Profiling of Mitochondrial DNA Heteroplasmy in a Prospective Oral Squamous Cell Carcinoma Study
Abstract
1. Introduction
2. Results
2.1. Coverage and Haplogroup Classification
2.2. Quality Control, NUMTS and Contamination Detection
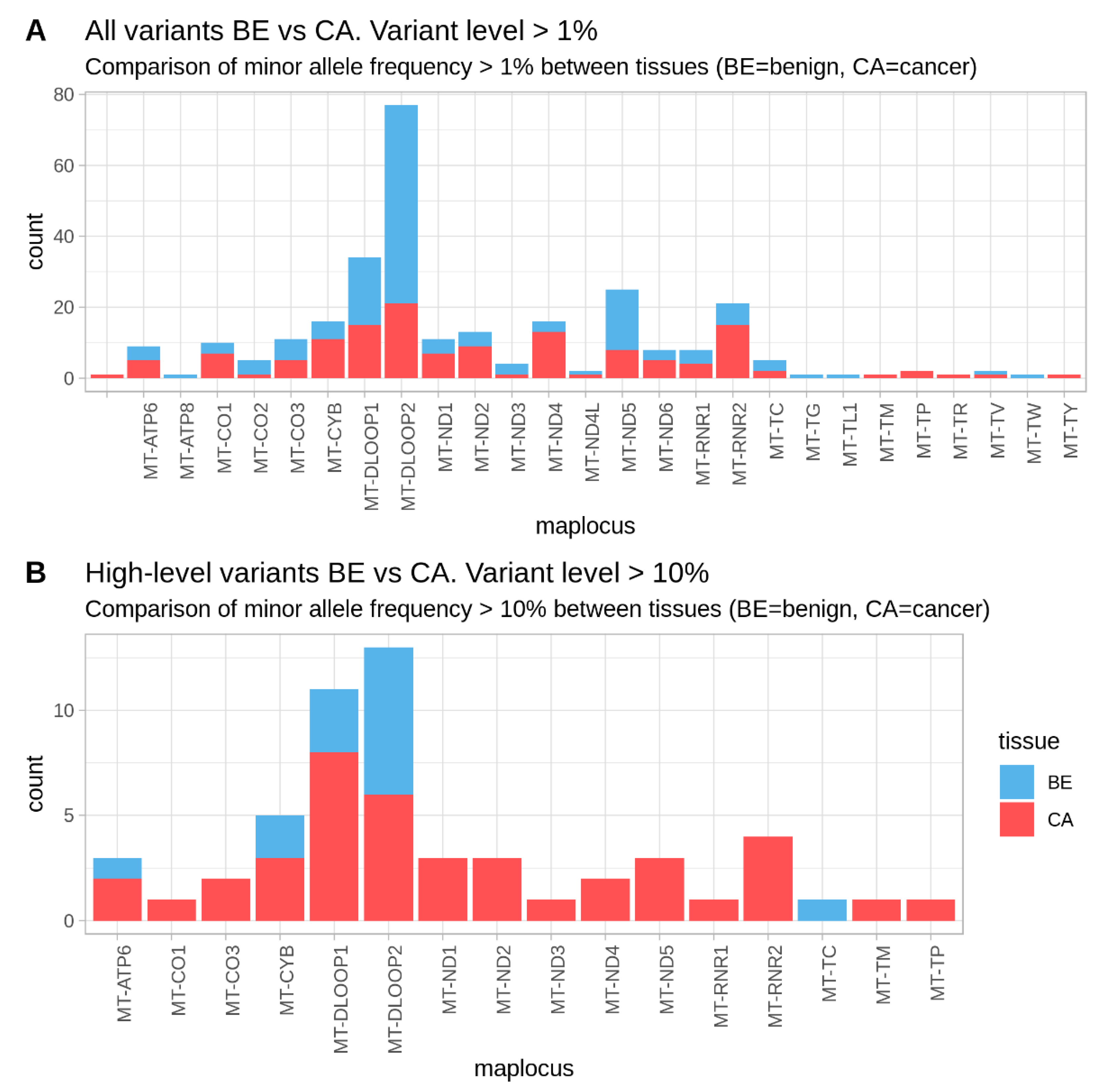
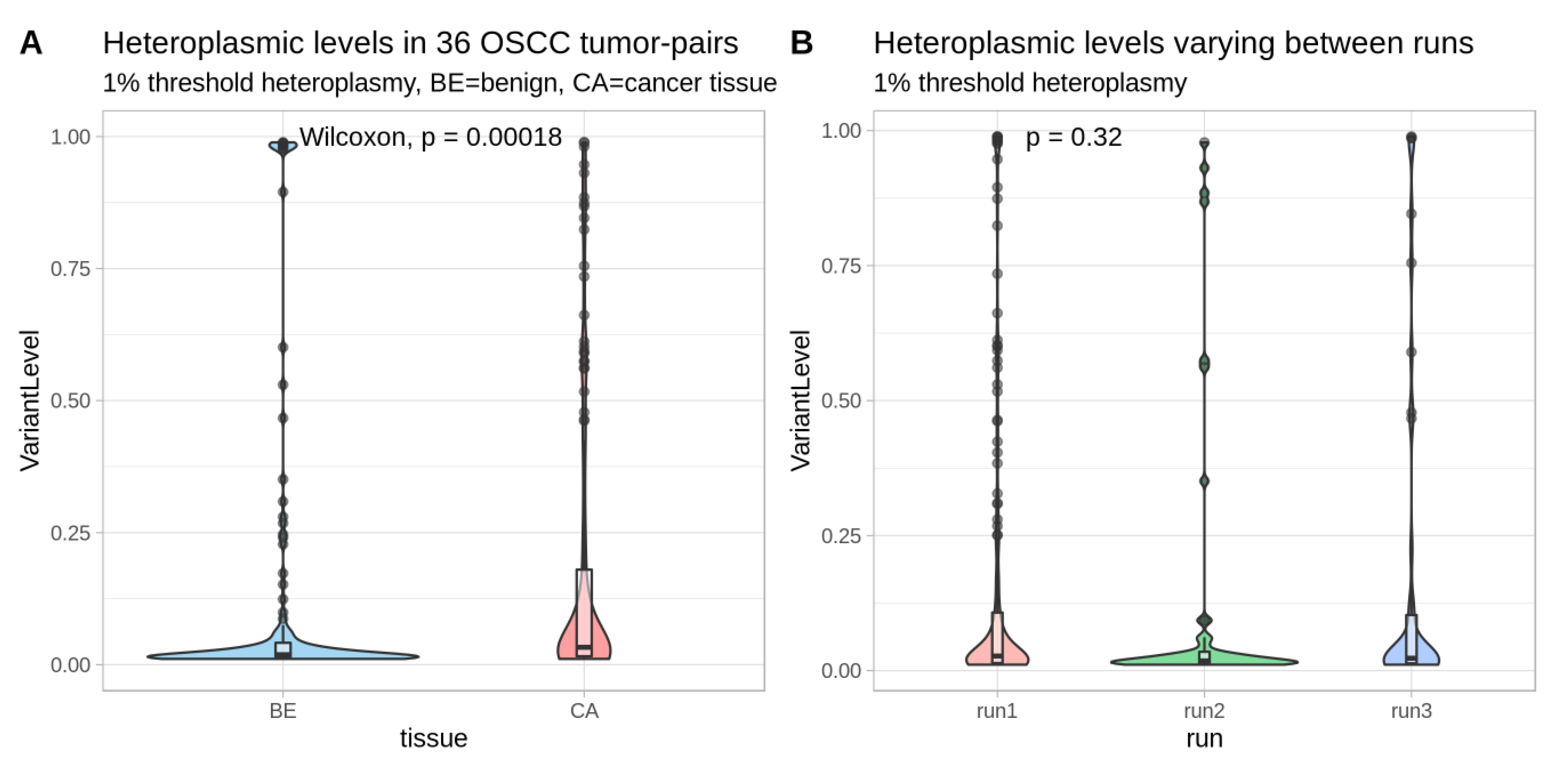
2.3. Mitochondrial DNA Low-Level Heteroplasmic Point Variants in Oral Cancer Sample Pairs
2.4. Mutations with High Heteroplasmy Difference Between Cancer and Benign and Their Clinical Impact
3. Discussion
Strenghts and Limitations
4. Materials and Methods
4.1. Samples
4.2. Clinical Endpoint and Follow-up
4.3. Extraction and Amplification of Mitochondrial DNA
4.4. Next-Generation Sequencing of Mitochondrial Genomes
4.5. Bioinformatic Analysis Pipeline for NGS mtDNA Sequencing Data
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sun, J. RASSF-1A modulates proliferation-mediated oral squamous cell carcinoma progression. Cancer Cell Int. 2019, 19, 213. [Google Scholar] [CrossRef] [PubMed]
- Hema, K.; Smitha, T.; Sheethal, H.; Mirnalini, S.A. Epigenetics in oral squamous cell carcinoma. J. Oral Maxillofac. Pathol. 2017, 21, 252. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Tomita, H.; Nakashima, T.; Hirata, A.; Tanaka, T.; Shibata, T.; Hara, A. Current mouse models of oral squamous cell carcinoma: Genetic and chemically induced models. Oral Oncol. 2017, 73, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Máximo, V.; Soares, P.; Lima, J.; Cameselle-Teijeiro, J.; Sobrinho-Simões, M. Mitochondrial DNA somatic mutations (point mutations and large deletions) and mitochondrial DNA variants in human thyroid pathology: A study with emphasis on Hürthle cell tumors. Am. J. Pathol. 2002, 160, 1857–1865. [Google Scholar] [CrossRef]
- Loogväli, E.L.; Kivisild, T.; Margus, T.; Villems, R. Explaining the imperfection of the molecular clock of hominid mitochondria. PLoS ONE 2009, 4, e8260. [Google Scholar] [CrossRef] [PubMed]
- Kivisild, T.; Shen, P.; Wall, D.P.; Do, B.; Sung, R.; Davis, K.; Passarino, G.; Underhill, P.A.; Scharfe, C.; Torroni, A.; et al. The role of selection in the evolution of human mitochondrial genomes. Genetics 2006, 172, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Kivisild, T.; Reidla, M.; Metspalu, E.; Rosa, A.; Brehm, A.; Pennarun, E.; Parik, J.; Geberhiwot, T.; Usanga, E.; Villems, R. Ethiopian mitochondrial DNA heritage: Tracking gene flow across and around the gate of tears. Am. J. Hum. Genet. 2004, 75, 752–770. [Google Scholar] [CrossRef]
