CN133, a Novel Brain-Penetrating Histone Deacetylase Inhibitor, Hampers Tumor Growth in Patient-Derived Pediatric Posterior Fossa Ependymoma Models
Abstract
1. Introduction
2. Results
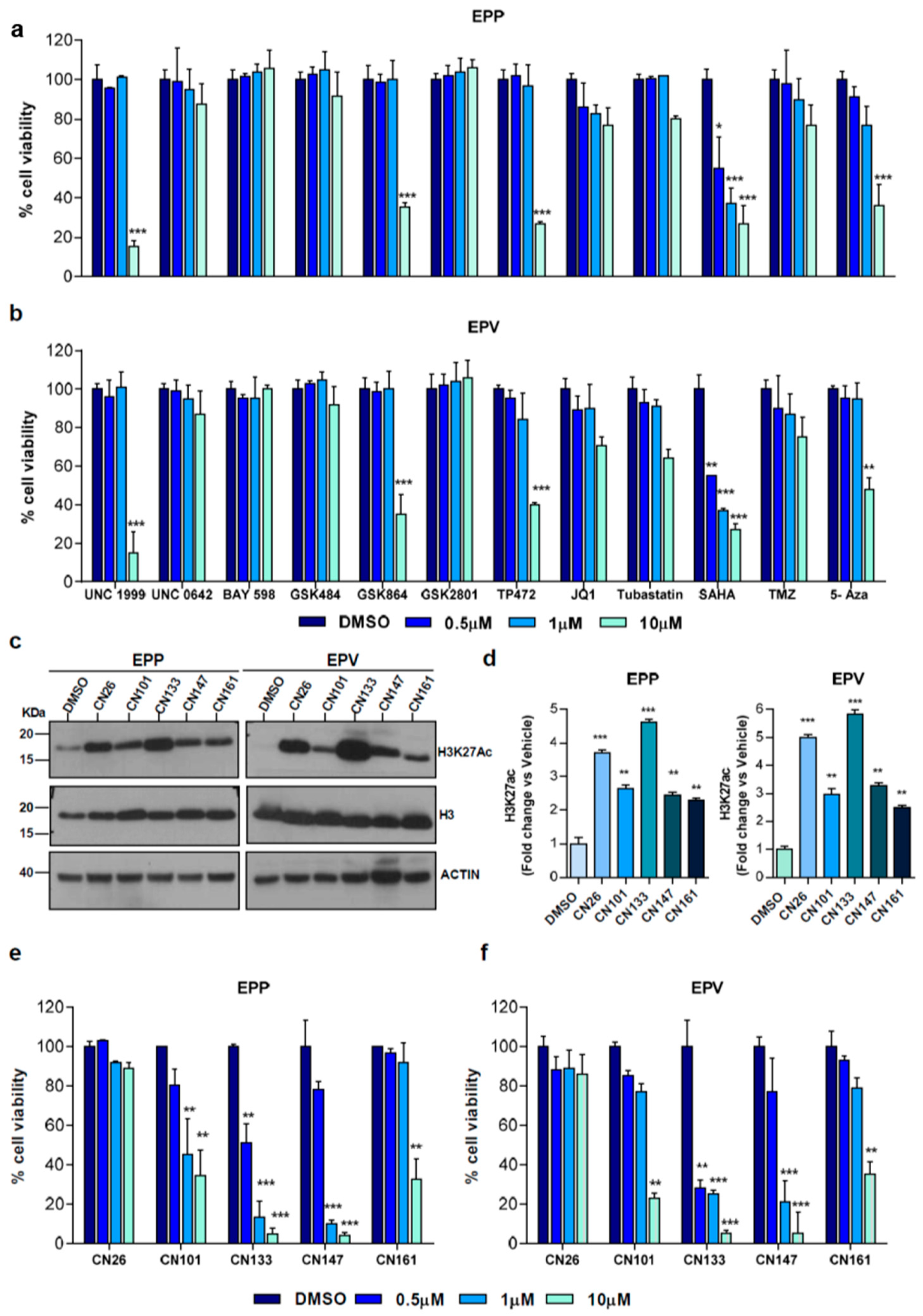
2.1. HDACi Are the Epigenetic Compounds with the Highest Therapeutic Potential in PF-EPN-A Cell Lines
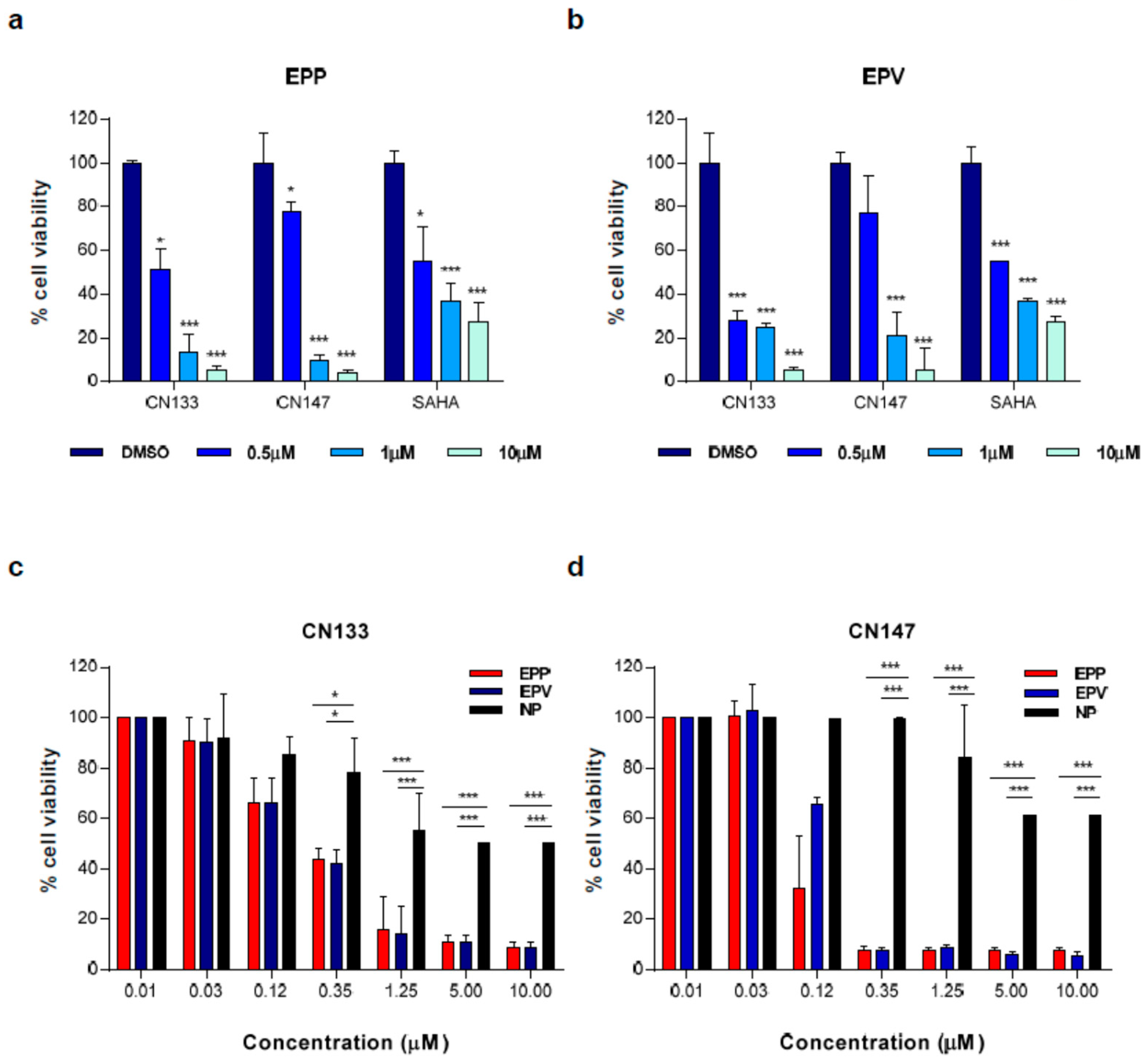
2.2. CN133 and CN147 HDACis Induce Apoptosis in EPN Cell Lines
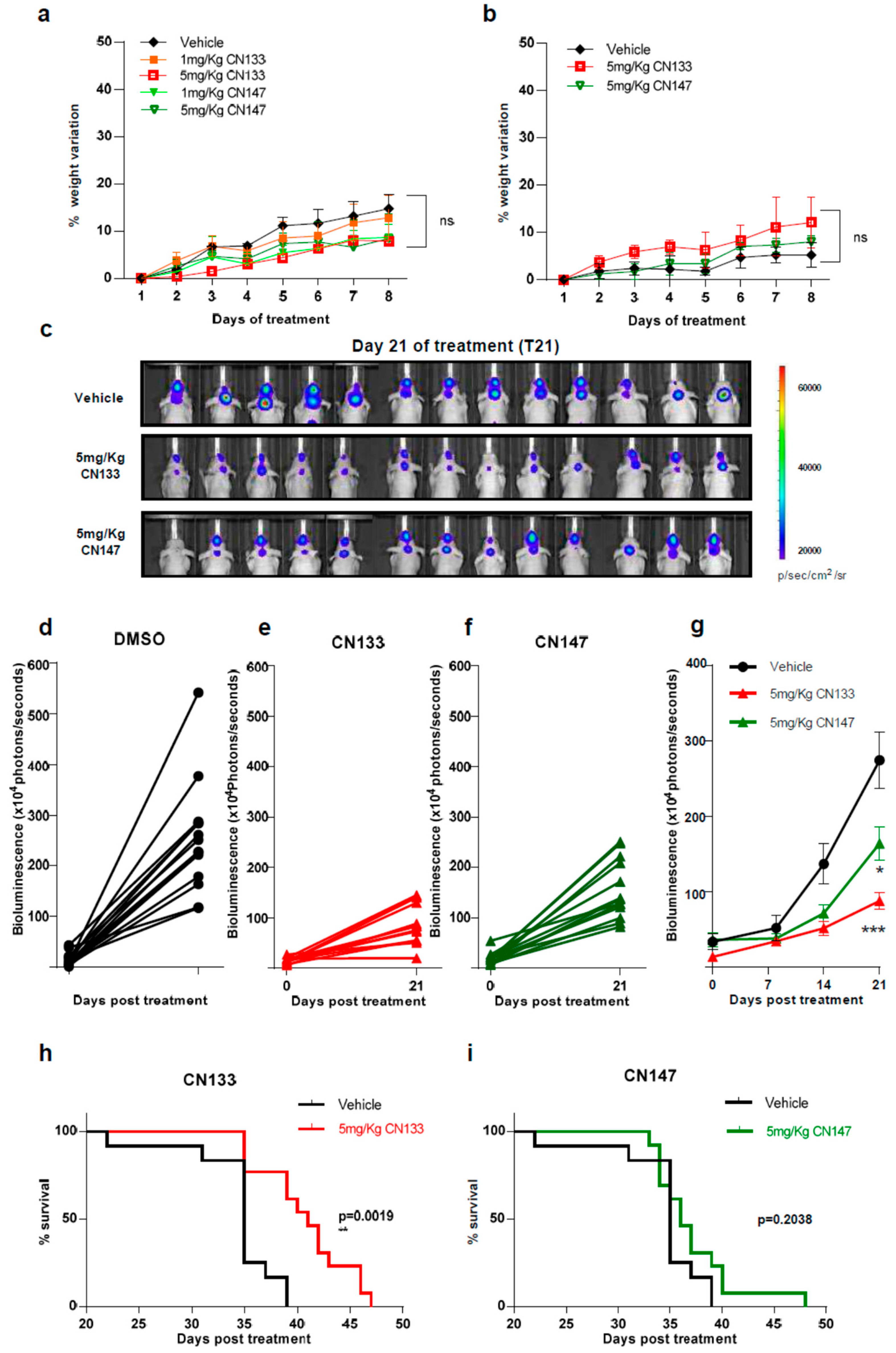
2.3. CN133 Impairs Tumour Growth and Extends Survival of Mice Bearing EPN Orthotopic Xenografts
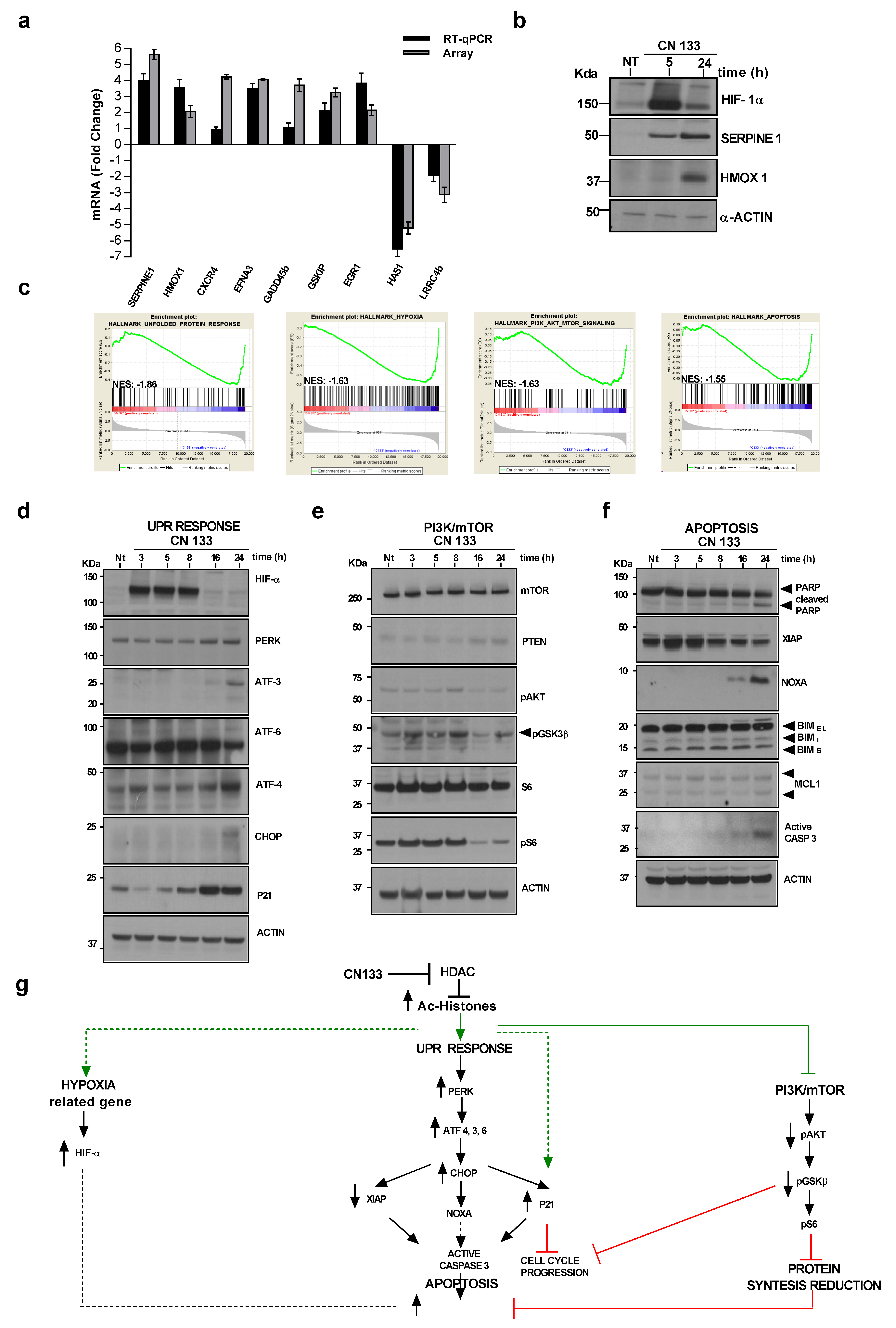
2.4. CN133 Modulates the Expression of Unfolded Protein Response, PI3K/AKT/mTOR, and Apoptosis-Related Genes in EPN Cell Lines
3. Discussion
4. Materials and Methods
4.1. PF-EPN-A Cell Lines
4.2. Chemical Compounds
4.3. Cell Viability Assay
4.4. Western Blot
4.5. Cell Death Assay
4.6. Annexin-V FITC/PI Assay
4.7. Ependymoma Orthotopic Mouse Xenografts
4.8. Transcriptome Analyses
4.9. Real-Time Quantitative PCR
4.10. siRNA Transfection
4.11. Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gatta, G.; Botta, L.; Rossi, S.; Aareleid, T.; Bielska-Lasota, M.; Clavel, J.; Dimitrova, N.; Jakab, Z.; Kaatsch, P.; Lacour, B.; et al. Childhood cancer survival in Europe 1999–2007: Results of EUROCARE-5—A population-based study. Lancet Oncol. 2014, 15, 35–47. [Google Scholar] [CrossRef]
- Cage, T.A.; Clark, A.J.; Aranda, D.; Gupta, N.; Sun, P.P.; Parsa, A.T.; Auguste, K.I. A systematic review of treatment outcomes in pediatric patients with intracranial ependymomas. J. Neurosurg. Pediatr. 2013, 11, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Pajtler, K.W.; Pfister, S.M.; Kool, M. Molecular dissection of ependymomas. Oncoscience 2015, 2, 827–828. [Google Scholar] [CrossRef] [PubMed]
