Abstract
Background: Gastric cancer is the fourth most common cause of cancer-related death. Currently, it is broadly accepted that the molecular complexity and heterogeneity of gastric cancer, both inter- and intra-tumor, display important barriers for finding specific biomarkers for the early detection and diagnosis of this malignancy. Early-onset gastric cancer is not as prevalent as conventional gastric carcinoma, but it is a preferable model for studying the genetic background, as young patients are less exposed to environmental factors, which influence cancer development. Aim: The main objective of this study was to reveal age-dependent genotypic characteristics of gastric cancer subtypes, as well as conduct mutation profiling for the most frequent alterations in gastric cancer development, using targeted next-generation sequencing technology. Patients and methods: The study group included 53 patients, consisting of 18 patients with conventional gastric cancer and 35 with an early-onset subtype. The DNA of all index cases was used for next-generation sequencing, employing a panel of 94 genes and 284 single nucleotide polymorphisms (SNPs) (TruSight Cancer Panel, Illumina), which is characteristic for common and rare types of cancer. Results: From among the 53 samples processed for sequencing, we were able to identify seven candidate genes (STK11, RET, FANCM, SLX4, WRN, MEN1, and KIT) and nine variants among them: one splice_acceptor, four synonymous, and four missense variants. These were selected for the age-dependent differentiation of gastric cancer subtypes. We found four variants with C-Score ≥ 10, as 10% of the most deleterious substitutions: rs1800862 (RET), rs10138997 (FANCM), rs2230009 (WRN), and rs2959656 (MEN1). We identified 36 different variants, among 24 different genes, which were the most frequent genetic alterations among study subjects. We found 16 different variants among the genes that were present in 100% of the total cohort: SDHB (rs2746462), ALK (rs1670283), XPC (rs2958057), RECQL4 (rs4925828; rs11342077, rs398010167; rs2721190), DDB2 (rs326212), MEN1 (rs540012), AIP (rs4930199), ATM (rs659243), HNF1A (rs1169305), BRCA2 (rs206075; rs169547), ERCC5 (rs9514066; rs9514067), and FANCI (rs7183618). Conclusions: The technology of next-generation sequencing is a useful tool for studying the development and progression of gastric carcinoma in a high-throughput way. Our study revealed that early-onset gastric cancer has a different mutation frequency profile in certain genes compared to conventional subtype.
1. Introduction
Gastric cancer (GC) is a heterogeneous disease with a wide range of molecular and genetic alterations that contribute to the development and progression of this malignancy [1,2]. Statistical investigations have revealed that GC is the fourth most common cause of cancer-related death worldwide, with a median overall survival of ≤12 months for an advanced stage [3]. Early-onset gastric cancer (EOGC) (onset in individuals ≤45 years old) is not as common as conventional gastric carcinoma (CGC); besides, fewer than 10% of patients are affected by GC before 45 years of age [4]. Recent investigations on EOGC have attempted to clarify the genetic background of GC, as young patients are less susceptible to environmental agents that predispose individuals to cancer [5].
GC expands through a round of well-established and distinguishable steps: inflammation, atrophy, intestinal metaplasia, dysplasia, and carcinogenesis. GC pathogenesis is closely related to the diet, environment, H. pylori infection, and genetic alterations [6,7,8,9]. The recognizable patterns of GC incorporate genetic alterations among various factors: cell cycle regulators, agents that regulate apoptosis, microsatellite instability, multidrug resistance proteins, factors that influence cell membrane properties, the module of HER2 expression, and agents with an impact on the progression of GC and peritoneal metastasis [10,11,12,13].
The most popular classification of GC is Laurén classification, which distinguishes between GC-diffuse and intestinal GC. These two types of GC are distinguished by their distinct characteristics, including their clinical features, morphology, genetics, and progression [14]. The intestinal type of GC primarily occurs in older patients and consists of tubular or glandular components with multiple stages of differentiation. The diffuse type of GC is comprised of weakly cohesive single cells without gland formation and mostly affects young patients; hereditary agents play the primary role in this type of GC [15].
The development of next-generation sequencing (NGS) has provided a high-throughput and systematic approach for discovering genetic alterations, mostly mutations in the cancer genome. Although some driver genes have been found, the molecular background of GC is still not completely understood. NGS studies have discovered several novel driver mutations with well-known driver genes in GC development [16,17,18]. Gene mutations that either predispose individuals to or induce GC can be organized based on different biological pathways, such as cell adhesion, genome integrity, chromatin remodeling, the Wnt pathway, the RAS family, the MAPK pathway, RTKs, or the PIK pathway [19].