- Behar, D.M.; Villems, R.; Soodyall, H.; Blue-Smith, J.; Pereira, L.; Metspalu, E.; Scozzari, R.; Makkan, H.; Tzur, S.; Comas, D.; et al. The Dawn of Human Matrilineal Diversity. Am. J. Hum. Genet. 2008, 82, 1130–1140. [Google Scholar] [CrossRef]
- Salas, A.; Yao, Y.G.; Macaulay, V.; Vega, A.; Carracedo, Á.; Bandelt, H.J. A critical reassessment of the role of mitochondria in tumorigenesis. PLoS Med. 2005, 2, e296. [Google Scholar] [CrossRef]
- Bussard, K.M.; Siracusa, L.D. Understanding Mitochondrial Polymorphisms in Cancer. Cancer Res. 2017, 77, 6051–6059. [Google Scholar] [CrossRef]
- Kloss-Brandstätter, A.; Schäfer, G.; Erhart, G.; Hüttenhofer, A.; Coassin, S.; Seifarth, C.; Summerer, M.; Bektic, J.; Klocker, H.; Kronenberg, F. Somatic mutations throughout the entire mitochondrial genome are associated with elevated PSA levels in prostate cancer patients. Am. J. Hum. Genet. 2010, 87, 802–812. [Google Scholar] [CrossRef]
- Fendt, L.; Niederstätter, H.; Huber, G.; Zelger, B.; Dünser, M.; Seifarth, C.; Röck, A.; Schäfer, G.; Klocker, H.; Parson, W. Accumulation of mutations over the entire mitochondrial genome of breast cancer cells obtained by tissue microdissection. Breast Cancer Res. Treat. 2011, 128, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Schöpf, B.; Weissensteiner, H.; Schäfer, G.; Fazzini, F.; Charoentong, P.; Naschberger, A.; Rupp, B.; Fendt, L.; Bukur, V.; Giese, I.; et al. OXPHOS remodeling in high-grade prostate cancer involves mtDNA mutations and increased succinate oxidation. Nat. Commun. 2020, 11, 1487. [Google Scholar] [CrossRef] [PubMed]
- Kloss-Brandstätter, A.; Weissensteiner, H.; Erhart, G.; Schäfer, G.; Forer, L.; Schönherr, S.; Pacher, D.; Seifarth, C.; Stöckl, A.; Fendt, L.; et al. Validation of Next-Generation Sequencing of Entire Mitochondrial Genomes and the Diversity of Mitochondrial DNA Mutations in Oral Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0135643. [Google Scholar] [CrossRef] [PubMed]
- Challen, C.; Brown, H.; Cai, C.; Betts, G.; Paterson, I.; Sloan, P.; West, C.; Birch-Machin, M.; Robinson, M. Mitochondrial DNA mutations in head and neck cancer are infrequent and lack prognostic utility. Br. J. Cancer 2011, 104, 1319–1324. [Google Scholar] [CrossRef]