- Pajtler, K.W.; Witt, H.; Sill, M.; Jones, D.T.; Hovestadt, V.; Kratochwil, F.; Wani, K.; Tatevossian, R.; Punchihewa, C.; Johann, P.; et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 2015, 27, 728–743. [Google Scholar] [CrossRef]
- Parker, M.; Mohankumar, K.M.; Punchihewa, C.; Weinlich, R.; Dalton, J.D.; Li, Y.; Lee, R.; Tatevossian, R.G.; Phoenix, T.N.; Thiruvenkatam, R.; et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature 2014, 506, 451–455. [Google Scholar] [CrossRef]
- Ozawa, T.; Arora, S.; Szulzewsky, F.; Juric-Sekhar, G.; Miyajima, Y.; Bolouri, H.; Yasui, Y.; Barber, J.; Kupp, R.; Dalton, J.; et al. A De Novo Mouse Model of C11orf95-RELA Fusion-Driven Ependymoma Identifies Driver Functions in Addition to NF-kappaB. Cell Rep. 2018, 23, 3787–3797. [Google Scholar] [CrossRef]
- Witt, H.; Mack, S.C.; Ryzhova, M.; Bender, S.; Sill, M.; Isserlin, R.; Benner, A.; Hielscher, T.; Milde, T.; Remke, M.; et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 2011, 20, 143–157. [Google Scholar] [CrossRef]
- Mack, S.C.; Witt, H.; Piro, R.M.; Gu, L.; Zuyderduyn, S.; Stutz, A.M.; Wang, X.; Gallo, M.; Garzia, L.; Zayne, K.; et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 2014, 506, 445–450. [Google Scholar] [CrossRef]
- Ramaswamy, V.; Taylor, M.D. Treatment implications of posterior fossa ependymoma subgroups. Chin. J. Cancer 2016, 35, 93. [Google Scholar] [CrossRef]