The molecular classification of GC based on NGS data was recently established in a publication by Cristescu et al. (2015) [17], where gene expression data of 300 primary gastric tumors was explored. Four molecular subtypes of GC were selected: MSS/TP53+, MSS/TP53−, MSI, and MSS/EMT subtypes. They were assigned to different patterns of molecular alterations, the prognosis of GC patients, progression, and cancer prognosis. Currently, it is crucial to expand knowledge about the molecular classification of GC, which is mostly based on NGS data, as this might be a valuable approach for developing targeted therapy suitable for particular patients. The molecular profiling of GC is not only of value for uncovering the molecular basis of the disease, but also for identifying genes of clinical utility for therapeutic targets for GC.
In this study, an attempt was made to compare age-dependent genotypic and phenotypic characteristics of GC subtypes, with mutation profiling for the most frequent alterations in GC development, using high-throughput sequencing of both EOGC and CGC subtypes.
2. Materials and Methods
2.1. Study Group
Our study materials included samples from patients with CGCs diagnosed in years 1993–2003 and samples of patients with EOGCs, which were collected from the Academic Medical Centre in Amsterdam, as well as other Institutions located in The Netherlands, Finland, and Poland. Tumor samples were classified according to Laurén classification [14] as either intestinal or diffuse gastric adenocarcinomas. The study material was used in a previous study [5], in which immunohistochemical labeling was conducted to look for different expression patterns of several GC markers in EOGCs compared to CGCs. We selected 53 GC patients for the sequencing analysis—35 with EOGC and 18 with CGC. EOGC cases were classified as diffuse, with the age range of patients being between 21 and 45 years. CGC samples included intestinal histology and the age range of cases was 47–86 years. The clinical data for each GC patient is available in Table S1. The CDH1 mutation status is described in Table S2, where the prediction for variant pathogenicity is presented. The group exhibited no pathogenic mutations in the CDH1 gene, which, according to current knowledge, is consistent with HDGC. The study was approved by the Medical University of Lublin Bioethical Committee (Opinion no. KE-0254/322/2019.) and was conducted in accordance with the Declaration of Helsinki.
DNA Extraction
The tissues were collected as previously described by Sitarz et al. (2008) [20]. They were stored in liquid nitrogen. The genomic DNA extraction from formalin-fixed paraffin-embedded (FFPE) tissues was performed using the QIAamp DNA Mini kit (Qiagen, Venlo, the Netherlands) or the Puregene DNA Isolation kit (Gentra, MN, USA), in accordance with the manufacturer’s instructions. Isolated DNA was kept at −80 °C. DNA quantity assessment was conducted with the Quantus™ Fluorometer with the QuantiFluor® dsDNA System, according to the protocol (Promega, Madison, WI, USA). The DNA quality was checked using the Agilent 2200 TapeStation and Genomic DNA ScreenTape (Agilent Technologies, Santa Clara, CA, USA). DNA samples were tested for high, middle, and low integrity. The DNA Integrity Number (DIN) algorithm was used to assess nucleic acid fragmentation. We selected samples with a DIN of at least 5 (where the scale is 1–10) for qPCR assessment. Further evaluation of the usefulness of the samples for sequencing was done with the Infinium HD FFPE QC Assay Protocol (Illumina, San Diego, CA, USA). Real-Time PCR by the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) was performed to check the amplification status in each sample. The average quantification cycle (Cq) values for the quality control template_standard (QCT_ST) were subtracted from the average Cq value for each sample to compute the Delta Cq value for each sample. All samples with a Delta Cq value below 5 were selected for library preparation.
2.2. Library Preparation for Next-Generation Sequencing
Libraries for sequencing experiments were prepared using the Trusight Rapid Capture Preparation Kit (Illumina, San Diego, CA, USA), according to the manufacturer’s protocol. DNA samples were standardized to 50 ng and underwent a tagmentation step. The libraries generated from input genomic DNA were amplified and adapter-tagged multiple short library fragments of 220–350 base pairs long. The libraries for each sample were combined to obtain proper complexity for a single run. Biotin-labeled probes were used to target regions of interest. Further steps of hybridization were conducted with custom-synthesized oligos that captured genes with the assigned predisposition towards multiple cancer types. The sequencing panel, the TruSight Cancer Panel (Illumina, San Diego, CA, USA), targeted 94 genes and 284 single nucleotide polymorphisms (SNPs) linked to common and rare cancers. The complete list of target genes and SNPs is available on the website of Illumina. Standard pools were enriched with regions of interest through streptavidin-coated beads that bound to biotinylated probes. Then, DNA fragments were eluted from the beads and another round of hybridization and enrichment was performed. Final amplified post-capture library concentrations were assessed with the Quantus™ Fluorometer, according to the manufacturer’s protocol. Libraries were quantified by qPCR using the KAPA Library Quantification kit (KAPA Biosystems, Boston, MA, USA). The size of the obtained library fragments was evaluated with the Agilent 2200 TapeStation and High Sensitivity D1000 ScreenTape System (Agilent Technologies, Santa Clara, CA, USA). Post-capture enriched libraries were sequenced on the MiSeq Sequencing Platform (Illumina, San Diego, CA, USA) with the manufacturer’s workflow. The concentration of loaded libraries amounted to 10 pM. The sequencing experiment was performed with the MiSeq Reagent Kit v2 (300 cycles).