- Uzawa, K.; Baba, T.; Uchida, F.; Yamatoji, M.; Kasamatsu, A.; Sakamoto, Y.; Ogawara, K.; Shiiba, M.; Bukawa, H.; Tanzawa, H. Circulating tumor-derived mutant mitochondrial DNA: A predictive biomarker of clinical prognosis in human squamous cell carcinoma. Oncotarget 2012, 3, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.H.; Huang, S.F.; Liao, C.T.; Chen, I.H.; Wang, H.M.; Hsieh, L.L. Clinical Significance in Oral Cavity Squamous Cell Carcinoma of Pathogenic Somatic Mitochondrial Mutations. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Mondal, R.; Ghosh, S.K. Accumulation of mutations over the complete mitochondrial genome in tobacco-related oral cancer from northeast India. Mitochondrial DNA 2013, 24, 432–439. [Google Scholar] [CrossRef]
- Gissi, D.B.; Tarsitano, A.; Leonardi, E.; Gabusi, A.; Neri, F.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P.; Morandi, L. Clonal analysis as a prognostic factor in multiple oral squamous cell carcinoma. Oral Oncol. 2017, 67, 131–137. [Google Scholar] [CrossRef]
- Morandi, L.; Tarsitano, A.; Gissi, D.; Leonardi, E.; Balbi, T.; Marchetti, C.; Montebugnoli, L.; Foschini, M.P. Clonality analysis in primary oral squamous cell carcinoma and related lymph-node metastasis revealed by TP53 and mitochondrial DNA next generation sequencing analysis. J. Cranio-Maxillofac. Surg. 2015, 43, 208–213. [Google Scholar] [CrossRef]
- Yuan, R.T.; Sun, Y.; Bu, L.X.; Jia, M.Y. Gene mutations in the D-loop region of mitochondrial DNA in oral squamous cell carcinoma. Mol. Med. Rep. 2015, 11, 4496–4500. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Wang, C.C.; Jiang, R.S.; Wang, W.Y.; Liu, S.A. Impact of somatic mutations in the D-Loop of mitochondrial DNA on the survival of oral squamous cell carcinoma patients. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Palodhi, A.; Ghosh, S.; Biswas, N.K.; Basu, A.; Majumder, P.P.; Maitra, A. Profiling of genomic alterations of mitochondrial DNA in gingivobuccal oral squamous cell carcinoma: Implications for disease progress. Mitochondrion 2019, 46, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Schubert, A.D.; Channah Broner, E.; Agrawal, N.; London, N.; Pearson, A.; Gupta, A.; Wali, N.; Seiwert, T.Y.; Wheelan, S.; Lingen, M.; et al. Somatic mitochondrial mutation discovery using ultra-deep sequencing of the mitochondrial genome reveals spatial tumor heterogeneity in head and neck squamous cell carcinoma. Cancer Lett. 2020, 471, 49–60. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Salas, A. Contamination and sample mix-up can best explain some patterns of mtDNA instabilities in buccal cells and oral squamous cell carcinoma. BMC Cancer 2009, 9, 113. [Google Scholar] [CrossRef]
