Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review
Abstract
:1. Introduction
2. Results
2.1. Eligible Studies and Characteristics
2.2. Applications of Deep Learning in Colorectal Cancer Pathology Image Analysis
2.2.1. Gland Segmentation
2.2.2. Tumor Classification
2.2.3. Tumor Microenvironment Analysis
2.2.4. Prognosis Prediction
3. Discussion
3.1. Challenges in Pathological Diagnosis of CRC
3.1.1. Small Tissue Artifacts
3.1.2. Regenerative Atypia
3.1.3. Inter-Observer Variation in Adenoma Grading and Subtype Classification
3.1.4. Importance of Tumor Microenvironment in CRCs
3.1.5. Prognostic Prediction in CRCs
3.2. Application of Deep Learning Models in CRC Pathological Diagnosis
3.2.1. Gland Segmentation
3.2.2. Tumor Classification
3.2.3. Tumor Microenvironment Analysis
3.2.4. Prognosis Prediction
3.3. Limitation of This Study
4. Materials and Methods
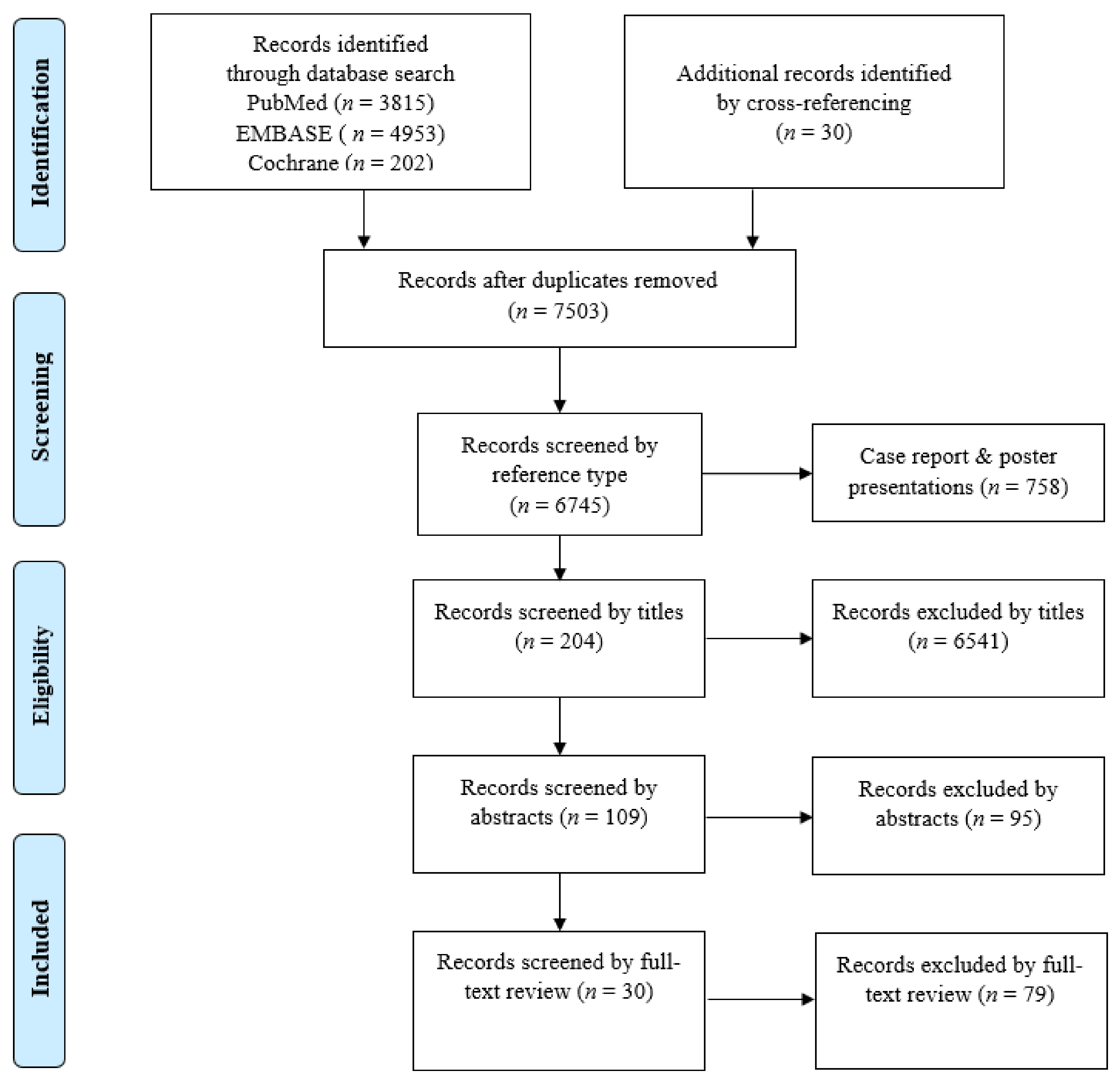
4.1. Literature Search
4.2. Study Selection, Reviewing, and Data Retrieval
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Center, M.M.; Jemal, A.; Ward, E. International Trends in Colorectal Cancer Incidence Rates. Cancer Epidemiol. Biomark. Amp Amp Prev. 2009, 18, 1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, R.; Sauvaget, C.; Sankaranarayanan, R. Mass screening for colorectal cancer is not justified in most developing countries. Int. J. Cancer 2009, 125, 253–256. [Google Scholar] [CrossRef]
- Joseph, D.A.; Meester, R.G.S.; Zauber, A.G.; Manninen, D.L.; Winges, L.; Dong, F.B.; Peaker, B.; van Ballegooijen, M. Colorectal cancer screening: Estimated future colonoscopy need and current volume and capacity. Cancer 2016, 122, 2479–2486. [Google Scholar] [CrossRef]
- Van den Bent, M.J. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician’s perspective. Acta Neuropathol. 2010, 120, 297–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, C.A.; Nesi, G.; Messerini, L.; Zampi, G.C.; Mandai, K.; Itabashi, M.; Takubo, K. The Vienna classification applied to colorectal adenomas. J. Gastroenterol. Hepatol. 2006, 21, 1697–1703. [Google Scholar] [CrossRef]
- Japanese Society for Cancer of the Colon and Rectum. Japanese Classification of Colorectal, Appendiceal, and Anal Carcinoma: The 3d English Edition [Secondary Publication]. J. Anus Rectum Colon 2019, 3, 175–195. [Google Scholar] [CrossRef] [Green Version]
- Schlemper, R.J.; Itabashi, M.; Kato, Y.; Lewin, K.J.; Riddell, R.H.; Shimoda, T.; Sipponen, P.; Stolte, M.; Watanabe, H. Differences in the diagnostic criteria used by japanese and western pathologists to diagnose colorectal carcinoma. Cancer 1998, 82, 60–69. [Google Scholar] [CrossRef] [Green Version]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Nam, S.; Chong, Y.; Jung, C.K.; Kwak, T.-Y.; Lee, J.Y.; Park, J.; Rho, M.J.; Go, H. Introduction to digital pathology and computer-aided pathology. J. Pathol. Transl. Med. 2020, 54, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Amisha; Malik, P.; Pathania, M.; Rathaur, V.K. Overview of artificial intelligence in medicine. J. Fam. Med. Prim. Care 2019, 8, 2328–2331. [Google Scholar] [CrossRef] [PubMed]