2.3. Data Processing
Results from each sample were mapped to the human reference genome GRCh37, also known as Human Genome version 19 (hg19), using the Burrows–Wheeler Aligner (BWA-mem, version 0.7.5). Readings with a low mapping quality score, unmapped readings, and duplicates were filtered out with Samtools (version 0.1.19) [21]. The local realignment of readings around indels (insertion or deletion) and detection of systematic errors in base quality scores were performed with the Genome Analysis Toolkit (GATK) [22]. Readings mapped outside the target region were removed. Variant calling for germline SNPs and indels was performed with the GATK HaplotypeCaller tool [23]. The callsets of SNPs and small insertions and deletions were separated for further filtering. The hard filters applied to variant callsets were, for SNPs, QD < 2.0, MQ < 40.0, FS > 60.0, HaplotypeScore > 13.0, QRankSum < −12.5, and ReadPosRankSum < −8.0, and for INDELs, QD < 2.0, ReadPosRankSum < −20.0, and FS > 200.0. Filtered variants were concatenated into one record (VCF file) and the discovered variants were then annotated with SnpEff (version 4.2) using GEMINI (GEnome MINIng 0.18.3) and loaded into the SQLite database [24].
2.4. In Silico Estimation of the Detected Variants
We attempted to provide an appropriate explanation for variants, which were prioritized as statistically significant for the age-dependent diversification of GC. There are several online annotations that provide variant interpretation, such as CADD and DANN scoring, FATHMM-XF, PROVEAN, and SIFT predictions, which are useful tools for estimating variants with a potentially high risk of GC development. Nevertheless, they also have several limitations. Firstly, factors of annotations differ in various aspects, from constitutions to functions. Secondly, each tool has a unique metric, which is hard to compare with the others. Thirdly, combined annotations might only deliver overlapping importance. In our study, we used the CADD scoring system, which is a framework employed for estimating the relative pathogenicity of human genetic variants, by incorporating multiple, different annotations into a single quantitative score.
3. Age-Dependent Genotypic and Phenotypic Characteristics of Gastric Cancer Subtypes
In the study cohort of 53 patients with GC, which included 18 cases with CGC and 35 with EOGC, we identified seven candidate genes for discriminating these two age-associated subtypes of GC (Table 1). Among them, we detected nine variants, which passed our selection criteria, including one splice_acceptor, four synonymous, and four missense variants. The frequency of each variant is presented in Table 1, separating EOGC and CGC groups, including the genotype, gene and chromosome location, type of alteration, and p-value (indicating the variant diversification among groups).

Table 1.
Candidate genes employed for age-dependent classification of gastric carcinoma.
Three of the described variants were assigned to patients with the EOGC subtype. The missense variant (rs1799939) was found in the RET gene. The alteration frequency distribution among groups was, respectively, 46.2% of cases heterozygous for this variant in EOGC, 3.8% of EOGC cases homozygous for this variant, and 5.9% of CGC heterozygous for this variant. Interestingly, the homozygous missense variant (rs2959656) in the Menin 1 gene (MEN1) was observed in 100% of the EOGCs and 82.4% of CGC cases. The heterozygous variant was very rare, presenting in 5.9% of CGC subtypes. The synonymous variant rs55986963 was only detected in KIT Proto-Oncogene Receptor Kinase (KIT) in the heterozygous form in 20.6% of EOGC cases. In CGC cases, the frequency of the homozygous variant was 5.6%.
In Silico Estimation
In our study, we used different scoring and prediction systems for the relative assessment of the variant pathogenicity of a particular variant. The results are shown in Table 2.

Table 2.
Relative assessment of the variant pathogenicity.