- Weissensteiner, H.; Forer, L.; Fuchsberger, C.; Schöpf, B.; Kloss-Brandstätter, A.; Specht, G.; Kronenberg, F.; Schönherr, S. mtDNA-Server: Next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res. 2016, 44, W64–W69. [Google Scholar] [CrossRef]
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstätter, A.; Forer, L.; Specht, G.; Bandelt, H.-J.; Kronenberg, F.; Salas, A.; Schönherr, S. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef]
- Weissensteiner, H.; Forer, L.; Fendt, L.; Kheirkhah, A.; Salas, A.; Kronenberg, F.; Schoenherr, S. Haplocheck: Phylogeny-based Contamination Detection in Mitochondrial and Whole-Genome Sequencing Studies. bioRxiv 2020. [Google Scholar] [CrossRef]
- Van Oven, M. PhyloTree Build 17: Growing the human mitochondrial DNA tree. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, 9–11. [Google Scholar] [CrossRef]
- Van Oven, M.; Kayser, M.; van Oven, M.; Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009, 30, E386–E394. [Google Scholar] [CrossRef]
- Wei, W.; Pagnamenta, A.T.; Gleadall, N.; Sanchis-Juan, A.; Stephens, J.; Broxholme, J.; Tuna, S.; Odhams, C.A.; Fratter, C.; Turro, E.; et al. Nuclear-mitochondrial DNA segments resemble paternally inherited mitochondrial DNA in humans. Nat. Commun. 2020, 11, 1740. [Google Scholar] [CrossRef] [PubMed]
- Salas, A.; Schönherr, S.; Bandelt, H.-J.; Gómez-Carballa, A.; Weissensteiner, H. Extraordinary claims require extraordinary evidence in asserted mtDNA biparental inheritance. Forensic Sci. Int. Genet. 2020, 47, 102274. [Google Scholar] [CrossRef] [PubMed]
- Balciuniene, J.; Balciunas, D. A Nuclear mtDNA Concatemer (Mega-NUMT) Could Mimic Paternal Inheritance of Mitochondrial Genome. Front. Genet. 2019, 10, 518. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C.; Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a021220. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.; Sabelnykova, V.; Weischenfeldt, J.; Simon, R.; Aguiar, J.; Alkallas, R.; Heisler, L.; Zhang, J.; Watson, J.; Chua, M.; et al. Mitochondrial mutations drive prostate cancer aggression. Nat. Commun. 2017, 2017, 1–7. [Google Scholar] [CrossRef]
- Pereira, L.; Soares, P.; Radivojac, P.; Li, B.; Samuels, D.C. Comparing Phylogeny and the Predicted Pathogenicity of Protein Variations Reveals Equal Purifying Selection across the Global Human mtDNA Diversity. Am. J. Hum. Genet. 2011, 88, 433–439. [Google Scholar] [CrossRef]
- Vigneswaran, N.; Williams, M.D. Epidemiologic Trends in Head and Neck Cancer and Aids in Diagnosis. Oral Maxillofac. Surg. Clin. North Am. 2014, 26, 123–141. [Google Scholar] [CrossRef]
