Telomerase Biogenesis and Activities from the Perspective of Its Direct Interacting Partners
Abstract
1. Introduction
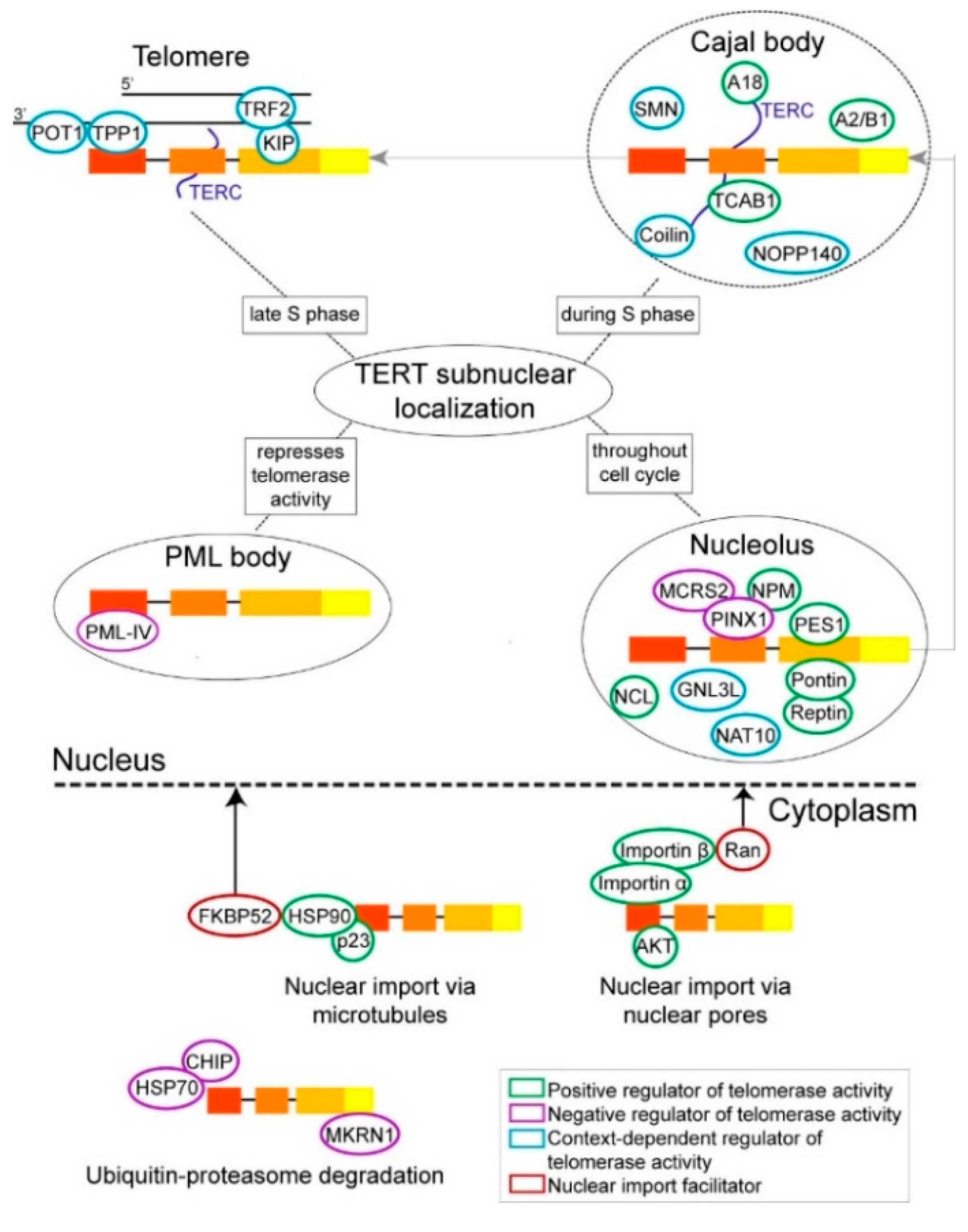
2. TERT Protein Interactions Implicated in Its Trafficking between the Cytoplasm and the Nucleus
3. TERT Protein Interactions Implicated in Its Trafficking within the Nucleus
3.1. TERT Protein Interactions in the Nucleolus
3.2. TERT Protein Interactions in the Cajal Bodies
3.3. TERT Protein Interactions at the Telomeres
4. TERT Protein Interactions and Non-Telomeric Activities in the Context of Cancer
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shay, J.W.; Wright, W.E. Telomeres and Telomerase: Three Decades of Progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. Shelterin: The Protein Complex That Shapes and Safeguards Human Telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. A Loopy View of Telomere Evolution. Front. Genet. 2015, 6, 321. [Google Scholar] [CrossRef] [PubMed]
- Veverka, P.; Janovič, T.; Hofr, C. Quantitative Biology of Human Shelterin and Telomerase: Searching for the Weakest Point. Int. J. Mol. Sci. 2019, 20, 3186. [Google Scholar] [CrossRef]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres Shorten during Ageing of Human Fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Aubert, G.; Lansdorp, P.M. Telomeres and Aging. Physiol. Rev. 2008, 88, 557–579. [Google Scholar] [CrossRef] [PubMed]
- Hemann, M.T.; Strong, M.A.; Hao, L.Y.; Greider, C.W. The Shortest Telomere, Not Average Telomere Length, Is Critical for Cell Viability and Chromosome Stability. Cell 2001, 107, 67–77. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. The Telomere Terminal Transferase of Tetrahymena Is a Ribonucleoprotein Enzyme with Two Kinds of Primer Specificity. Cell 1987, 51, 887–898. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. A Telomeric Sequence in the RNA of Tetrahymena Telomerase Required for Telomere Repeat Synthesis. Nature 1989, 337, 331–337. [Google Scholar] [CrossRef]
- Wright, W.E.; Piatyszek, M.A.; Rainey, W.E.; Byrd, W.; Shay, J.W. Telomerase Activity in Human Germline and Embryonic Tissues and Cells. Dev. Genet. 1996, 18, 173–179. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Telomeres and Telomerase in Normal and Cancer Stem Cells. FEBS Lett. 2010, 584, 3819–3825. [Google Scholar] [CrossRef] [PubMed]
- Aubert, G. Telomere Dynamics and Aging. Prog. Mol. Biol. Trans. 2014, 125, 89–111. [Google Scholar]
- Armanios, M.; Blackburn, E.H. The Telomere Syndromes. Nat. Rev. Genet. 2012, 693–704. [Google Scholar] [CrossRef] [PubMed]
- De Vitis, M.; Berardinelli, F.; Sgura, A. Telomere Length Maintenance in Cancer: At the Crossroad between Telomerase and Alternative Lengthening of Telomeres (ALT). Int. J. Mol. Sci. 2018, 19, 606. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.C.; Cech, T.R. Human Telomerase: Biogenesis, Trafficking, Recruitment, and Activation. Genes Dev. 2015, 1095–1105. [Google Scholar] [CrossRef]
- Hukezalie, K.R.; Wong, J.M.Y. Structure-Function Relationship and Biogenesis Regulation of the Human Telomerase Holoenzyme. FEBS J. 2013, 280, 3194–3204. [Google Scholar] [CrossRef]
- Wang, C.; Query, C.C.; Meier, U.T. Immunopurified Small Nucleolar Ribonucleoprotein Particles Pseudouridylate RRNA Independently of Their Association with Phosphorylated Nopp140. Mol. Cell. Biol. 2002, 22, 8457–8466. [Google Scholar] [CrossRef]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse Transcriptase Motifs in the Catalytic Subunit of Telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms Underlying the Activation of TERT Transcription and Telomerase Activity in Human Cancer: Old Actors and New Players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef]
- Ouellette, M.M.; Liao, M.; Herbert, B.S.; Johnson, M.; Holt, S.E.; Liss, H.S.; Shay, J.W.; Wright, W.E. Subsenescent Telomere Lengths in Fibroblasts Immortalized by Limiting Amounts of Telomerase. J. Biol. Chem. 2000, 275, 10072–10076. [Google Scholar] [CrossRef]