We defined variants with C-Score ≥10 (top 10% in the ranking for pathogenicity) as 10% of the most deleterious substitutions. This criterion was fulfilled by the following four variants: rs1800862, rs10138997, rs2230009, and rs2959656. In practice, we cannot define pathogenicity as “deleterious” by only employing the C-score, as functional and/or clinical evidence is mandatory to confirm pathogenicity. The gnomAD database displayed variants with different frequencies among populations. The variants with CADD score higher than 10 were distributed respectively: rs1800862 (MAF = 0.04899), rs10138997 (MAF = 0.05851), rs2230009 (MAF = 0.05857), and rs2959656 (MAF = 0.9960).
4. High-Throughput Mutation Profiling Identifies the Most Frequent Mutations in EOGC and CGC Samples
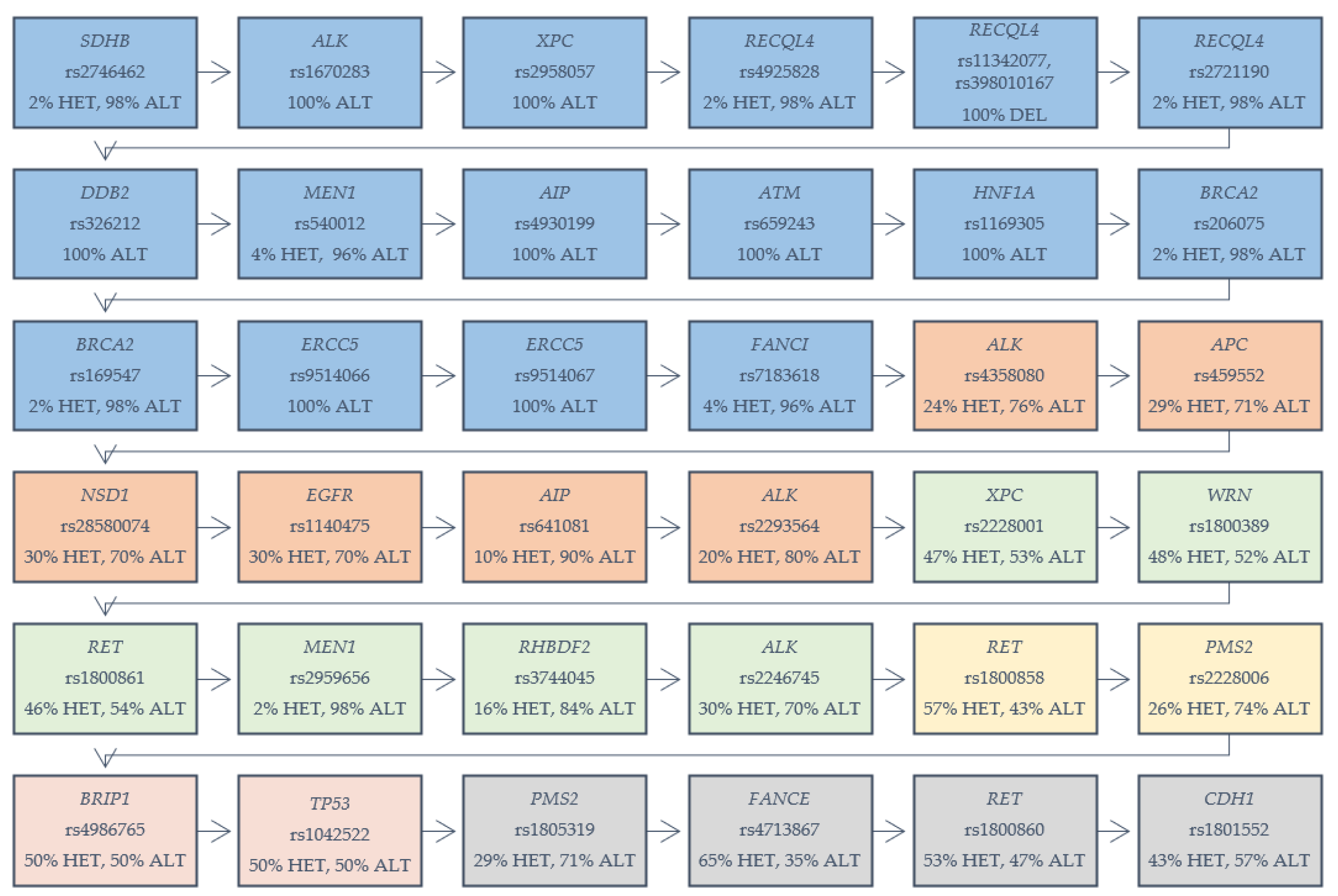
In our cohort of 53 patients with GC, we identified 36 different variants, among 24 various genes, which are presented in Figure 1. Our selection criterion was the frequency of chosen variants among the analyzed group (both EOGC and CGC). The variants were selected based on the number of homozygous reference variants in analyzed cases. We determined the range of 0–5 numbers of homozygous reference variants in analyzed cases. We found 16 different variants among genes which were present in 100% of the total cohort, as heterozygous or alternative homozygous variants: SDHB (rs2746462), ALK (rs1670283), XPC (rs2958057), RECQL4 (rs4925828; rs11342077, rs398010167; rs2721190), DDB2 (rs326212), MEN1 (rs540012), AIP (rs4930199), ATM (rs659243), HNF1A (rs1169305), BRCA2 (rs206075; rs169547), ERCC5 (rs9514066; rs9514067), and FANCI (rs7183618). Six variants with one or two homozygous reference variants were detected in several genes: ALK (rs4358080; rs2293564), APC (rs459552), NSD1 (rs28580074), EGFR (rs1140475), AIP (rs641081) and XPC (rs2228001), WRN (rs1800389), RET (rs1800861), MEN1 (rs2959656), RHBDF2 (rs3744045), and