- Ebner, S.; Mangge, H.; Langhof, H.; Halle, M.; Siegrist, M.; Aigner, E.; Paulmichl, K.; Paulweber, B.; Datz, C.; Sperl, W.; et al. Mitochondrial haplogroup T is associated with obesity in Austrian juveniles and adults. PLoS ONE 2015, 10, e0135622. [Google Scholar] [CrossRef]
- Ye, K.; Lu, J.; Ma, F.; Keinan, A.; Gu, Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl. Acad. Sci. USA 2014, 111, 10654–10659. [Google Scholar] [CrossRef] [PubMed]
- Skonieczna, K.; Malyarchuk, B.; Jawień, A.; Marszałek, A.; Banaszkiewicz, Z.; Jarmocik, P.; Grzybowski, T. Mitogenomic differences between the normal and tumor cells of colorectal cancer patients. Hum. Mutat. 2018, 39, 691–701. [Google Scholar] [CrossRef]
- Carinci, F.; Pelucchi, S.; Farina, A.; De Franciscis, G.; Calearo, C. Extension as a prognostic factor in oropharyngeal cancer: Largest mucosal dimension compared with number of (sub)sites involved. Br. J. Oral Maxillofac. Surg. 1998, 36, 440–445. [Google Scholar] [CrossRef]
- McMahon, J.; O’Brien, C.J.; Pathak, I.; Hamill, R.; McNeil, E.; Hammersley, N.; Gardiner, S.; Junor, E. Influence of condition of surgical margins on local recurrence and disease-specific survival in oral and oropharyngeal cancer. Br. J. Oral Maxillofac. Surg. 2003, 41, 224–231. [Google Scholar] [CrossRef]
- Byers, R.M.; El-Naggar, A.K.; Lee, Y.-Y.; Rao, B.; Fornage, B.; Terry, N.H.A.; Sample, D.; Hankins, P.; Smith, T.L.; Wolf, P.J. Can we detect or predict the presence of occult nodal metastases in patients with squamous carcinoma of the oral tongue? Head Neck 1998, 20, 138–144. [Google Scholar] [CrossRef]
- Woolgar, J.A.; Rogers, S.N.; Lowe, D.; Brown, J.S.; Vaughan, E.D. Cervical lymph node metastasis in oral cancer: The importance of even microscopic extracapsular spread. Oral Oncol. 2003, 39, 130–137. [Google Scholar] [CrossRef]
- Suoglu, Y.; Erdamar, B.; Karatay, M.C.; Katircioglu, O.S.; Sunay, T. Extracapsular Spread in Ipsilateral Neck and Contralateral Neck Metastases in Laryngeal Cancer. Ann. Otol. Rhinol. Laryngol. 2002, 111, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Enepekides, D.J.; Sultanem, K.; Nguyen, C.; Shenouda, G.; Black, M.J.; Rochon, L. Occult Cervical Metastases: Immunoperoxidase Analysis of the Pathologically Negative Neck. Otolaryngol. Neck Surg. 1999, 120, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-S.; Siddiq, F.; Talwar, H.S.; Chen, W.; Voichita, C.; Draghici, S.; Jeyapalan, G.; Chatterjee, M.; Fribley, A.; Yoo, G.H.; et al. Serum prognostic biomarkers in head and neck cancer patients. Laryngoscope 2014, 124, 1819–1826. [Google Scholar] [CrossRef]