- Thompson, C.A.H.; Wong, J.M.Y. Non-Canonical Functions of Telomerase Reverse Transcriptase: Emerging Roles and Biological Relevance. Curr. Top. Med. Chem. 2020, 20, 498–507. [Google Scholar] [CrossRef]
- Fleisig, H.B.; Hukezalie, K.R.; Thompson, C.A.H.; Au-Yeung, T.T.T.; Ludlow, A.T.; Zhao, C.R.; Wong, J.M.Y. Telomerase Reverse Transcriptase Expression Protects Transformed Human Cells against DNA-Damaging Agents, and Increases Tolerance to Chromosomal Instability. Oncogene 2016, 35, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Ludlow, A.T.; Slusher, A.L.; Sayed, M.E. Insights into Telomerase/HTERT Alternative Splicing Regulation Using Bioinformatics and Network Analysis in Cancer. Cancers 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.E.; Aisner, D.L.; Baur, J.; Tesmer, V.M.; Dy, M.; Ouellette, M.; Trager, J.B.; Morin, G.B.; Toft, D.O.; Shay, J.W.; et al. Functional Requirement of P23 and Hsp90 in Telomerase Complexes. Genes Dev. 1999, 13, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.Y.; Her, J.; Oh, S.Y.; Chung, I.K. Hsp90-Binding Immunophilin FKBP52 Modulates Telomerase Activity by Promoting the Cytoplasmic Retrotransport of HTERT. Biochem. J. 2016, 473, 3517–3532. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.S.; Kwon, T.; Kwon, D.Y.; Do, S.I. Akt Protein Kinase Enhances Human Telomerase Activity through Phosphorylation of Telomerase Reverse Transcriptase Subunit. J. Biol. Chem. 1999, 274, 13085–13090. [Google Scholar] [CrossRef]
- Haendeler, J.; Hoffmann, J.; Rahman, S.; Zeiher, A.M.; Dimmeler, S. Regulation of Telomerase Activity and Anti-Apoptotic Function by Protein-Protein Interaction and Phosphorylation. FEBS Lett. 2003, 536, 180–186. [Google Scholar] [CrossRef]
- Jeong, S.A.; Kim, K.; Lee, J.H.; Cha, J.S.; Khadka, P.; Cho, H.-S.; Chung, I.K.; Seok Cha, J. Akt-Mediated Phosphorylation Increases the Binding Affinity of HTERT for Importin α to Promote Nuclear Translocation. J. Cell Sci. 2015, 128, 2951. [Google Scholar] [CrossRef][Green Version]
- Xi, P.; Zhou, L.; Wang, M.; Liu, J.-P.; Cong, Y.-S. Serine/Threonine-Protein Phosphatase 2A Physically Interacts With Human Telomerase Reverse Transcriptase HTERT and Regulates Its Subcellular Distribution. J. Cell. Biochem. 2013, 114, 409–417. [Google Scholar] [CrossRef]
- Seimiya, H.; Sawada, H.; Muramatsu, Y.; Shimizu, M.; Ohko, K.; Yamane, K.; Tsuruo, T. Involvement of 14-3-3 Proteins in Nuclear Localization of Telomerase. EMBO J. 2000, 19, 2652–2661. [Google Scholar] [CrossRef]
- Zhang, P.; Chan, S.L.; Fu, W.; Mendoza, M.; Mattson, M.P. TERT Suppresses Apoptotis at a Premitochondrial Step by a Mechanism Requiring Reverse Transcriptase Activity and 14-3-3 Protein-Binding Ability. FASEB J. 2003, 17, 767–769. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, S.M.; Kang, M.R.; Oh, S.Y.; Lee, T.H.; Muller, M.T.; Chung, I.K. Ubiquitin Ligase MKRN1 Modulates Telomere Length Homeostasis through a Proteolysis of HTERT. Genes Dev. 2005, 19, 776–781. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Khadka, P.; Baek, S.H.; Chung, I.K. CHIP Promotes Human Telomerase Reverse Transcriptase Degradation and Negatively Regulates Telomerase Activity. J. Biol. Chem. 2010, 285, 42033–42045. [Google Scholar] [CrossRef] [PubMed]
- Perera, O.N.; Sobinoff, A.P.; Teber, E.T.; Harman, A.; Maritz, M.F.; Yang, S.F.; Pickett, H.A.; Cesare, A.J.; Arthur, J.W.; MacKenzie, K.L.; et al. Telomerase Promotes Formation of a Telomere Protective Complex in Cancer Cells. Sci. Adv. 2019, 5, eaav4409. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.L.; Blackburn, E.H. Telomerase and ATM/Tel1p Protect Telomeres from Nonhomologous End Joining. Mol. Cell 2003, 11, 1379–1387. [Google Scholar] [CrossRef]