- Azuaje, F. Artificial intelligence for precision oncology: Beyond patient stratification. NPJ Precis. Oncol. 2019, 3, 6. [Google Scholar] [CrossRef]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; Consortium, a.t.C. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Ertosun, M.G.; Rubin, D.L. Automated Grading of Gliomas using Deep Learning in Digital Pathology Images: A modular approach with ensemble of convolutional neural networks. AMIA Annu. Symp. Proc. AMIA Symp. 2015, 2015, 1899–1908. [Google Scholar] [PubMed]
- Teramoto, A.; Tsukamoto, T.; Kiriyama, Y.; Fujita, H. Automated Classification of Lung Cancer Types from Cytological Images Using Deep Convolutional Neural Networks. Biomed Res. Int. 2017, 2017, 4067832. [Google Scholar] [CrossRef] [PubMed]
- Meier, A.; Nekolla, K.; Earle, S.; Hewitt, L.; Aoyama, T.; Yoshikawa, T.; Schmidt, G.; Huss, R.; Grabsch, H.I. 77PEnd-to-end learning to predict survival in patients with gastric cancer using convolutional neural networks. Ann. Oncol. 2018, 29. [Google Scholar] [CrossRef]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-van de Kaa, C.; Bult, P.; van Ginneken, B.; van der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.Y.; Jung, C.K.; Woo, J.I.; Lee, S.; Cho, J.; Kim, S.W.; Kwak, T.-Y. Artificial Intelligence in Pathology. J. Pathol. Transl. Med. 2019, 53, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Yamashita, Y.; Shimazu, T.; Cosatto, E.; Kiyuna, T.; Taniguchi, H.; Sekine, S.; Ochiai, A. Automated histological classification of whole slide images of colorectal biopsy specimens. Oncotarget 2017, 8, 90719. [Google Scholar] [CrossRef] [Green Version]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.-A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef]
- Chen, H.; Qi, X.; Yu, L.; Heng, P.-A. DCAN: Deep Contour-Aware Networks for Accurate Gland Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; IEEE: Las Vegas, NV, USA, 2016; pp. 2487–2496. [Google Scholar] [CrossRef] [Green Version]
- Sirinukunwattana, K.; Pluim, J.P.W.; Chen, H.; Qi, X.; Heng, P.-A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BenTaieb, A.; Hamarneh, G. Topology aware fully convolutional networks for histology gland segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2016; pp. 460–468. [Google Scholar]
- Li, W.; Manivannan, S.; Akbar, S.; Zhang, J.; Trucco, E.; McKenna, S.J. Gland Segmentation in Colon Histology Images Using Hand-Crafted Features And Convolutional Neural Networks. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI); IEEE: Prague, Czech Republic, 2016; pp. 1405–1408. [Google Scholar]
- Yang, L.; Zhang, Y.; Chen, J.; Zhang, S.; Chen, D.Z. Suggestive Annotation: A Deep Active Learning Framework for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017; pp. 399–407. [Google Scholar]