- Lin, H.-S.; Talwar, H.S.; Tarca, A.L.; Ionan, A.; Chatterjee, M.; Ye, B.; Wojciechowski, J.; Mohapatra, S.; Basson, M.D.; Yoo, G.H.; et al. Autoantibody Approach for Serum-Based Detection of Head and Neck Cancer. Cancer Epidemiol. Biomark. Prev. 2007, 16, 2396–2405. [Google Scholar] [CrossRef]
- Gottschlich, S.; Maune, S.; Maass, J.D.; Görögh, T.; Hoffmann, M.; Hoffmann-Fazel, A.; Meyer, J.; Werner, J.A.; Rudert, H. Serum p53 Autoantibodies in the Follow-Up of Head and Neck Cancer Patients. Oncology 2000, 59, 31–35. [Google Scholar] [CrossRef]
- Dasgupta, S.; Koch, R.; Westra, W.H.; Califano, J.A.; Ha, P.K.; Sidransky, D.; Koch, W.M. Mitochondrial DNA Mutation in Normal Margins and Tumors of Recurrent Head and Neck Squamous Cell Carcinoma Patients. Cancer Prev. Res. 2010, 3, 1205–1211. [Google Scholar] [CrossRef]
- Kumar, M.; Srivastava, S.; Singh, S.A.; Das, A.K.; Das, G.C.; Dhar, B.; Ghosh, S.K.; Mondal, R. Cell-free mitochondrial DNA copy number variation in head and neck squamous cell carcinoma: A study of non-invasive biomarker from Northeast India. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Cicchillitti, L.; Corrado, G.; De Angeli, M.; Mancini, E.; Baiocco, E.; Patrizi, L.; Zampa, A.; Merola, R.; Martayan, A.; Conti, L.; et al. Circulating cell-free DNA content as blood based biomarker in endometrial cancer. Oncotarget 2017, 8, 115230–115243. [Google Scholar] [CrossRef] [PubMed]
- Creed, J.; Klotz, L.; Harbottle, A.; Maggrah, A.; Reguly, B.; George, A.; Gnanapragasm, V. A single mitochondrial DNA deletion accurately detects significant prostate cancer in men in the PSA ‘grey zone’. World J. Urol. 2017, 36, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Kirches, E. MtDNA As a Cancer Marker: A Finally Closed Chapter? Curr. Genom. 2017, 18, 255–267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Schröder, R.; Ni, S.; Madea, B.; Stoneking, M. Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc. Natl. Acad. Sci. USA 2015, 112, 2491–2496. [Google Scholar] [CrossRef]
- Aoki, K.; Tanaka, H.; Kawahara, T. Multiplexed Microsphere Suspension-Array Assay for Urine Mitochondrial DNA Typing by C-Stretch Length in Hypervariable Regions. J. Clin. Med. Res. 2018, 10, 552–561. [Google Scholar] [CrossRef][Green Version]
- Fendt, L.; Zimmermann, B.; Daniaux, M.; Parson, W. Sequencing strategy for the whole mitochondrial genome resulting in high quality sequences. BMC Genom. 2009, 10, 139. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Dayama, G.; Emery, S.B.; Kidd, J.M.; Mills, R.E. The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res. 2014, 42, 12640–12649. [Google Scholar] [CrossRef] [PubMed]