- Haendeler, J.; Hoffmann, J.; Brandes, R.P.; Zeiher, A.M.; Dimmeler, S. Hydrogen Peroxide Triggers Nuclear Export of Telomerase Reverse Transcriptase via Src Kinase Family-Dependent Phosphorylation of Tyrosine 707. Mol. Cell. Biol. 2003, 23, 4598–4610. [Google Scholar] [CrossRef]
- Jakob, S.; Schroeder, P.; Lukosz, M.; Büchner, N.; Spyridopoulos, I.; Altschmied, J.; Haendeler, J. Nuclear Protein Tyrosine Phosphatase Shp-2 Is One Important Negative Regulator of Nuclear Export of Telomerase Reverse Transcriptase. J. Biol. Chem. 2008, 283, 33155–33161. [Google Scholar] [CrossRef]
- Santos, J.H.; Meyer, J.N.; Skorvaga, M.; Annab, L.A.; Van Houten, B. Mitochondrial HTERT Exacerbates Free-Radical-Mediated MtDNA Damage. Aging Cell 2004, 3, 399–411. [Google Scholar] [CrossRef]
- Haendeler, J.; Dröse, S.; Büchner, N.; Jakob, S.; Altschmied, J.; Goy, C.; Spyridopoulos, I.; Zeiher, A.M.; Brandt, U.; Dimmeler, S. Mitochondrial Telomerase Reverse Transcriptase Binds to and Protects Mitochondrial DNA and Function from Damage. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 929–935. [Google Scholar] [CrossRef]
- Singhapol, C.; Pal, D.; Czapiewski, R.; Porika, M.; Nelson, G.; Saretzki, G.C. Mitochondrial Telomerase Protects Cancer Cells from Nuclear DNA Damage and Apoptosis. PLoS ONE 2013, 8, e52989. [Google Scholar] [CrossRef]
- Kovalenko, O.A.; Caron, M.J.; Ulema, P.; Medrano, C.; Thomas, A.P.; Kimura, M.; Bonini, M.G.; Herbig, U.; Santos, J.H. A Mutant Telomerase Defective in Nuclear-Cytoplasmic Shuttling Fails to Immortalize Cells and Is Associated with Mitochondrial Dysfunction. Aging Cell 2010, 9, 203–219. [Google Scholar] [CrossRef]
- Kovalenko, O.A.; Kaplunov, J.; Herbig, U.; deToledo, S.; Azzam, E.I.; Santos, J.H. Expression of NES-HTERT in Cancer Cells Delays Cell Cycle Progression and Increases Sensitivity to Genotoxic Stress. PLoS ONE 2010, 5, e10812. [Google Scholar] [CrossRef][Green Version]
- Richard, P.; Darzacq, X.; Bertrand, E.; Jády, B.E.; Verheggen, C.; Kiss, T. A Common Sequence Motif Determines the Cajal Body-Specific Localization of Box H/ACA ScaRNAs. EMBO J. 2003, 22, 4283–4293. [Google Scholar] [CrossRef]
- Venteicher, A.S.; Artandi, S.E. TCAB1: Driving Telomerase to Cajal Bodies. Cell Cycle 2009, 8, 1329–1331. [Google Scholar] [CrossRef] [PubMed]
- Collins, K. Physiological Assembly and Activity of Human Telomerase Complexes. Mech. Ageing Dev. 2008, 129, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, Y.S.; Jeong, S.A.; Khadka, P.; Roth, J.; Chung, I.K. Catalytically Active Telomerase Holoenzyme Is Assembled in the Dense Fibrillar Component of the Nucleolus during S Phase. Histochem. Cell Biol. 2014, 141, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, R.L.; Ziegler, T.D.; Supakorndej, T.; Terns, R.M.; Terns, M.P. Cell Cycle-Regulated Trafficking of Human Telomerase to Telomeres. Mol. Biol. Cell 2006, 17, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, Y.; Zhang, C.; Huang, H.; Weissman, S.M. Nucleolar Localization of HTERT Protein Is Associated with Telomerase Function. Exp. Cell Res. 2002, 277, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Etheridge, K.T.; Banik, S.S.R.; Armbruster, B.N.; Zhu, Y.; Terns, R.M.; Terns, M.P.; Counter, C.M. The Nucleolar Localization Domain of the Catalytic Subunit of Human Telomerase. J. Biol. Chem. 2002, 277, 24764–24770. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Jin, R.; Zhang, B.; Chen, H.; Bai, Y.X.; Yang, P.X.; Han, S.W.; Xie, Y.H.; Huang, P.T.; Huang, C.; et al. Nucleolar Localization of TERT Is Unrelated to Telomerase Function in Human Cells. J. Cell Sci. 2008, 121, 2169–2176. [Google Scholar] [CrossRef][Green Version]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A Human Telomerase Holoenzyme Protein Required for Cajal Body Localization and Telomere Synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tomlinson, R.L.; Lukowiak, A.A.; Terns, R.M.; Terns, M.P. Telomerase RNA Accumulates in Cajal Bodies in Human Cancer Cells. Mol. Biol. Cell 2004, 15, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Platani, M.; Goldberg, I.; Swedlow, J.R.; Lamond, A.I. In Vivo Analysis of Cajal Body Movement, Separation, and Joining in Live Human Cells. J. Cell Biol. 2000, 151, 1561–1574. [Google Scholar] [CrossRef] [PubMed]