- Thorp, N.; Gandola, L. Management of Ependymoma in Children, Adolescents and Young Adults. Clin. Oncol. (R. Coll. Radiol.) 2019, 31, 162–170. [Google Scholar] [CrossRef]
- Yang, X.J.; Seto, E. HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007, 26, 5310–5318. [Google Scholar] [CrossRef]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 8, 1414. [Google Scholar] [CrossRef] [PubMed]
- Galanis, E.; Jaeckle, K.A.; Maurer, M.J.; Reid, J.M.; Ames, M.M.; Hardwick, J.S.; Reilly, J.F.; Loboda, A.; Nebozhyn, M.; Fantin, V.R.; et al. Phase II trial of vorinostat in recurrent glioblastoma multiforme: A north central cancer treatment group study. J. Clin. Oncol. 2009, 27, 2052–2058. [Google Scholar] [CrossRef] [PubMed]
- Hanson, J.E.; La, H.; Plise, E.; Chen, Y.H.; Ding, X.; Hanania, T.; Sabath, E.V.; Alexandrov, V.; Brunner, D.; Leahy, E.; et al. SAHA enhances synaptic function and plasticity in vitro but has limited brain availability in vivo and does not impact cognition. PLoS ONE 2013, 8, e69964. [Google Scholar] [CrossRef]
- Meco, D.; Servidei, T.; Lamorte, G.; Binda, E.; Arena, V.; Riccardi, R. Ependymoma stem cells are highly sensitive to temozolomide in vitro and in orthotopic models. Neuro Oncol. 2014, 16, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, F.A.; Wang, C.; Van de Bittner, G.C.; Neelamegam, R.; Takakura, W.R.; Karunakaran, A.; Wey, H.Y.; Reis, S.A.; Gale, J.; Zhang, Y.L.; et al. PET imaging demonstrates histone deacetylase target engagement and clarifies brain penetrance of known and novel small molecule inhibitors in rat. ACS Chem. Neurosci. 