| Sample ID | Age at Diagnosis | Gender | Material Used | Smoking Status | Time to Death after Diagnosis (Months) | Follow Up Time (Months) | T | N | M | R | Grading | Coding-Region Mutations | Haplogroup |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MKG1 | 32 | F | Primary tumor | no-smoker | - | 128 | 2 | 0 | 0 | 0 | 3 | yes | H1 |
| MKG2 | 71 | F | Primary tumor | no-smoker | - | 118 | 1 | 0 | 0 | 0 | 2 | no | U5 |
| MKG4 | 70 | M | Primary tumor | active smoker | 64 | 64 | 4a | 1 | 0 | 1 | 3 | no | H16 |
| MKG5 | 73 | M | Primary tumor | former smoker | 107 | 107 | 4a | 0 | NA | 1 | 2 | no | K1 |
| MKG6 | 58 | M | Primary tumor | active smoker | 27 | 27 | 4a | 1 | 1 | 0 | 2 | yes | H1 |
| MKG7 | 59 | M | Primary tumor | former smoker | - | 119 | 4a | 0 | 0 | 0 | 2 | no | H27 |
| MKG8 | 51 | F | Recurrence | active smoker | 38 | 38 | 4 | 0 | 0 | 1 | 2 | yes | H11 |
| MKG9 | 62 | F | Primary tumor | no-smoker | - | 114 | 1 | 0 | 0 | 0 | 1 | no | J1 |
| MKG10 | 60 | M | Recurrence | former smoker | 47 | 47 | 4a | 0 | 0 | 0 | 2 | no | H13 |
| MKG11 | 53 | M | Primary tumor | former smoker | - | 116 | 4 | 0 | 0 | 0 | 3 | no | J1 |
| MKG12 | 50 | M | Primary tumor | no-smoker | - | 54 | 2 | 1 | 0 | 0 | 2 | no | H11 |
| MKG13 | 64 | F | Recurrence | no-smoker | 83 | 83 | 4a | 0 | 0 | 0 | 2 | no | T1 |
| MKG14 | 63 | M | Primary tumor | active smoker | - | 111 | 4a | 0 | 0 | 1 | NA | no | H1 |
| MKG15 | 64 | M | Primary tumor | active smoker | 6 | 6 | 1 | 2a | 0 | 0 | 3 | yes | H3 |
| MKG16 | 49 | M | Primary tumor | former smoker | - | 98 | 1 | 0 | 0 | 0 | 3 | no | U4 |
| MKG17 | 68 | F | Primary tumor | no-smoker | - | 43 | 2 | 0 | 0 | 0 | 2 | no | H5 |
| MKG18 | 68 | M | Primary tumor | former smoker | - | 97 | 2 | 1 | 0 | 0 | 2 | no | T2 |
| MKG19 | 37 | M | Recurrence | no-smoker | 36 | 36 | 4a | 0 | 0 | 0 | 3 | no | H1 |
| MKG20 | 57 | M | Primary tumor | active smoker | 7 | 7 | 4a | 1 | 0 | 1 | 3 | yes | H |
| MKG21 | 75 | F | Primary tumor | no-smoker | 23 | 23 | 2 | 0 | 0 | 0 | 2 | no | U5 |
| MKG22 | 67 | M | Primary tumor | former smoker | - | 103 | 2 | 0 | 0 | 0 | 2 | yes | U5 |
| MKG23 | 56 | M | Primary tumor | former smoker | 42 | 42 | 2 | 1 | 0 | 0 | 3 | no | W1 |
| MKG24 | 45 | M | Primary tumor | active smoker | 13 | 13 | 4a | 0 | 0 | 0 | 2 | no | H5 |
| MKG25 | 66 | F | Primary tumor | former smoker | 1 | 1 | 2 | 0 | 0 | 0 | 2 | yes | H3 |