- Trinkle-Mulcahy, L.; Sleeman, J.E. The Cajal Body and the Nucleolus: “In a Relationship” or “It’s Complicated”? RNA Biol. 2017, 14, 739–751. [Google Scholar] [CrossRef]
- Hearst, S.M.; Gilder, A.S.; Negi, S.S.; Davis, M.D.; George, E.M.; Whittom, A.A.; Toyota, C.G.; Husedzinovic, A.; Gruss, O.J.; Hebert, M.D. Cajal-Body Formation Correlates with Differential Coilin Phosphorylation in Primary and Transformed Cell Lines. J. Cell Sci. 2009, 122, 1872–1881. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Zaug, A.J.; Cech, T.R. Live Cell Imaging Reveals the Dynamics of Telomerase Recruitment to Telomeres. Cell 2016, 166, 1188–1197. [Google Scholar] [CrossRef]
- Lallemand-Breitenbach, V.; de Thé, H. PML Nuclear Bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000661. [Google Scholar] [CrossRef]
- Oh, W.; Ghim, J.; Lee, E.W.; Yang, M.R.; Kim, E.T.; Ahn, J.H.; Song, J. PML-IV Functions as a Negative Regulator of Telomerase by Interacting with TERT. J. Cell Sci. 2009, 122, 2613–2622. [Google Scholar] [CrossRef]
- Zhou, X.Z.; Lu, K.P. The Pin2/TRF1-Interacting Protein PinX1 Is a Potent Telomerase Inhibitor. Cell 2001, 107, 347–359. [Google Scholar] [CrossRef]
- Banik, S.S.R.; Counter, C.M. Characterization of Interactions between PinX1 and Human Telomerase Subunits HTERT and HTR. J. Biol. Chem. 2004, 279, 51745–51748. [Google Scholar] [CrossRef] [PubMed]
- Cheung, D.H.C.; Ho, S.T.; Lau, K.F.; Jin, R.; Wang, Y.N.; Kung, H.F.; Huang, J.J.; Shaw, P.C. Nucleophosmin Interacts with PIN2/TERF1-Interacting Telomerase Inhibitor 1 (PinX1) and Attenuates the PinX1 Inhibition on Telomerase Activity. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Ho, S.T.; Jin, R.; Cheung, D.H.C.; Huang, J.J.; Shaw, P.C. The PinX1/NPM Interaction Associates with HTERT in Early-S Phase and Facilitates Telomerase Activation. Cell Biosci. 2019, 9, 47. [Google Scholar] [CrossRef]
- Song, H.; Li, Y.; Chen, G.; Xing, Z.; Zhao, J.; Yokoyama, K.K.; Li, T.; Zhao, M. Human MCRS2, a Cell-Cycle-Dependent Protein, Associates with LPTS/PinX1 and Reduces the Telomere Length. Biochem. Biophys. Res. Commun. 2004, 316, 1116–1123. [Google Scholar] [CrossRef] [PubMed]
- Berger, C.M.; Gaume, X.; Bouvet, P. The Roles of Nucleolin Subcellular Localization in Cancer. Biochimie. 2015, 113, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Khurts, S.; Masutomi, K.; Delgermaa, L.; Arai, K.; Oishi, N.; Mizuno, H.; Hayashi, N.; Hahn, W.C.; Murakami, S. Nucleolin Interacts with Telomerase. J. Biol. Chem. 2004, 279, 51508–51515. [Google Scholar] [CrossRef] [PubMed]
- Huber, O.; Ménard, L.; Haurie, V.; Nicou, A.; Taras, D.; Rosenbaum, J. Pontin and Reptin, Two Related ATPases with Multiple Roles in Cancer. Cancer Res. 2008, 68, 6873–6876. [Google Scholar] [CrossRef] [PubMed]
- Venteicher, A.S.; Meng, Z.; Mason, P.J.; Veenstra, T.D.; Artandi, S.E. Identification of ATPases Pontin and Reptin as Telomerase Components Essential for Holoenzyme Assembly. Cell 2008, 132, 945–957. [Google Scholar] [CrossRef]
- Her, J.; Chung, I.K. The AAA-ATPase NVL2 Is a Telomerase Component Essential for Holoenzyme Assembly. Biochem. Biophys. Res. Commun. 2012, 417, 1086–1092. [Google Scholar] [CrossRef]
- Fu, D.; Collins, K. Purification of Human Telomerase Complexes Identifies Factors Involved in Telomerase Biogenesis and Telomere Length Regulation. Mol. Cell 2007, 28, 773–785. [Google Scholar] [CrossRef]
- Cheng, L.; Yuan, B.; Ying, S.; Niu, C.; Mai, H.; Guan, X.; Yang, X.; Teng, Y.; Lin, J.; Huang, J.; et al. PES1 Is a Critical Component of Telomerase Assembly and Regulates Cellular Senescence. Sci. Adv. 2019, 5, eaav1090. [Google Scholar] [CrossRef]