2014, 5, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Wey, H.Y.; Gilbert, T.M.; Zurcher, N.R.; She, A.; Bhanot, A.; Taillon, B.D.; Schroeder, F.A.; Wang, C.; Haggarty, S.J.; Hooker, J.M. Insights into neuroepigenetics through human histone deacetylase PET imaging. Sci. Transl. Med. 2016, 8, 351ra106. [Google Scholar] [CrossRef]
- Lee, P.; Murphy, B.; Miller, R.; Menon, V.; Banik, N.L.; Giglio, P.; Lindhorst, S.M.; Varma, A.K.; Vandergrift, W.A., 3rd; Patel, S.J.; et al. Mechanisms and clinical significance of histone deacetylase inhibitors: Epigenetic glioblastoma therapy. Anticancer Res. 2015, 35, 615–625. [Google Scholar]
- Tran, T.H.; Shah, A.T.; Loh, M.L. Precision Medicine in Pediatric Oncology: Translating Genomic Discoveries into Optimized Therapies. Clin. Cancer Res. 2017, 23, 5329–5338. [Google Scholar] [CrossRef][Green Version]
- Gore, L.; Triche, T.J., Jr.; Farrar, J.E.; Wai, D.; Legendre, C.; Gooden, G.C.; Liang, W.S.; Carpten, J.; Lee, D.; Alvaro, F.; et al. A multicenter, randomized study of decitabine as epigenetic priming with induction chemotherapy in children with AML. Clin. Epigenetics 2017, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Italiano, A.; Soria, J.C.; Toulmonde, M.; Michot, J.M.; Lucchesi, C.; Varga, A.; Coindre, J.M.; Blakemore, S.J.; Clawson, A.; Suttle, B.; et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: A first-in-human, open-label, phase 1 study. Lancet Oncol. 2018, 19, 649–659. [Google Scholar] [CrossRef]