- Kainz, P.; Pfeiffer, M.; Urschler, M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ 2017, 5, e3874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, L.; Chen, J.; Fredericksen, M.; Hughes, D.P.; Chen, D.Z. Deep Adversarial Networks for Biomedical Image Segmentation Utilizing Unannotated Images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017; pp. 408–416. [Google Scholar]
- Xu, Y.; Li, Y.; Wang, Y.; Liu, M.; Fan, Y.; Lai, M.; Eric, I.; Chang, C. Gland instance segmentation using deep multichannel neural networks. IEEE Trans. Biomed. Eng. 2017, 64, 2901–2912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, S.; Chen, H.; Gamper, J.; Dou, Q.; Heng, P.-A.; Snead, D.; Tsang, Y.W.; Rajpoot, N. MILD-Net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Med. Image Anal. 2019, 52, 199–211. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Yang, X.; Cheng, K.-T.T. A Deep Model with Shape-Preserving Loss for Gland Instance Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018; pp. 138–146. [Google Scholar]
- Manivannan, S.; Li, W.; Zhang, J.; Trucco, E.; McKenna, S.J. Structure prediction for gland segmentation with hand-crafted and deep convolutional features. IEEE Trans. Med. Imaging 2017, 37, 210–221. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Li, J.; Xu, X. Segnet-Based Gland Segmentation from Colon Cancer Histology Images. In Proceedings of the 2018 33rd Youth Academic Annual Conference of Chinese Association of Automation (YAC); IEEE: Prague, Czech Republic, 2018; pp. 1078–1082. [Google Scholar]
- Ding, H.; Pan, Z.; Cen, Q.; Li, Y.; Chen, S. Multi-scale fully convolutional network for gland segmentation using three-class classification. Neurocomputing 2020, 380, 150–161. [Google Scholar] [CrossRef]
- Raza, S.E.A.; Cheung, L.; Epstein, D.; Pelengaris, S.; Khan, M.; Rajpoot, N.M. Mimonet: Gland Segmentation Using Multi-Input-Multi-Output Convolutional Neural Network. In Proceedings of the Annual Conference on Medical Image Understanding and Analysis; Springer: Cham, Switzerland, 2017; pp. 698–706. [Google Scholar]
- Liu, L.; Wu, J.; Li, D.; Senhadji, L.; Shu, H. Fractional Wavelet Scattering Network and Applications. IEEE Trans. Biomed. Eng. 2019, 66, 553–563. [Google Scholar] [CrossRef]
- Binder, T.; Tantaoui, E.M.; Pati, P.; Catena, R.; Set-Aghayan, A.; Gabrani, M. Multi-organ gland segmentation using deep learning. Front. Med. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Khvostikov, A.; Krylov, A.; Mikhailov, I.; Kharlova, O.; Oleynikova, N.; Malkov, P. Automatic mucous glands segmentation in histological images. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-2/W12, 103–109. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Jia, Z.; Wang, L.B.; Ai, Y.; Zhang, F.; Lai, M.; Chang, E.I. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 281. [Google Scholar] [CrossRef] [Green Version]
- Korbar, B.; Olofson, A.M.; Miraflor, A.P.; Nicka, C.M.; Suriawinata, M.A.; Torresani, L.; Suriawinata, A.A.; Hassanpour, S. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J. Pathol. Inf. 2017, 8, 30. [Google Scholar] [CrossRef]