- Enwerem, I.I.; Velma, V.; Broome, H.J.; Kuna, M.; Begum, R.A.; Hebert, M.D. Coilin Association with Box C/D ScaRNA Suggests a Direct Role for the Cajal Body Marker Protein in ScaRNP Biogenesis. Biol. Open 2014, 3, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Bachand, F.; Boisvert, F.-M.; Côté, J.; Richard, S.; Autexier, C. The Product of the Survival of Motor Neuron (SMN) Gene Is a Human Telomerase-Associated Protein. Mol. Biol. Cell 2002, 13, 3192–3202. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.R.; Hebert, M.D. SMN and Coilin Negatively Regulate Dyskerin Association with Telomerase RNA. Biol. Open 2016, 5, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Dreyfuss, G.; Kim, V.N.; Kataoka, N. Messenger-RNA-Binding Proteins and the Messages They Carry. Nat. Rev Mol. Cell Biol. 2002, 3, 195–205. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The HnRNP Family: Insights into Their Role in Health and Disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Mizuno, H.; Honda, M.; Shirasaki, T.; Yamashita, T.; Yamashita, T.; Mizukoshi, E.; Kaneko, S. Heterogeneous Nuclear Ribonucleoprotein A2/B1 in Association with HTERT Is a Potential Biomarker for Hepatocellular Carcinoma. Liver Int. 2012, 32, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tang, M.L.; Zeng, Z.X.; Wu, R.Y.; Xue, Y.; Hao, Y.H.; Pang, D.W.; Zhao, Y.; Tan, Z. Telomere- and Telomerase-Interacting Protein That Unfolds Telomere G-Quadruplex and Promotes Telomere Extension in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20413–20418. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Mao, P.; Li, F.; Han, X.; Zhang, Y.; Jiang, S.; Chen, Y.; Huang, J.; Liu, D.; et al. Cold-Inducible RNA-Binding Protein CIRP/HnRNP A18 Regulates Telomerase Activity in a Temperature-Dependent Manner. Nucleic Acids Res. 2016, 44, 761–775. [Google Scholar] [CrossRef]
- Stern, J.L.; Zyner, K.G.; Pickett, H.A.; Cohen, S.B.; Bryan, T.M. Telomerase Recruitment Requires Both TCAB1 and Cajal Bodies Independently. Mol. Cell. Biol. 2012, 32, 2384–2395. [Google Scholar] [CrossRef] [PubMed]
- Vogan, J.M.; Zhang, X.; Youmans, D.T.; Regalado, S.G.; Johnson, J.Z.; Hockemeyer, D.; Collins, K. Minimized Human Telomerase Maintains Telomeres and Resolves Endogenous Roles of H/ACA Proteins, TCAB1, and Cajal Bodies. Elife 2016, 5, e18221. [Google Scholar] [CrossRef] [PubMed]
- Na, J.H.; Lee, W.K.; Yu, Y.G. How Do We Study the Dynamic Structure of Unstructured Proteins: A Case Study on Nopp140 as an Example of a Large, Intrinsically Disordered Protein. Int. J. Mol Sci. 2018, 19, 381. [Google Scholar] [CrossRef]
- Isaac, C.; Yang, Y.; Meier, U.T. Nopp140 Functions as a Molecular Link between the Nucleolus and the Coiled Bodies. J. Cell Biol. 1998, 142, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Bizarro, J.; Bhardwaj, A.; Smith, S.; Thomas Meiera, U. Nopp140-Mediated Concentration of Telomerase in Cajal Bodies Regulates Telomere Length. Mol. Biol. Cell 2019, 30, 3136–3150. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.L.; Batista, L.F.Z.; Freund, A.; Pech, M.F.; Venteicher, A.S.; Artandi, S.E. TPP1 OB-Fold Domain Controls Telomere Maintenance by Recruiting Telomerase to Chromosome Ends. Cell 2012, 150, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.; Tesmer, V.M.; Nandakumar, J. The N Terminus of the OB Domain of Telomere Protein TPP1 Is Critical for Telomerase Action. Cell Rep. 2018, 22, 1132–1140. [Google Scholar] [CrossRef]
- Gun, E.L.; Eun, Y.Y.; Chae, H.C.; Lee, J.; Muller, M.T.; In, K.C. DNA-Protein Kinase Catalytic Subunit-Interacting Protein KIP Binds Telomerase by Interacting with Human Telomerase Reverse Transcriptase. J. Biol. Chem. 2004, 279, 34750–34755. [Google Scholar]
- Khadka, P.; Lee, J.H.; Baek, S.H.; Oh, S.Y.; Chung, I.K. DNA-PKcs-Interacting Protein KIP Binding to TRF2 Is Required for the Maintenance of Functional Telomeres. Biochem. J. 2014, 463, 19–30. [Google Scholar] [CrossRef]