- Karahoca, M.; Momparler, R.L. Pharmacokinetic and pharmacodynamic analysis of 5-aza-2′-deoxycytidine (decitabine) in the design of its dose-schedule for cancer therapy. Clin. Epigenetics 2013, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Milde, T.; Kleber, S.; Korshunov, A.; Witt, H.; Hielscher, T.; Koch, P.; Kopp, H.G.; Jugold, M.; Deubzer, H.E.; Oehme, I.; et al. A novel human high-risk ependymoma stem cell model reveals the differentiation-inducing potential of the histone deacetylase inhibitor Vorinostat. Acta Neuropathol. 2011, 122, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Reichert, N.; Choukrallah, M.A.; Matthias, P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell Mol. Life Sci. 2012, 69, 2173–2187. [Google Scholar] [CrossRef]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef]
- Komata, T.; Kanzawa, T.; Nashimoto, T.; Aoki, H.; Endo, S.; Kon, T.; Takahashi, H.; Kondo, S.; Tanaka, R. Histone deacetylase inhibitors, N-butyric acid and trichostatin A, induce caspase-8- but not caspase-9-dependent apoptosis in human malignant glioma cells. Int. J. Oncol. 2005, 26, 1345–1352. [Google Scholar] [CrossRef]
- Bangert, A.; Hacker, S.; Cristofanon, S.; Debatin, K.M.; Fulda, S. Chemosensitization of glioblastoma cells by the histone deacetylase inhibitor MS275. Anticancer Drugs 2011, 22, 494–499. [Google Scholar] [CrossRef]
- Egler, V.; Korur, S.; Failly, M.; Boulay, J.L.; Imber, R.; Lino, M.M.; Merlo, A. Histone deacetylase inhibition and blockade of the glycolytic pathway synergistically induce glioblastoma cell death. Clin. Cancer Res. 2008, 14, 3132–3140. [Google Scholar] [CrossRef]
- Richon, V.M.; Sandhoff, T.W.; Rifkind, R.A.; Marks, P.A. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. USA 2000, 97, 10014–10019. [Google Scholar] [CrossRef]
