnexal marginal zone B-cell lymphomas. Hematol. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, L.; Noerenberg, D.; Young, E.; Mylonas, E.; Abdulla, M.; Frick, M.; Asmar, F.; Ljungstro m, V.; Schneider, M.; Yoshida, K.; et al. Frequent NFKBIE deletions are associated with poor outcome in primary mediastinal B-cell lymphoma. Blood 2016, 128, 2666–2670. [Google Scholar] [CrossRef] [Green Version]
- Levine, D.A.; The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Allen, E.M.; Wagle, N.; Sucker, A.; Treacy, D.J.; Johannessen, C.M.; Goetz, E.M.; Place, C.S.; Taylor-Weiner, A.; Whittaker, S.; Kryukov, G.V.; et al. The Genetic Landscape of Clinical Resistance to RAF Inhibition in Metastatic Melanoma. Cancer Discov. 2014, 4, 94–109. [Google Scholar] [CrossRef] [Green Version]
- Robertson, A.G.; Kim, J.; Guo, G.; Laird, P.W.; Akbani, R.; Gordenin, D.A.; Shukla, S.A.; Sanchez-Vega, F.; Czerniak, B.A.; de Sa Carvalho, B.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556.e25. [Google Scholar] [CrossRef]
- Morin, R.D.; Mungall, K.; Pleasance, E.; Mungall, A.J.; Goya, R.; Huff, R.D.; Scott, D.W.; Ding, J.; Roth, A.; Chiu, R.; et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 2013, 122, 1256–1265. [Google Scholar] [CrossRef] [Green Version]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Lohr, J.G.; Stojanov, P.; Carter, S.L.; Cruz-Gordillo, P.; Lawrence, M.S.; Auclair, D.; Sougnez, C.; Knoechel, B.; Gould, J.; Saksena, G.; et al. Widespread Genetic Heterogeneity in Multiple Myeloma: Implications for Targeted Therapy. Cancer Cell 2014, 25, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Puente, X.S.; Beà, S.; Valdés-Mas, R.; Villamor, N.; Gutiérrez-Abril, J.; Martín-Subero, J.I.; Munar, M.; Rubio-Pérez, C.; Jares, P.; Aymerich, M.; et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015, 526, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.; Rabionet, R.; Espinet, B.; Zapata, L.; Puiggros, A.; Melero, C.; Puig, A.; Sarria-Trujillo, Y.; Ossowski, S.; Garcia-Muret, M.P.; et al. Identification of Gene Mutations and Fusion Genes in Patients with Sézary Syndrome. J. Invest. Dermatol. 2016, 136, 1490–1499. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Wang, H.; Lin, Q.; Fan, F.; Zhang, F.; Peng, X.; Fang, X.; Liu, J.; Ouyang, K. Loss of IP 3Receptor–Mediated Ca2+ Release in Mouse B Cells Results in Abnormal B Cell Development and Function. J. Immunol. 2017, 199, 570–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, F.; Morita, M.; Monkawa, T.; Michikawa, T.; Furuichi, T.; Mikoshiba, K. Mutational Analysis of the Ligand Binding Site of the Inositol 1,4,5-Trisphosphate Receptor. J. Biol. Chem. 1996, 271, 18277–18284. [Google Scholar] [CrossRef] [Green Version]