ALK (rs2246745). Two variants in genes—RET (rs1800858) and PMS2 (rs2228006) were present among the samples, with three homozygous reference variants. Two variants were detected with four references-in genes BRIP1 (rs4986765) and TP53 (rs1042522). Genes PMS2 (rs1805319), FANCE (rs4713867), RET (rs1800860), and CDH1 (rs1801552)—each with a variant showing five references among the analyzed samples.
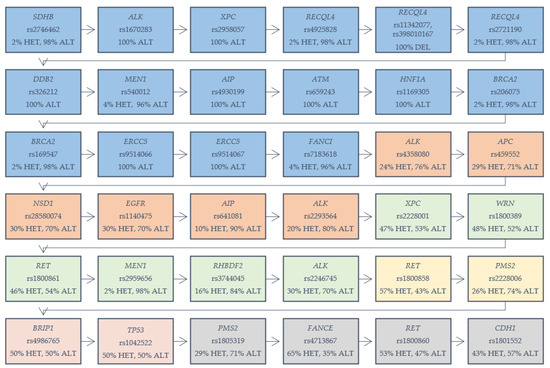
Figure 1.
Variants in genes detected in gastric cancer samples (both early-onset gastric cancer (EOGC) and conventional gastric carcinoma (CGC)). HET: heterozygous variant; ALT: alternative homozygous variant. Five colored boxes are displayed: blue: 100% percent of cases with the mentioned variant; orange: one reference homozygote in the analyzed cases; green: two reference homozygotes; yellow: three reference homozygotes; red: four reference homozygotes; gray: five reference homozygotes.
Among the thirty-six variants identified in our patient cohort, it is expected that sixteen of them (fifteen missense and one frameshift) will result in amino acid substitution (Table 3). Nine variants according to the CADD scoring system, with C-Score ≥ 10, were described as 10% of the most deleterious substitutions and thus most likely affect gene or protein function. Twenty of the detected variants were synonymous substitutions in which the produced amino acid sequence was not modified.

Table 3.
In silico estimation of the variant’s pathogenicity. NA: not applicable.
Nine of the mentioned variants with C-Score ≥ 10 were sequences of nine different genes: AIP, BRCA2, ERCC5, APC, XPC, RET, MEN1, RHBDF2, and PMS2. These genes are known to have roles in different processes and syndromes, such as DNA damage responses, DNA repair, multiple endocrine neoplasia type 1 syndrome, the aryl hydrocarbon receptor pathway, the Wnt signaling pathway, and ERK signaling. All of the variants are listed in Table 3.