- Huang, H.L.; Peng, C.Y.; Lai, M.J.; Chen, C.H.; Lee, H.Y.; Wang, J.C.; Liou, J.P.; Pan, S.L.; Teng, C.M. Novel oral histone deacetylase inhibitor, MPT0E028, displays potent growth-inhibitory activity against human B-cell lymphoma in vitro and in vivo. Oncotarget 2015, 6, 4976–4991. [Google Scholar] [CrossRef]
- Chen, Y.; Tsai, Y.H.; Tseng, S.H. RECK regulated endoplasmic reticulum stress response and enhanced cisplatin-induced cell death in neuroblastoma cells. Surgery 2013, 154, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Chueh, A.C.; Tse, J.W.T.; Dickinson, M.; Ioannidis, P.; Jenkins, L.; Togel, L.; Tan, B.; Luk, I.; Davalos-Salas, M.; Nightingale, R.; et al. ATF3 Repression of BCL-XL Determines Apoptotic Sensitivity to HDAC Inhibitors across Tumor Types. Clin. Cancer Res. 2017, 23, 5573–5584. [Google Scholar] [CrossRef] [PubMed]
- Bangert, A.; Cristofanon, S.; Eckhardt, I.; Abhari, B.A.; Kolodziej, S.; Hacker, S.; Vellanki, S.H.; Lausen, J.; Debatin, K.M.; Fulda, S. Histone deacetylase inhibitors sensitize glioblastoma cells to TRAIL-induced apoptosis by c-myc-mediated downregulation of cFLIP. Oncogene 2012, 31, 4677–4688. [Google Scholar] [CrossRef]
- Francisco, R.; Perez-Perarnau, A.; Cortes, C.; Gil, J.; Tauler, A.; Ambrosio, S. Histone deacetylase inhibition induces apoptosis and autophagy in human neuroblastoma cells. Cancer Lett. 2012, 318, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Munster, P.N.; Marchion, D.; Thomas, S.; Egorin, M.; Minton, S.; Springett, G.; Lee, J.H.; Simon, G.; Chiappori, A.; Sullivan, D.; et al. Phase I