- Haj-Hassan, H.; Chaddad, A.; Harkouss, Y.; Desrosiers, C.; Toews, M.; Tanougast, C. Classifications of Multispectral Colorectal Cancer Tissues Using Convolution Neural Network. J. Pathol. Inf. 2017, 8, 1. [Google Scholar] [CrossRef]
- Yoon, H.; Lee, J.; Oh, J.E.; Kim, H.R.; Lee, S.; Chang, H.J.; Sohn, D.K. Tumor Identification in Colorectal Histology Images Using a Convolutional Neural Network. J. Digit. Imaging 2019, 32, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Ponzio, F.; Macii, E.; Ficarra, E.; Di Cataldo, S. Colorectal Cancer Classification Using Deep Convolutional Networks. In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies 2018, Funchal, Portugal, 19–21 January 2018; Volume 2, pp. 58–66. [Google Scholar]
- Sena, P.; Fioresi, R.; Faglioni, F.; Losi, L.; Faglioni, G.; Roncucci, L. Deep learning techniques for detecting preneoplastic and neoplastic lesions in human colorectal histological images. Oncol. Lett. 2019, 18, 6101–6107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iizuka, O.; Kanavati, F.; Kato, K.; Rambeau, M.; Arihiro, K.; Tsuneki, M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci. Rep. 2020, 10, 1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Microscopic nuclei classification, segmentation and detection with improved Deep Convolutional Neural Network (DCNN) approaches. arXiv, 2018; arXiv:1811.03447. [Google Scholar]
- Swiderska-Chadaj, Z.; Pinckaers, H.; van Rijthoven, M.; Balkenhol, M.; Melnikova, M.; Geessink, O.; Manson, Q.; Sherman, M.; Polonia, A.; Parry, J.; et al. Learning to detect lymphocytes in immunohistochemistry with deep learning. Med. Image Anal. 2019, 58. [Google Scholar] [CrossRef]
- Shapcott, M.; Hewitt, K.J.; Rajpoot, N. Deep Learning With Sampling in Colon Cancer Histology. Front. Bioeng. Biotechnol. 2019, 7, 52. [Google Scholar] [CrossRef] [Green Version]
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018, 8, 3395. [Google Scholar] [CrossRef]
- The Gland Segmentation in Colon Histology Images (GlaS) Challenge. Available online: https://warwick.ac.uk/fac/sci/dcs/research/tia/glascontest/ (accessed on 15 May 2020).
- Montgomery, E.; Voltaggio, L. Biopsy Interpretation of the Gastrointestinal Tract Mucosa, 3rd ed.; Wolters Kluwer: Philadelphia, PA, USA, 2018; pp. 10–12. [Google Scholar]
- Schlemper, R.J.; Riddell, R.H.; Kato, Y.; Borchard, F.; Cooper, H.S.; Dawsey, S.M.; Dixon, M.F.; Fenoglio-Preiser, C.M.; Fléjou, J.F.; Geboes, K.; et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut 2000, 47, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhao, Y.; Dai, Y.; Cheng, J.-N.; Gong, Z.; Feng, Y.; Sun, C.; Jia, Q.; Zhu, B. Immune Landscape of Colorectal Cancer Tumor Microenvironment from Different Primary Tumor Location. Front. Immunol. 2018, 9, 1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idos, G.E.; Kwok, J.; Bonthala, N.; Kysh, L.; Gruber, S.B.; Qu, C. The Prognostic Implications of Tumor Infiltrating Lymphocytes in Colorectal Cancer: A Systematic Review and Meta-Analysis. Sci. Rep. 2020, 10, 3360. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ge, X.; He, J.; Cheng, Y.; Wang, Z.; Wang, J.; Sun, L. The prognostic value of tumor-infiltrating lymphocytes in colorectal cancer differs by anatomical subsite: A systematic review and meta-analysis. World J. Surg. Oncol. 2019, 17, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogino, S.; Nosho, K.; Irahara, N.; Meyerhardt, J.A.; Baba, Y.; Shima, K.; Glickman, J.N.; Ferrone, C.R.; Mino-Kenudson, M.; Tanaka, N.; et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 6412–6420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, A.F.; Bretes, L.; Furtado, I. Review of PD-1/PD-L1 Inhibitors in Metastatic dMMR/MSI-H Colorectal Cancer. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- European guidelines for quality assurance in colorectal cancer screening and diagnosis: Overview and introduction to the full Supplement publication. Endoscopy 2013, 45, 51–59. [CrossRef] [Green Version]
- Van Putten, P.G.; Hol, L.; van Dekken, H.; Han van Krieken, J.; van Ballegooijen, M.; Kuipers, E.J.; van Leerdam, M.E. Inter-observer variation in the histological diagnosis of polyps in colorectal cancer screening. Histopathology 2011, 58, 974–981. [Google Scholar] [CrossRef] [Green Version]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Compton, C.C. Optimal Pathologic Staging: Defining Stage II Disease. Clin. Cancer Res. 2007, 13, 6862s–6870s. [Google Scholar] [CrossRef] [Green Version]
- Nauta, R.; Stablein, D.M.; Holyoke, E.D. Survival of Patients With Stage B2 Colon Carcinoma: The Gastrointestinal Tumor Study Group Experience. Arch. Surg. 1989, 124, 180–182. [Google Scholar] [CrossRef]
- Wang, S.; Yang, D.M.; Rong, R.; Zhan, X.; Fujimoto, J.; Liu, H.; Minna, J.; Wistuba, I.I.; Xie, Y.; Xiao, G. Artificial intelligence in lung cancer pathology image analysis. Cancers 2019, 11, 1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153–173. [Google Scholar] [CrossRef] [PubMed]
- Klaver, C.E.L.; Bulkmans, N.; Drillenburg, P.; Grabsch, H.I.; van Grieken, N.C.T.; Karrenbeld, A.; Koens, L.; van Lijnschoten, I.; Meijer, J.; Nagtegaal, I.D.; et al. Interobserver, intraobserver, and interlaboratory variability in reporting pT4a colon cancer. Virchows Arch. 2020, 476, 219–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, M.S.; Chan, L.; Tse, G.; Tang, M.; Deng, J.; Norouzi, S.; Rowsell, C.; Plataniotis, K.N.; Damaskinos, S. Atlas of Digital Pathology: A Generalized Hierarchical Histological Tissue Type-Annotated Database for Deep Learning. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 11739–11748. [Google Scholar]
- Maier-Hein, L.; Eisenmann, M.; Reinke, A.; Onogur, S.; Stankovic, M.; Scholz, P.; Arbel, T.; Bogunovic, H.; Bradley, A.P.; Carass, A.; et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 2018, 9, 5217. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Model No. | Author/Team | Dataset | Base Model | Performance Metrics | Year | Country | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1-Score | Object Dice | Object Hausdorff | ||||||||||