- Mohiuddin, I.S.; Kang, M.H. DNA-PK as an Emerging Therapeutic Target in Cancer. Front. Oncol. 2019, 9, 635. [Google Scholar] [CrossRef]
- Rice, C.; Shastrula, P.K.; Kossenkov, A.V.; Hills, R.; Baird, D.M.; Showe, L.C.; Doukov, T.; Janicki, S.; Skordalakes, E. Structural and Functional Analysis of the Human POT1-TPP1 Telomeric Complex. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Zaug, A.J.; Podell, E.R.; Cech, T.R. Human POT1 Disrupts Telomeric G-Quadruplexes Allowing Telomerase Extension in Vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 10864–10869. [Google Scholar] [CrossRef]
- Loayza, D.; De Lange, T. POT1 as a Terminal Transducer of TRF1 Telomere Length Control. Nature 2003, 423, 1013–1018. [Google Scholar] [CrossRef]
- Chen, L.Y.; Redon, S.; Lingner, J. The Human CST Complex Is a Terminator of Telomerase Activity. Nature 2012, 488, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Kiselar, J.; Whited, T.L.; Hernandez-Sanchez, W.; Taylor, D.J. POT1-TPP1 Differentially Regulates Telomerase via POT1 His266 and as a Function of Single-Stranded Telomere DNA Length. Proc. Natl. Acad. Sci. USA 2019, 116, 23527–23533. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.J.; Zaug, A.J.; Kim, H.J.; Cech, T.R. Reconstitution of Human Shelterin Complexes Reveals Unexpected Stoichiometry and Dual Pathways to Enhance Telomerase Processivity. Nat. Commun. 2017, 8, 1075. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.; Löbrich, M. The Pendulum of the Ku-Ku Clock. DNA Repair (Amst.) 2018, 71, 164–171. [Google Scholar] [CrossRef]
- Chai, W.; Ford, L.P.; Lenertz, L.; Wright, W.E.; Shay, J.W. Human Ku70/80 Associates Physically with Telomerase through Interaction with HTERT. J. Biol. Chem. 2002, 277, 47242–47247. [Google Scholar] [CrossRef]
- Ting, N.S.Y.; Yu, Y.; Pohorelic, B.; Lees-Miller, S.P.; Beattie, T.L. Human Ku70/80 Interacts Directly with HTR, the RNA Component of Human Telomerase. Nucleic Acids Res. 2005, 33, 2090–2098. [Google Scholar] [CrossRef]
- Pfingsten, J.S.; Goodrich, K.J.; Taabazuing, C.; Ouenzar, F.; Chartrand, P.; Cech, T.R. Mutually Exclusive Binding of Telomerase RNA and DNA by Ku Alters Telomerase Recruitment Model. Cell 2012, 148, 922–932. [Google Scholar] [CrossRef]
- Cao, Y.; Li, H.; Deb, S.; Liu, J.P. TERT Regulates Cell Survival Independent of Telomerase Enzymatic Activity. Oncogene 2002, 21, 3130–3138. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Beck, S.; Sohn, Y.-W.; Kim, J.-K.; Kim, S.-H.; Yin, J.; Pian, X.; Kim, S.-C.; Choi, Y.-J.; Kim, H. Human Telomerase Catalytic Subunit (HTERT) Suppresses P53-Mediated Anti-Apoptotic Response via Induction of Basic Fibroblast Growth Factor. Exp. Mol. Med. 2010, 42, 574–582. [Google Scholar] [CrossRef]
- Pleschke, J.M.; Kleczkowska, H.E.; Strohm, M.; Althaus, F.R. Poly(ADP-Ribose) Binds to Specific Domains in DNA Damage Checkpoint Proteins. J. Biol. Chem. 2000, 275, 40974–40980. [Google Scholar] [CrossRef]
- Jin, Y.; You, L.; Kim, H.J.; Lee, H.W. Telomerase Reverse Transcriptase Contains a BH3-like Motif and Interacts with BCL-2 Family Members. Mol. Cells 2018, 41, 684–694. [Google Scholar] [PubMed]
- Park, J.I.; Venteicher, A.S.; Hong, J.Y.; Choi, J.; Jun, S.; Shkreli, M.; Chang, W.; Meng, Z.; Cheung, P.; Ji, H.; et al. Telomerase Modulates Wnt Signalling by Association with Target Gene Chromatin. Nature 2009, 460, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.J.; Grandori, C.; Amacker, M.; Simon-Vermot, N.; Polack, A.; Lingner, J.; Dalla-Favera, R. Direct Activation of TERT Transcription by C-MYC. Nat. Genet. 1999, 21, 220–224. [Google Scholar] [CrossRef]