trial of vorinostat and doxorubicin in solid tumours: Histone deacetylase 2 expression as a predictive marker. Br. J. Cancer 2009, 101, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Krauze, A.V.; Myrehaug, S.D.; Chang, M.G.; Holdford, D.J.; Smith, S.; Shih, J.; Tofilon, P.J.; Fine, H.A.; Camphausen, K. A Phase 2 Study of Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients With Glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 2015, 92, 986–992. [Google Scholar] [CrossRef]
- Servidei, T.; Meco, D.; Trivieri, N.; Patriarca, V.; Vellone, V.G.; Zannoni, G.F.; Lamorte, G.; Pallini, R.; Riccardi, R. Effects of epidermal growth factor receptor blockade on ependymoma stem cells in vitro and in orthotopic mouse models. Int. J. Cancer 2012, 131, E791–E803. [Google Scholar] [CrossRef]
- Panwalkar, P.; Clark, J.; Ramaswamy, V.; Hawes, D.; Yang, F.; Dunham, C.; Yip, S.; Hukin, J.; Sun, Y.; Schipper, M.J.; et al. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 2017, 134, 705–714. [Google Scholar] [CrossRef]
- Bayliss, J.; Mukherjee, P.; Lu, C.; Jain, S.U.; Chung, C.; Martinez, D.; Sabari, B.; Margol, A.S.; Panwalkar, P.; Parolia, A.; et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci. Transl. Med. 2016, 8, 366ra161. [Google Scholar] [CrossRef]
- Rogers, H.A.; Chapman, R.; Kings, H.; Allard, J.; Barron-Hastings, J.; Pajtler, K.W.; Sill, M.; Pfister, S.; Grundy, R.G. Limitations of current in vitro models for testing the clinical potential of epigenetic inhibitors for treatment of pediatric ependymoma. Oncotarget 2018, 9, 36530–36541. [Google