| Part A | Part B | Part A | Part B | Part A | Part B | |||||||
| 1 * | CUMed Vision2 | WQD | DCAN | 0.912 | 0.716 | 0.897 | 0.781 | 45.4 | 160.3 | 2015 | Hong Kong | [21] |
| 2 * | ExB1 | WQD | Two path CNN | 0.891 | 0.703 | 0.882 | 0.786 | 57.4 | 145.6 | 2015 | Germany | [22] |
| 3 * | ExB3 | WQD | Two path CNN | 0.896 | 0.719 | 0.886 | 0.765 | 57.4 | 159.9 | 2015 | Germany | [22] |
| 4 * | Freiberg2 | WQD | U-Net | 0.87 | 0.695 | 0.876 | 0.786 | 57.1 | 148.5 | 2015 | Germany | [22] |
| 5 * | CUMed Vision1 | WQD | FCN | 0.868 | 0.769 | 0.867 | 0.8 | 74.6 | 153.6 | 2015 | Hong Kong | [21] |
| 6 * | ExB2 | WQD | Two path CNN | 0.892 | 0.686 | 0.884 | 0.754 | 54.8 | 187.4 | 2015 | Germany | [22] |
| 7 * | Freiburg1 | WQD | U-Net | 0.834 | 0.605 | 0.875 | 0.783 | 57.2 | 146.6 | 2015 | Germany | [22] |
| 8 * | CVML | WQD | CNN | 0.652 | 0.541 | 0.644 | 0.654 | 155.4 | 176.2 | 2015 | UK | [22] |
| 9 * | LIB | WQD | K-means/naïve Bayesian | 0.777 | 0.306 | 0.781 | 0.617 | 112.7 | 190.4 | 2015 | France | [22] |
| 10 * | Vision4GlaS | WQD | Object-Net/Separator-Net | 0.635 | 0.527 | 0.737 | 0.61 | 107.5 | 210.1 | 2015 | Austria | [22] |
| 11 ǂ | BenTaieb | WQD | FCN+Smoothness+Topology | NA | NA | 0.80 ± 0.12 | NA | NA | 2016 | Canada | [23] | |
| 12 ǂ | Li | 85 WQD | CNN+HC-SVM | NA | NA | 0.87 ± 0.08 | NA | NA | 2016 | UK | [24] | |
| 13 ǂ | Yang | WQD | FCN | 0.921 | 0.855 | 0.904 | 0.858 | 44.7 | 97.0 | 2017 | USA | [25] |
| 14 ǂ | Kainz | WQD | Object-Net | 0.670 | 0.570 | 0.70 | 0.620 | 137.4 | 216.4 | 2017 | Austria | [26] |
| Separator-Net | 0.680 | 0.610 | 0.750 | 0.650 | 103.5 | 187.8 | ||||||
| 15 ǂ | Zhang | WQD | DAN | 0.916 | 0.855 | 0.903 | 0.838 | 45.3 | 105.0 | 2017 | USA | [27] |
| 16 ǂ | Xu | WQD | FCN/DCAN/RPN/CNN | 0.893 | 0.843 | 0.908 | 0.833 | 44.1 | 116.8 | 2017 | China | [28] |
| 17 ǂ | Graham | WQD /16 CRAG WSIs | MILD-Net | 0.914 | 0.844 | 0.913 | 0.836 | 41.5 | 105.9 | 2018 | UK | [29] |
| 18 ǂ | Yan | WQD | Holistically-nested networks | 0.924 | 0.844 | 0.902 | 0.840 | 49.9 | 106.1 | 2018 | Hong Kong | [30] |
| 19 ǂ | Manivannan | WQD | FCN | 0.892 | 0.801 | 0.887 | 0.853 | 51.2 | 87.0 | 2018 | UK | [31] |
| 20 ǂ | Tang | WQD | Segnet | NA | NA | 0.882 | 0.836 | 106.6 | 102.6 | 2018 | China | [32] |
| 21 ǂ | Ding | WQD/213 CRAG images | TCC-MSFCN | 0.914 | 0.85 | 0.913 | 0.858 | 39.8 | 93.2 | 2019 | China | [33] |
| 22 ǂ | Raza | WQD | MIMO-Net | 0.913 | 0.724 | 0.906 | 0.785 | 49.2 | 134.0 | 2019 | UK | [34] |
| 23 ǂ | Liu | WQD | Wavelet Scattering Network | 0.874 | 0.71 | 0.875 | 0.791 | 56.6 | 146.6 | 2019 | China | [35] |
| 24 ǂ | Binder | WQD | U-Net | NA | NA | 0.920 | 11.0 | 2019 | France | [36] | ||
| 25 ǂ | Khvostikov | WQD/20 PATH- DT-MSU images | U-Net | NA | NA | 0:880 | NA | NA | 2019 | Russia | [37] | |
| Model No. | Feature | Task | Dataset | External Cross-Validation | Base Model | Performance | Author/Team | Year | Country | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Tumor Classification | 6 classes (cancer subtypes): NL/ADC/MC/SC/PC/CCTA | 717 patches | Not done | AlexNet | Accuracy—97.5% | Xu | 2017 | China | [38] |