- Strong, M.A.; Vidal-Cardenas, S.L.; Karim, B.; Yu, H.; Guo, N.; Greider, C.W. Phenotypes in MTERT+/- and MTERT-/- Mice Are Due to Short Telomeres, Not Telomere-Independent Functions of Telomerase Reverse Transcriptase. Mol. Cell. Biol. 2011, 31, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Listerman, I.; Gazzaniga, F.S.; Blackburn, E.H. An Investigation of the Effects of the Core Protein Telomerase Reverse Transcriptase on Wnt Signaling in Breast Cancer Cells. Mol. Cell. Biol. 2014, 34, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, N.; Yasukawa, M.; Nguyen, C.; Kasim, V.; Maida, Y.; Possemato, R.; Shibata, T.; Ligon, K.L.; Fukami, K.; Hahn, W.C.; et al. Maintenance of Tumor Initiating Cells of Defined Genetic Composition by Nucleostemin. Proc. Natl. Acad. Sci. USA 2011, 108, 20388–20393. [Google Scholar] [CrossRef]
- Maida, Y.; Yasukawa, M.; Okamoto, N.; Ohka, S.; Kinoshita, K.; Totoki, Y.; Ito, T.K.; Minamino, T.; Nakamura, H.; Yamaguchi, S.; et al. Involvement of Telomerase Reverse Transcriptase in Heterochromatin Maintenance. Mol. Cell. Biol. 2014, 34, 1576–1593. [Google Scholar] [CrossRef]
- Lassmann, T.; Maida, Y.; Tomaru, Y.; Yasukawa, M.; Ando, Y.; Kojima, M.; Kasim, V.; Simon, C.; Daub, C.O.; Carninci, P.; et al. Telomerase Reverse Transcriptase Regulates MicroRNAs. Int. J. Mol. Sci. 2015, 16, 1192–1208. [Google Scholar] [CrossRef]
- Ghosh, A.; Saginc, G.; Leow, S.C.; Khattar, E.; Shin, E.M.; Yan, T.D.; Wong, M.; Zhang, Z.; Li, G.; Sung, W.K.; et al. Telomerase Directly Regulates NF-B-Dependent Transcription. Nat. Cell Biol. 2012, 14, 1270–1281. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Q.L.; Sun, W.; Chandrasekharan, P.; Cheng, H.S.; Ying, Z.; Lakshmanan, M.; Raju, A.; Tenen, D.G.; Cheng, S.Y.; et al. Non-Canonical NF-ΚB Signalling and ETS1/2 Cooperatively Drive C250T Mutant TERT Promoter Activation. Nat. Cell Biol. 2015, 17, 1327–1338. [Google Scholar] [CrossRef]
- Ding, D.; Xi, P.; Zhou, J.; Wang, M.; Cong, Y.S. Human Telomerase Reverse Transcriptase Regulates MMP Expression Independently of Telomerase Activity via NF-ΚB-Dependent Transcription. FASEB J. 2013, 27, 4375–4383. [Google Scholar] [CrossRef]
- Qin, Y.; Tang, B.; Hu, C.J.; Xiao, Y.F.; Xie, R.; Yong, X.; Wu, Y.Y.; Dong, H.; Yang, S.M. An HTERT/ZEB1 Complex Directly Regulates E-Cadherin to Promote Epithelial-to-Mesenchymal Transition (EMT) in Colorectal Cancer. Oncotarget 2016, 7, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Khattar, E.; Kumar, P.; Liu, C.Y.; Can Akincilar, S.; Raju, A.; Lakshmanan, M.; Maury, J.J.P.; Qiang, Y.; Li, S.; Tan, E.Y.; et al. Telomerase Reverse Transcriptase Promotes Cancer Cell Proliferation by Augmenting TRNA Expression. J. Clin. Investig. 2016, 126, 4045–4060. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ding, D.; Hao, W.; Yang, F.; Wu, X.; Wang, M.; Xu, X.; Ju, Z.; Liu, J.-P.; Song, Z.; et al. HTERT Promotes Tumor Angiogenesis by Activating VEGF via Interactions with the Sp1 Transcription Factor. Nucleic Acids Res. 2016, 44, 8693–8703. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Cech, T.R. Inventory of Telomerase Components in Human Cells Reveals Multiple Subpopulations of HTR and HTERT. Nucleic Acids Res. 2014, 42, 8565–8577. [Google Scholar] [CrossRef] [PubMed]


| Protein(s) | TERT-Related Function(s) | Reference(s) | |
|---|---|---|---|
| Participate in TERT nuclear localization | HSP90—p23 | Chaperones, facilitate TERT nuclear import via microtubules, protect TERT from degradation in cytoplasm | [24,33] |
| FKBP52 | Immunophilin, facilitates TERT nuclear import via microtubules | [25] | |
| Importin α | Karyopherin, facilitates TERT nuclear import via nuclear pores | [28] | |
| Ran | GTPase, facilitates TERT nuclear import (and export) via nuclear pores | [28] | |
| 14-3-3 | Phosphoprotein-binding protein, promotes TERT nuclear localization by impairing TERT-CRM1 interaction, may participate in TERT anti-apoptotic function in the mitochondria | [30,31] | |
| AKT | Kinase, promotes TERT nuclear import via importin α mechanism by phosphorylating residue S227 | [28] | |