| MKG26 | 62 | M | Primary tumor | former smoker | - | 45 | 2 | 2b | 0 | 0 | 2 | no | H |
| MKG27 | 59 | F | Primary tumor | active smoker | 15 | 15 | 2 | 2b | 0 | 0 | 2 | no | H3 |
| MKG28 | 73 | M | Recurrence | active smoker | 27 | 27 | 2 | 0 | 0 | 0 | 2 | no | H1 |
| KT000 | 60 | M | Recurrence | former smoker | - | 139 | 1 | 0 | 0 | 0 | 2 | yes | H65 |
| KT001 | 54 | F | Primary tumor | active smoker | 1 | 1 | 4b | 2c | 1 | NA | 2 | yes | J1 |
| KT003 | 72 | M | Primary tumor | active smoker | - | 70 | 2 | 2b | 0 | 1 | 2 | no | K2 |
| KT007 | 45 | M | Primary tumor | active smoker | 16 | 16 | 2 | 0 | 0 | 0 | 2 | no | H3 |
| KT008 | 83 | F | Primary tumor | no-smoker | - | 65 | 1 | 0 | 0 | 0 | 0 | no | U2 |
| KT012 | 50 | M | Primary tumor | active smoker | - | 52 | 2 | 0 | 0 | 0 | 3 | no | H3 |
| KT013 | 73 | M | Recurrence | no-smoker | - | 104 | 1 | 0 | 0 | 0 | 2 | no | H1 |
| KT014 | 57 | F | Primary tumor | active smoker | - | 41 | 1 | 0 | 0 | 0 | 2 | yes | K2 |
| KT016 | 62 | M | Primary tumor | no-smoker | - | 39 | 4a | 1 | 0 | 0 | 3 | yes | H10 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fendt, L.; Fazzini, F.; Weissensteiner, H.; Bruckmoser, E.; Schönherr, S.; Schäfer, G.; Losso, J.L.; Streiter, G.A.; Lamina, C.; Rasse, M.; et al. Profiling of Mitochondrial DNA Heteroplasmy in a Prospective Oral Squamous Cell Carcinoma Study. Cancers 2020, 12, 1933. https://doi.org/10.3390/cancers12071933
Fendt L, Fazzini F, Weissensteiner H, Bruckmoser E, Schönherr S, Schäfer G, Losso JL, Streiter GA, Lamina C, Rasse M, et al. Profiling of Mitochondrial DNA Heteroplasmy in a Prospective Oral Squamous Cell Carcinoma Study. Cancers. 2020; 12(7):1933. https://doi.org/10.3390/cancers12071933
Chicago/Turabian StyleFendt, Liane, Federica Fazzini, Hansi Weissensteiner, Emanuel Bruckmoser, Sebastian Schönherr, Georg Schäfer, Jamie Lee Losso, Gertraud A. Streiter, Claudia Lamina, Michael Rasse, and et al. 2020. "Profiling of Mitochondrial DNA Heteroplasmy in a Prospective Oral Squamous Cell Carcinoma Study" Cancers 12, no. 7: 1933. https://doi.org/10.3390/cancers12071933
APA StyleFendt, L., Fazzini, F., Weissensteiner, H., Bruckmoser, E., Schönherr, S., Schäfer, G., Losso, J. L., Streiter, G. A., Lamina, C., Rasse, M., Klocker, H., Kofler, B., Kloss-Brandstätter, A., Huck, C. W., Kronenberg, F., & Laimer, J. (2020). Profiling of Mitochondrial DNA Heteroplasmy in a Prospective Oral Squamous Cell Carcinoma Study. Cancers, 12(7), 1933. https://doi.org/10.3390/cancers12071933