| 2 | 5 classes (polyp subtypes): | 2074 patches 936 WSI | Not done | ResNet | Accuracy—93.0% | Korbar | 2017 | USA | [39] | |
| NL/HP/SSP/TSA/TA/TVA-VA, | ||||||||||
| 3 | 3 classes: NL/AD/ADC | 30 multispectral image patches | Not done | CNN | Accuracy—99.2% | Haj-Hassan | 2017 | France | [40] | |
| 4 | 2 classes: NL/Tumor | 57 WSI (10,280 patches) | Not done | VGG | Accuracy—93.5%, | Yoon | 2018 | South Korea | [41] | |
| Sensitivity—95.1% | ||||||||||
| Specificity—92.8% | ||||||||||
| 5 | 3 classes: NL/AD/ADC | 27 WSI (13,500 patches) | Not done | VGG16 | Accuracy—96 % | Ponzio | 2018 | Italy | [42] | |
| 6 | 4 classes: NL/HP/AD/ADC | 393 WSI | Not done | CNN | Accuracy—80% | Sena | 2019 | Italy | [43] | |
| (12,565 patches) | ||||||||||
| 7 | 3 classes: NL/AD/ADC | 4036 WSI | Not done | CNN/RNN | AUCs—0.96 (ADC) | Iizuka | 2020 | Japan | [44] | |
| 0.99 (AD) | ||||||||||
| 8. | 2 classes: NL/Tumor | 94 WSI, | Done using 378 DACHS data | ResNet18 | AUC > 0.99 | Kather | 2019 | Germany | [45] | |
| 370 TCGA-KR, | ||||||||||
| (60,894 patches) | ||||||||||
| 378 TCGA-DX, | ||||||||||
| (93,408 patches) | ||||||||||
| 9 | Tumor Microenvironment Analysis | Classification, Segmentation and Detection: EC/IC/FC/MC | 21,135 patches | Not done | DCRN/R2U-Net | Classification | Alom | 2018 | USA | [46] |
| F1-score—0.81 | ||||||||||
| AUC—0.96 | ||||||||||
| Accuracy—91.1% | ||||||||||
| Segmentation | ||||||||||
| Accuracy—92.1% | ||||||||||
| Detection | ||||||||||
| F1 score—0.831 | ||||||||||
| 10 | Detection of immune cell CD3+, CD8+ | 28 WSI IHC | Not done | FCN/LSM/U-Net | FI score—0.80 Sensitivity—74.0% Precision—86 | Swiderska-Chadaj | 2019 | Netherland | [47] | |
| 11 | Detection and classification EC/IC/FC/MC | 853 patches & 142 TCGA images | Not done | CNN | Detection Accuracy—65% Classification Accuracy—76 % | Shapcott | 2019 | UK | [48] | |
| 12 | Classification of 9 cell types ADI, BAC, DEB, LYM, MUC, SM, NL, SC and EC | 86 WSI (100,000) NCT&UMM | Not done | VGG19 | Accuracy—94–99% | Kather | 2019 | Germany | [20] | |
| 13 | Prognosis Prediction | 5-year disease-specific survival | 420 TMA | Not done | LSTM | AUC—0.69 | Bychkov | 2018 | Finland | [49] |
| 14 | Survival predictions | 25 DACHS WSI | Not done | VGG19 | Accuracy—94–99% | Kather | 2019 | Germany | [20] | |
| 862 TCGA WSI | ||||||||||
| 409 DACHS WSI | ||||||||||
| 15 | MSI predictions | 360 TCGA- DX (93,408 patches) 378 TCGA- KR (60,894 patches) | Done using 378 DACHS data | ResNet18 | AUC TCGA-DX—0.77 TCGA-KR—0.84 | Kather | 2019 | Germany | [45] |
| Model No. | Author/Team | Performance Metrics | Rank-Sum | Rank-Sum Rank | Year | Ref. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1-Score | Object Dice | Object Hausdorff | |||||||||||||||
| Part A | Rank | Part B | Rank | Part A | Rank | Part B | Rank | Part B | Rank | Part B | Rank | ||||||