5. Discussion
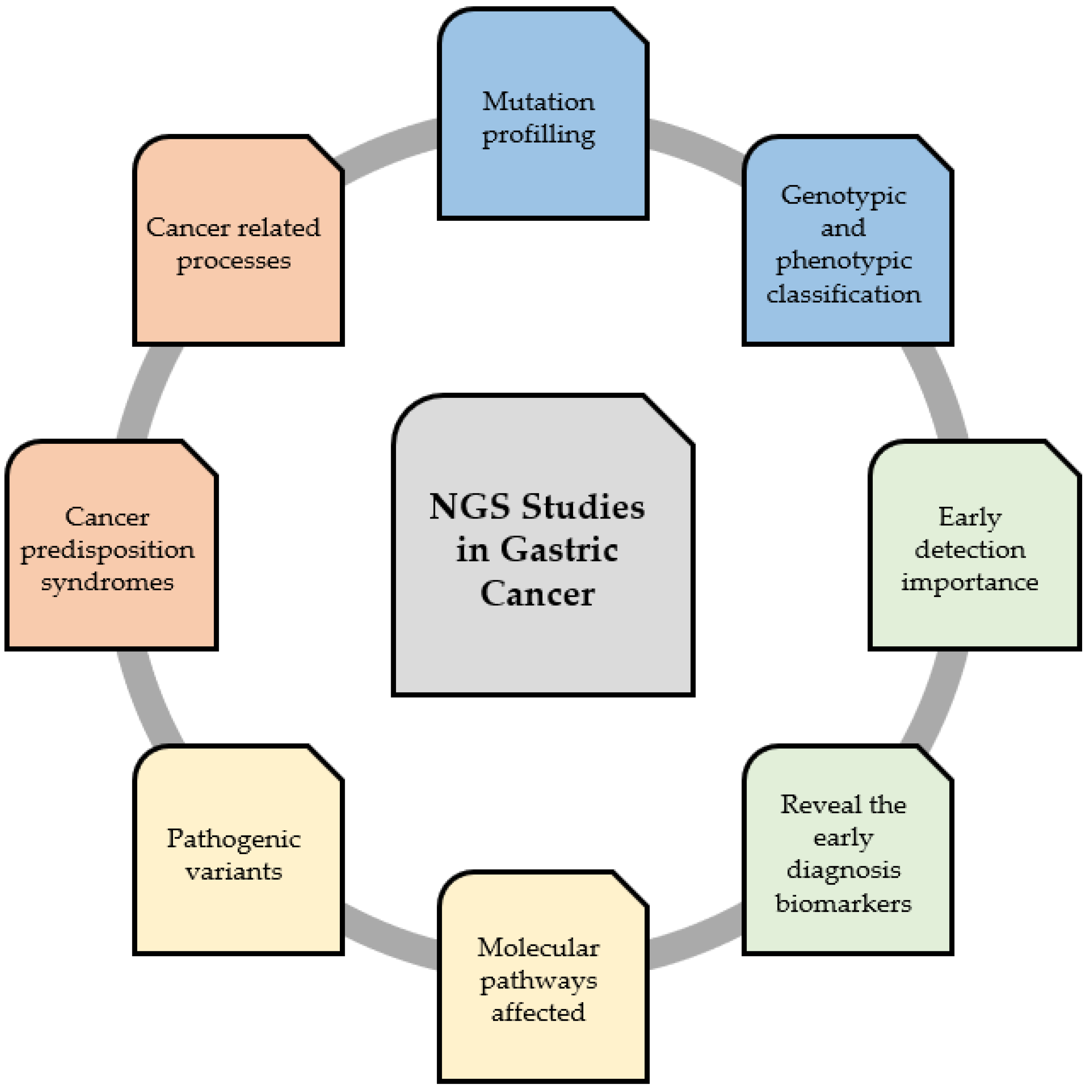
GC is one of the most common cancers and one of the most frequent causes of cancer-related deaths [25]. The prognosis and 5-year survival rate of patients with GC are still very poor. NGS is a useful tool for revealing the mutation profiling, importance of early and late development, and genotypic and phenotypic classification of GC. This technology helps to identify specific biomarkers, tumor suppressor genes, and carcinogens, as well as facilitate the understanding of mechanisms and affected pathways of GC tumorigenesis [26]. Therefore, this might be applied in the future for early diagnosis or personal treatment by either determining the particular GC biomarker or a specific drug-resistant gene [27]. Figure 2 summarizes the conclusions that can be drawn from the obtained results in the context of applying NGS studies as a useful tool for studying GC development in a high-throughput way.
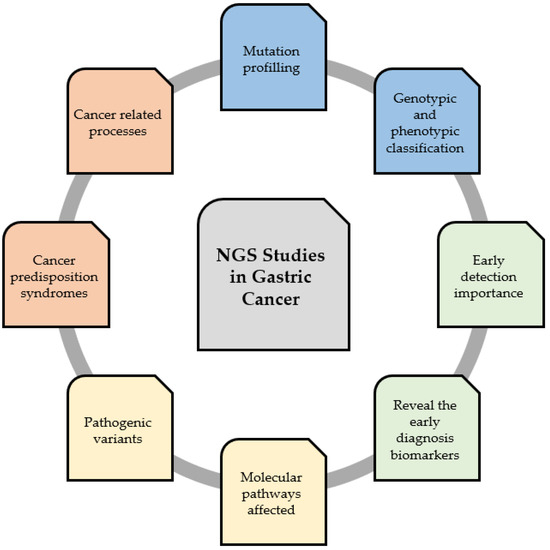
Figure 2.
Next-generation sequencing (NGS) as a useful tool for studying the development of gastric carcinoma in a high-throughput way.