- Clipson, A.; Wang, M.; de Leval, L.; Ashton-Key, M.; Wotherspoon, A.; Vassiliou, G.; Bolli, N.; Grove, C.; Moody, S.; Escudero-Ibarz, L.; et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia 2014, 29, 1177–1185. [Google Scholar] [CrossRef] [Green Version]
- Martínez, N.; Almaraz, C.; Vaqué, J.P.; Varela, I.; Derdak, S.; Beltran, S.; Mollejo, M.; Campos-Martin, Y.; Agueda, L.; Rinaldi, A.; et al. Whole-exome sequencing in splenic marginal zone lymphoma reveals mutations in genes involved in marginal zone differentiation. Leukemia 2013, 28, 1334–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiel, M.J.; Velusamy, T.; Betz, B.L.; Zhao, L.; Weigelin, H.G.; Chiang, M.Y.; Huebner-Chan, D.R.; Bailey, N.G.; Yang, D.T.; Bhagat, G.; et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J. Exp. Med. 2012, 209, 1553–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyeon, J.; Lee, B.; Shin, S.-H.; Yoo, H.Y.; Kim, S.J.; Kim, W.S.; Park, W.-Y.; Ko, Y.-H. Targeted deep sequencing of gastric marginal zone lymphoma identified alterations of TRAF3 and TNFAIP3 that were mutually exclusive for MALT1 rearrangement. Mod. Pathol. 2018, 31, 1418–1428. [Google Scholar] [CrossRef]
- Jung, H.; Yoo, H.Y.; Lee, S.H.; Shin, S.; Kim, S.-C.; Lee, S.; Joung, J.-G.; Nam, J.-Y.; Ryu, D.; Yun, J.W.; et al. The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP. Oncotarget 2017, 8, 17038–17049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, V.; Lu, R. IRF4 and IRF8: Governing the virtues of B lymphocytes. Front. Biol. 2014, 9, 269–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonetti, G.; Carette, A.; Silva, K.; Wang, H.; De Silva, N.S.; Heise, N.; Siebel, C.W.; Shlomchik, M.J.; Klein, U. IRF4 controls the positioning of mature B cells in the lymphoid microenvironments by regulating NOTCH2 expression and activity. J. Exp. Med. 2013, 210, 2887–2902. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Papenhausen, P.; Shao, H. The Role of c-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes 2017, 8, 116. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Guo, L.; Liu, H.; Zheng, B.; Ying, J.; Lv, N. C-MYC overexpression predicts aggressive transformation and a poor outcome in mucosa-associated lymphoid tissue lymphomas. Int. J. Clin. Exp. Pathol. 2014, 7, 5634–5644. [Google Scholar]
- Nathwani, B.N.; Anderson, J.R.; Armitage, J.O.; Cavalli, F.; Diebold, J.; Drachenberg, M.R.; Harris, N.L.; MacLennan, K.A.; Muller-Hermelink, H.-K.; Ullrich, F.A.; et al. Marginal zone B-cell lymphoma: A clinical comparison of nodal and mucosa-associated lymphoid tissue types. Non-Hodgkin’s Lymphoma Classification Project. J. Clin. Oncol. 1999, 17, 2486–2492. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 2nd ed.; International Agency for Research on Cancer: Lyon, France, 2017. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2013, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]