Scholar] [CrossRef][Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonelli, R.; Jiménez, C.; Riley, M.; Servidei, T.; Riccardi, R.; Soriano, A.; Roma, J.; Martínez-Saez, E.; Martini, M.; Ruggiero, A.; et al. CN133, a Novel Brain-Penetrating Histone Deacetylase Inhibitor, Hampers Tumor Growth in Patient-Derived Pediatric Posterior Fossa Ependymoma Models. Cancers 2020, 12, 1922. https://doi.org/10.3390/cancers12071922
Antonelli R, Jiménez C, Riley M, Servidei T, Riccardi R, Soriano A, Roma J, Martínez-Saez E, Martini M, Ruggiero A, et al. CN133, a Novel Brain-Penetrating Histone Deacetylase Inhibitor, Hampers Tumor Growth in Patient-Derived Pediatric Posterior Fossa Ependymoma Models. Cancers. 2020; 12(7):1922. https://doi.org/10.3390/cancers12071922
Chicago/Turabian StyleAntonelli, Roberta, Carlos Jiménez, Misha Riley, Tiziana Servidei, Riccardo Riccardi, Aroa Soriano, Josep Roma, Elena Martínez-Saez, Maurizio Martini, Antonio Ruggiero, and et al. 2020. "CN133, a Novel Brain-Penetrating Histone Deacetylase Inhibitor, Hampers Tumor Growth in Patient-Derived Pediatric Posterior Fossa Ependymoma Models" Cancers 12, no. 7: 1922. https://doi.org/10.3390/cancers12071922
APA StyleAntonelli, R., Jiménez, C., Riley, M., Servidei, T., Riccardi, R., Soriano, A., Roma, J., Martínez-Saez, E., Martini, M., Ruggiero, A., Moreno, L., Sánchez de Toledo, J., Gallego, S., Bové, J., Hooker, J. M., & Segura, M. F. (2020). CN133, a Novel Brain-Penetrating Histone Deacetylase Inhibitor, Hampers Tumor Growth in Patient-Derived Pediatric Posterior Fossa Ependymoma Models. Cancers, 12(7), 1922. https://doi.org/10.3390/cancers12071922