We used the high-throughput sequencing of GC patients to compare and characterize age-dependent genotypic and phenotypic characteristics of GC subtypes. Based on the results obtained in this study, we identified potential candidate genes for distinguishing between EOGC and CGC. We were able to identify seven candidate genes, as well as nine variants, among them, which were statistically significantly different in these two subgroups. Variants, including rs1799939 (CADD = 8.26, MAF = 0.1847, p = 0.010), rs2959656 (CADD = 11.00, MAF = 0.9960, p = 0.041), and rs55986963 (CADD = 7.36, MAF = 0.03027, p = 0.052), were predominantly detected in EOGC patients. Interestingly, variant rs2959656 was found in 100% of cases of EOGC as homozygous. According to the NCBI ClinVar database, variant rs1799939 in the RET gene is associated with multiple endocrine neoplasia, hereditary cancer-predisposing syndrome, renal dysplasia, and pheochromocytoma. Activation of the RET proto-oncogene might constitute one of the molecular drivers of gastric inflammatory and neoplastic diseases [3]. Variant rs2959656 was detected in the MEN1 gene. Multiple Endocrine Neoplasia type 1 (MEN1) is a rare hereditary endocrine cancer syndrome, mainly leading to tumors of the parathyroid glands, endocrine gastroenteropancreatic tract, and the anterior pituitary [28]. Other endocrine and non-endocrine neoplasms are also observed, including GC [29]. Variant rs55986963 in the KIT gene, according to the NCBI ClinVar database, is associated with gastrointestinal stromal tumors (GISTs), mastocytosis, and partial albinism. Capelli et al. (2016) [30] also investigated exons 9, 11, 13, and 17 of the KIT gene by direct sequencing and showed that KIT mutations were observed in 53.8% of patients with gastric GISTs. Besides, KIT deletions in exon 11, mostly those involving codons 557, 558, and 559, were primarily associated with the more aggressive gastric GIST phenotype and a higher probability of death or relapse.
Variants that were most frequently detected among CGC included long deletion in the STK11 gene (p = 0.004), rs1800862 (CADD = 10.26, MAF = 0.04899, p = 0.006), rs10138997 (CADD = 14.99, MAF = 0.05851, p = 0.009), rs3810812 (CADD = 1.21, MAF = 0.5139, p = 0.018), rs2230009 (CADD = 13.29, MAF = 0.05857, p = 0.024), and rs28516461 (CADD = 0.14, MAF = 0.01392, p = 0.054). Variant rs1800862 (RET), according to NCBI ClinVar, is associated with multiple endocrine neoplasia, pheochromocytoma, and Hirschsprung disease, and is dominant. Variant rs10138997 (FANCM) and variants rs3810812 and rs28516461 (SLX4) are Fanconi anemia (FA) disease associated (NCBI ClinVar). The study performed by Swift et al. showed 102 deaths in relatives of eight FA families and a higher rate of leukemia and gastric, colorectal, and tongue cancer [31]. The variant rs2230009 (WRN) is associated with Werner syndrome (NCBI ClinVar). Patients with Werner syndrome present with an increased incidence of cancer, indicating that the lack of a proper WRN function affects tumorigenesis [32].
Our study of a cohort of 53 patients with GC allowed us to identify 36 different variants, among 24 various genes, which turned out to be the most probable genetic alterations, causing the development of GC. Importantly, we displayed 16 different variants that were detected in 100% of the total cohort. DNA damage appears to be the underlying cause of cancer [33] and deficiencies in DNA repair genes induce the development of different types of cancer [34]. If DNA repair is defective, DNA damage accumulates, increasing the number of mutations that occur due to error-prone translational synthesis, which consequently leads to the initiation of cancer development. Alterations in DNA double-strand break repair and DNA damage-response genes were detected in our study, such as variants rs2958057 (XPC); rs4925828, rs11342077, rs398010167 and rs2721190 (RECQL4); rs206075 and rs169547 (BRCA2); rs659243 (ATM); rs326212 (DDB2); rs9514066 and rs9514067 (ERCC5). It has been shown in different studies that the response to DNA damage plays an important role in the pathobiology of GCs [35,36,37]. The FA pathway constitutes a part of the DNA-damage network, including breast cancer-susceptibility proteins BRCA1 and BRCA2. The pathway is activated by ataxia telangiectasia and Rad3-related (ATR) kinase; however, the underlying mechanism remains unclear. A new study has demonstrated that the major switch activating the pathway is the ATR-dependent phosphorylation of FANCI [38]. Variant rs2746462 (SDHB) is related to paraganglioma and gastric stromal cell sarcoma, as well as the hereditary cancer-predisposing syndrome. Miettinen and Lasota (2014) described that approximately half of the patients with GISTs present SDH subunit gene mutations, mostly germline, with both alleles being inactivated in the tumor cells [39]. Variant rs1670283 (ALK) is associated with the hereditary cancer-predisposing syndrome and neuroblastoma 3 (NCBI ClinVar). Variant rs1169305 (HNF1A) is assigned to maturity-onset diabetes of young type 3 (MODY3) and might also result in hepatic adenomas (NCBI ClinVar). Variant rs4930199 (AIP) is associated with the hereditary cancer-predisposing syndrome and familial isolated pituitary adenomas.