| Discovery Set | Validation Set | Total | p | ||
|---|---|---|---|---|---|
| Age | Median (min–max) | 64 (42–78) | 63 (29–85) | 64 (29–85) | 0.902 |
| <60 years | 4 (50.0%) | 15 (50.0%) | 19 (50.0%) | 1.000 | |
| ≥60 years | 4 (50.0%) | 15 (50.0%) | 19 (50.0%) | ||
| Sex | Male | 4 (50.0%) | 16 (53.3%) | 20 (52.6%) | 1.000 |
| Female | 4 (50.0%) | 14 (46.7%) | 18 (47.4%) | ||
| B-symptoms | Absent | 7 (87.5%) | 26 (86.7%) | 33 (86.8%) | 1.000 |
| Present | 1 (12.5%) | 4 (13.3%) | 5 (13.2%) | ||
| ECOG PS | 0 | 5 (62.5%) | 15 (53.6%) | 20 (55.6%) | 0.454 |
| 1 | 3 (37.5%) | 10 (35.7%) | 13 (36.1%) | ||
| 2 | 0 (0.0%) | 3 (10.7%) | 3 (8.3%)) | ||
| LDH | No elevation | 4 (50.0%) | 16 (57.1%) | 20 (55.6%) | 1.000 |
| Elevation | 4 (50.0%) | 12 (42.9%) | 16 (44.4%) | ||
| Ann Arbor Stage | I | 1 (12.5%) | 5 (16.7%) | 6 (15.8%) | 0.250 |
| II | 0 (0.0%) | 9 (30.0%) | 9 (23.7%) | ||
| III | 3 (37.5%) | 5 (16.7%) | 8 (21.1%) | ||
| IV | 4 (50.0%) | 11 (36.7%) | 15 (39.5%) | ||
| IPI | 0 | 0 (0.0%) | 6 (21.4%) | 6 (16.7%) | 0.353 |
| 1 | 2 (25.0%) | 7 (25.0%) | 9 (25.0%) | ||
| 2 | 4 (50.0%) | 7 (25.0%) | 11 (30.6%) | ||
| 3 | 1 (12.5%) | 6 (21.4%) | 7 (19.4%) | ||
| 4 | 1 (12.5%) | 2 (7.1%) | 3 (8.3%) | ||
| BM involvement | Absent | 4 (50.0%) | 13 (59.1%) | 17 (56.7%) | 0.698 |
| Present | 4 (50.0%) | 9 (40.9%) | 13 (43.3%) | ||
| Progression | No | 5 (62.5%) | 25 (83.3%) | 30 (78.9%) | 0.327 |
| Yes | 3 (37.5%) | 5 (16.7%) | 8 (21.1%) | ||
| HBV or HCV infection | Absent | 5 (62.5%) | 26 (86.7%) | 31 (81.6%) | 0.146 |
| Present | 3 (37.5%) | 4 (13.3%) | 7 (18.4%) | ||
| Large cell component | <20% | 4 (50.0%) | 12 (40.0%) | 16 (42.1%) | 0.698 |
| ≥20% | 4 (50.0%) | 18 (60.0%) | 22 (57.9%) | ||
| Total | 8 (21.1%) | 30 (78.9%) | 38 (100.0%) | ||
| Subgroup 1 (Ki-67Low) | Subgroup 2 (Ki-67High) | Total | p | ||
|---|---|---|---|---|---|
| Age | Median (range, years) | 58 (29–79) | 70 (30–85) | 64 (29–85) | 0.043 |
| <60 years | 13 (68.4%) | 6 (31.6%) | 19 (50.0%) | 0.050 | |
| ≥60 years | 6 (31.6%) | 13 (68.4%) | 19 (50.0%) | ||
| Sex | Male | 11 (57.9%) | 9 (47.4%) | 20 (52.6%) | 0.516 |
| Female | 8 (42.1%) | 10 (52.6%) | 18 (47.4%) | ||
| B-symptoms | Absent | 18 (94.7%) | 15 (78.9%) | 33 (86.8%) | 0.340 |
| Present | 1 (5.3%) | 4 (21.1%) | 5 (13.2%) | ||
| ECOG PS | 0 | 13 (72.2%) | 7 (38.9%) | 20 (55.6%) | 0.022 |
| 1 | 5 (27.8%) | 8 (44.4%) | 13 (36.1%) | ||
| 2 | 0 (0.0%) | 3 (16.7%) | 3 (8.1%) | ||