| PP2A | Phosphatase whose activity antagonizes TERT nuclear import, may mediate TERT—14-3-3 interaction | [29] | |
| Participate in TERT nuclear export | CRM1 | Karyopherin, facilitates TERT nuclear export via nuclear pores | [30] |
| Src | Kinase, promotes TERT nuclear export by phosphorylating residue Y707 | [36] | |
| Shp2 | Phosphatase, prevents TERT nuclear export by dephosphorylating residue Y707 | [37] | |
| Participate in TERT degradation in cytoplasm | MKRN1 | Ubiquitin ligase, facilitates TERT degradation via the ubiquitin-proteasome pathway | [32] |
| HSP70—CHIP | Chaperones, promote TERT degradation by enhancing TERT-MKRN1 interaction | [33] | |
| Interact with TERT in PML bodies | PML IV | PML protein isoform 4, negative regulator of telomerase activity | [58] |
| Interact with TERT in the nucleolus | PINX1 | Tumor suppressor, negative regulator of telomerase activity | [59,60] |
| NPM | Phosphoprotein, positive regulator of telomerase activity | [61,62] | |
| MCRS2 | RNA-binding protein, negative regulator of telomerase activity | [63] | |
| NCL | Phosphoprotein, facilitates TERT nucleolar localization | [65] | |
| Pontin, reptin, NVL2 | ATPases, positive regulators of telomerase activity | [67,68] | |
| NAT10, GNL3L | NTPases, regulate telomerase activity in a context-/cell cycle-dependent manner | [69] | |
| PES1 | Positive regulator of telomerase activity, may participate in TERT pro-proliferative function | [70] | |
| Interact with TERT in Cajal bodies | TCAB1 | Chaperone, facilitates (TERC-mediated) TERT localization in Cajal bodies, delivers telomerase to telomeres during catalysis | [43,44,45,79,80] |
| Coilin, SMN | Constituents of Cajal bodies, may regulate telomerase activity in a context-dependent manner | [51,71,72,73] | |
| A2/B1, A18 | RNA-binding proteins, positive regulators of telomerase activity | [76,77,78] | |
| Interact with TERT at telomeres | TPP1 | Shelterin subunit, directly tethers telomerase to telomeric DNA during catalysis, forms a dimer with POT1 which regulates telomerase activity in a context-/cell cycle-dependent manner | [84,85,89,90,91,92,93] |
| KIP | Calcium-binding protein, tethers telomerase to shelterin subunit TRF2 during catalysis | [86,87] | |
| Participate in TERT non-telomeric activities | p53 | Tumor suppressor, contributes to TERT anti-apoptotic and pro-proliferative effects | [99,100] |
| PARP | DNA damage repair protein, forms a ternary complex with TERT and p53 | [99,101] | |
| MCL1, BCL-xL | Anti-apoptosis proteins, interact with TERT in the mitochondria | [102] | |
| BRG1 | Transcription factor, engages TERT at promoter region of Wnt pathway target genes | [103] | |
| NFκB p65 | Transcription factor, engages TERT at promoter region of selective NFκB target genes | [111] | |
| ZEB1 | Transcription repressor, engages TERT at promoter region of E-cadherin gene | [113] | |
| RPC32 | RNA polymerase III subunit, engages TERT at promoter regions of tRNA genes | [114] | |
| Sp1 | Transcription factor, engages TERT at promoter region of VEGF gene | [115] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, K.T.T.T.; Wong, J.M.Y. Telomerase Biogenesis and Activities from the Perspective of Its Direct Interacting Partners. Cancers 2020, 12, 1679. https://doi.org/10.3390/cancers12061679
Nguyen KTTT, Wong JMY. Telomerase Biogenesis and Activities from the Perspective of Its Direct Interacting Partners. Cancers. 2020; 12(6):1679. https://doi.org/10.3390/cancers12061679
Chicago/Turabian StyleNguyen, Kathryn T. T. T., and Judy M. Y. Wong. 2020. "Telomerase Biogenesis and Activities from the Perspective of Its Direct Interacting Partners" Cancers 12, no. 6: 1679. https://doi.org/10.3390/cancers12061679
APA StyleNguyen, K. T. T. T., & Wong, J. M. Y. (2020). Telomerase Biogenesis and Activities from the Perspective of Its Direct Interacting Partners. Cancers, 12(6), 1679. https://doi.org/10.3390/cancers12061679