| 19 ǂ | Ding | 0.914 | 4 | 0.850 | 3 | 0.913 | 1 | 0.858 | 1 | 39.8 | 1 | 93.2 | 2 | 12 | 1 | 2019 | [33] |
| 11 ǂ | Yang | 0.921 | 2 | 0.855 | 1 | 0.904 | 5 | 0.858 | 1 | 44.7 | 4 | 97.0 | 3 | 16 | 2 | 2017 | [25] |
| 15 ǂ | Graham | 0.914 | 4 | 0.844 | 4 | 0.913 | 1 | 0.836 | 6 | 41.5 | 2 | 105.9 | 5 | 22 | 3 | 2018 | [29] |
| 13 ǂ | Zhang | 0.916 | 3 | 0.855 | 1 | 0.903 | 6 | 0.838 | 5 | 45.3 | 5 | 105.0 | 4 | 24 | 4 | 2017 | [27] |
| 16 ǂ | Yan | 0.924 | 1 | 0.844 | 4 | 0.902 | 7 | 0.840 | 4 | 49.9 | 8 | 106.1 | 6 | 30 | 5 | 2018 | [30] |
| 14 ǂ | Xu | 0.893 | 9 | 0.843 | 6 | 0.908 | 3 | 0.833 | 7 | 44.1 | 3 | 116.8 | 7 | 35 | 6 | 2017 | [28] |
| 17 ǂ | Manivannan | 0.892 | 10 | 0.801 | 7 | 0.887 | 9 | 0.853 | 3 | 51.2 | 9 | 87.0 | 1 | 39 | 7 | 2018 | [31] |
| 20 ǂ | Raza | 0.913 | 6 | 0.724 | 9 | 0.906 | 4 | 0.785 | 12 | 49.2 | 7 | 134.0 | 8 | 46 | 8 | 2019 | [34] |
| 1 * | CUMedVision2 | 0.912 | 7 | 0.716 | 11 | 0.897 | 8 | 0.781 | 14 | 45.4 | 6 | 160.3 | 15 | 61 | 9 | 2015 | [21] |
| 21 ǂ | Liu | 0.874 | 13 | 0.710 | 12 | 0.875 | 14 | 0.791 | 9 | 56.6 | 11 | 146.6 | 10 | 69 | 10 | 2019 | [35] |
| 2 * | ExB1 | 0.891 | 12 | 0.703 | 13 | 0.882 | 12 | 0.786 | 10 | 57.4 | 15 | 145.6 | 9 | 71 | 11 | 2015 | [22] |
| 3 * | ExB3 | 0.896 | 8 | 0.719 | 10 | 0.886 | 10 | 0.765 | 15 | 57.4 | 14 | 159.9 | 14 | 71 | 11 | 2015 | [22] |
| 4 * | Freiberg2 | 0.870 | 14 | 0.695 | 14 | 0.876 | 13 | 0.786 | 10 | 57.1 | 12 | 148.5 | 12 | 75 | 13 | 2015 | [22] |
| 5 * | CUMedVision1 | 0.868 | 15 | 0.769 | 8 | 0.867 | 16 | 0.800 | 8 | 74.6 | 16 | 153.6 | 13 | 76 | 14 | 2015 | [21] |
| 6 * | ExB2 | 0.892 | 10 | 0.686 | 15 | 0.884 | 11 | 0.754 | 16 | 54.8 | 10 | 187.4 | 17 | 79 | 15 | 2015 | [22] |
| 7 * | Freiburg1 | 0.834 | 16 | 0.605 | 17 | 0.875 | 14 | 0.783 | 13 | 57.2 | 13 | 146.6 | 10 | 83 | 16 | 2015 | [22] |
| 12 ǂ | Kainz | 0.680 | 18 | 0.610 | 16 | 0.750 | 18 | 0.650 | 18 | 103.5 | 17 | 187.8 | 18 | 105 | 17 | 2017 | [26] |
| 8 * | CVML | 0.652 | 19 | 0.541 | 18 | 0.644 | 20 | 0.654 | 17 | 155.4 | 20 | 176.2 | 16 | 110 | 18 | 2015 | [22] |
| 9 * | LIB | 0.777 | 17 | 0.306 | 20 | 0.781 | 17 | 0.617 | 19 | 112.7 | 19 | 190.4 | 19 | 111 | 19 | 2015 | [22] |
| 10 * | Vision4GlaS | 0.635 | 20 | 0.527 | 19 | 0.737 | 19 | 0.610 | 20 | 107.5 | 18 | 210.1 | 20 | 116 | 20 | 2015 | [22] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakur, N.; Yoon, H.; Chong, Y. Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review. Cancers 2020, 12, 1884. https://doi.org/10.3390/cancers12071884
Thakur N, Yoon H, Chong Y. Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review. Cancers. 2020; 12(7):1884. https://doi.org/10.3390/cancers12071884
Chicago/Turabian StyleThakur, Nishant, Hongjun Yoon, and Yosep Chong. 2020. "Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review" Cancers 12, no. 7: 1884. https://doi.org/10.3390/cancers12071884
APA StyleThakur, N., Yoon, H., & Chong, Y. (2020). Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review. Cancers, 12(7), 1884. https://doi.org/10.3390/cancers12071884