| LDH | No elevation | 14 (82.4%) | 6 (31.6%) | 20 (55.6%) | 0.002 |
| Elevation | 3 (17.6%) | 13 (68.4%) | 16 (44.4%) | ||
| Ann Arbor Stage | I | 6 (31.6%) | 0 (0.0%) | 6 (15.8%) | 0.021 |
| II | 5 (26.3%) | 4 (21.1%) | 9 (23.7%) | ||
| III | 2 (10.5%) | 6 (31.6%) | 8 (21.1%) | ||
| IV | 6 (31.6%) | 9 (47.4%) | 15 (39.5%) | ||
| IPI | 0 | 5 (29.4%) | 1 (5.3%) | 6 (16.7%) | 0.001 |
| 1 | 6 (35.3%) | 3 (15.8%) | 9 (25.0%) | ||
| 2 | 6 (35.3%) | 5 (26.3%) | 11 (30.6%) | ||
| 3 | 0 (0.0%) | 7 (36.8%) | 7 (19.4%) | ||
| 4 | 0 (0.0%) | 3 (15.8%) | 3 (8.3%) | ||
| BM involvement | Absent | 10 (62.5%) | 7 (50.0%) | 17 (56.7%) | 0.491 |
| Present | 6 (37.5%) | 7 (50.0%) | 13 (43.3%) | ||
| HBV or HCV infection | Absent | 17 (89.5%) | 14 (73.7%) | 31 (81.6%) | 0.209 |
| Present | 2 (10.5%) | 5 (26.3%) | 7 (18.4%) | ||
| Progression | No | 17 (94.4%) | 12 (63.2%) | 29 (78.4%) | 0.021 |
| Yes | 1 (5.6%) | 7 (36.8%) | 8 (21.6%) | ||
| NFKBIE | Wild type | 19 (100.0%) | 16 (84.2%) | 35 (92.1%) | 0.230 |
| Mutant | 0 (0.0%) | 3 (15.8%) | 3 (7.9%) | ||
| ITPR2 | Wild type | 16 (84.2%) | 15 (88.2%) | 31 (86.1%) | 1.000 |
| Mutant | 3 (15.8%) | 2 (11.8%) | 5 (13.9%) | ||
| MYC IHC | <10% | 19 (100.0%) | 12 (63.2%) | 31 (81.6%) | 0.008 |
| ≥10% | 0 (0.0%) | 7 (36.8%) | 7 (18.4%) | ||
| Large cell component | <20% | 12 (63.2%) | 4 (21.1%) | 16 (42.1%) | 0.009 |
| ≥20% | 7 (36.8%) | 15 (78.9%) | 22 (57.9%) | ||
| Total | 19 (50.0%) | 19 (50.0%) | 38 (100.0%) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koh, J.; Jang, I.; Choi, S.; Kim, S.; Jang, I.; Ahn, H.K.; Lee, C.; Paik, J.H.; Kim, C.W.; Lim, M.S.; et al. Discovery of Novel Recurrent Mutations and Clinically Meaningful Subgroups in Nodal Marginal Zone Lymphoma. Cancers 2020, 12, 1669. https://doi.org/10.3390/cancers12061669
Koh J, Jang I, Choi S, Kim S, Jang I, Ahn HK, Lee C, Paik JH, Kim CW, Lim MS, et al. Discovery of Novel Recurrent Mutations and Clinically Meaningful Subgroups in Nodal Marginal Zone Lymphoma. Cancers. 2020; 12(6):1669. https://doi.org/10.3390/cancers12061669
Chicago/Turabian StyleKoh, Jiwon, Insoon Jang, Seongmin Choi, Sehui Kim, Ingeon Jang, Hyun Kyung Ahn, Cheol Lee, Jin Ho Paik, Chul Woo Kim, Megan S. Lim, and et al. 2020. "Discovery of Novel Recurrent Mutations and Clinically Meaningful Subgroups in Nodal Marginal Zone Lymphoma" Cancers 12, no. 6: 1669. https://doi.org/10.3390/cancers12061669
APA StyleKoh, J., Jang, I., Choi, S., Kim, S., Jang, I., Ahn, H. K., Lee, C., Paik, J. H., Kim, C. W., Lim, M. S., Kim, K., & Jeon, Y. K. (2020). Discovery of Novel Recurrent Mutations and Clinically Meaningful Subgroups in Nodal Marginal Zone Lymphoma. Cancers, 12(6), 1669. https://doi.org/10.3390/cancers12061669