Six of the variants with one and two homozygous reference variants were displayed in genes: ALK, APC, NSD1, EGFR, AIP and XPC, WRN, RET, MEN1, RHBDF2, and ALK. Two variants in genes—RET and PMS2—were detected with three homozygous reference variants. For BRIP1 and TP53 genes, two variants were detected with four references. Genes PMS2, FANCE, RET, and CDH1—each with variants showing five references among the analyzed group. Variant rs459552 (APC) is associated with familial adenomatous polyposis 1 and hereditary cancer-predisposing syndrome. The APC mutation is involved in the carcinogenesis of the intestinal type of GC and related to the LOH pathway in GC [40]. Variant rs28580074 (NSD1) is prevalent in Weaver syndrome and Sotos syndrome according to NCBI ClinVar. Variant rs1140475 is present in EGFR. EGFR, HER2, and MET signaling is important, especially in proximal non-diffuse tumors, and constitutes the logical targets for molecular therapy [41]. Variant rs3744045 in the RHBDF2 gene is associated with Howel–Evans syndrome. RHBDF2 seems to regulate oncogenic and non-canonical TGFB1 signaling. GC-associated fibroblasts increase their motility, via the expression of rhomboid 5 homolog 2, and the ability to induce the invasiveness of GC cells [42]. Variant rs2228006 (PMS2) is common for hereditary nonpolyposis colorectal cancer type 4 and mismatch repair cancer syndrome. BRIP1 and variant rs4986765 are related to familial cancer of the breast, neoplasms of the ovary, hereditary cancer-predisposing syndrome, and Fanconi anemia, which is also associated with the variant rs4713867 (FANCE) gene. The TP53 alteration (rs1042522) is related to Li–Fraumeni syndrome and hereditary cancer-predisposing syndrome. Besides, p53 alterations are displayed quite early in the development of GC, even being present in the non-neoplastic mucosa, and their incidence increases with the progression of GC [43]. Even though our study was limited due to a low number of samples, it provides a significant insight into genetic alterations that occur in GC, primarily EOGC and CGC. Further studies with larger groups of patients are needed, since the differences between EOGC and CGC cases may possibly be more significant.
6. Conclusions
We were able to compare the age-dependent genotypic and phenotypic profile of GC patients, focusing on the most frequent alterations in GC development, both EOGC and CGC, using high-throughput sequencing technology. We found variants among several genes, which might be considered for future studies on the early detection and diagnosis of GC. We displayed the processes and syndromes involved in EOGC and CGC development. Mutation prediction tools enable us to estimate potential candidate biomarkers with a pathogenic impact on disease development. This study primarily placed emphasis on EOGC development, which is less exposed to environmental factors and constitutes a good model for further studies on the genetic background of GC development.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/7/1981/s1: Table S1: A table containing the clinical characteristics of the studied group, Table S2: A table with in silico estimation of the CDH1 mutation status among GC patients.
Author Contributions
Conceptualization, P.W. and R.S.; methodology, J.M., F.H.M.M, G.J.A.O., and R.S.; software, P.K. and P.W.; formal analysis, J.M., P.K., and R.S.; investigation, J.M., P.K., J.B., F.H.M.M., P.W., R.M., G.J.A.O., and R.S.; writing—original draft preparation, J.M. and R.S.; writing—review and editing, J.M., P.K., J.B., F.H.M.M., P.W., R.M., G.J.A.O., and R.S.; supervision, P.W., G.J.A.O., and R.S.; funding acquisition, R.M. and R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partly supported by a grant from the Polish Ministry of Science and Higher Education, No. NN402423838.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Abbreviations
| CGC | conventional gastric carcinoma |
| EOGC | early-onset gastric cancer |
| FA | Fanconi anemia |
| FFPE | formalin-fixed paraffin-embedded |
| GC | gastric cancer |
| GIST | gastrointestinal stromal tumor |
| MEN1 | multiple endocrine neoplasia type 1 |
| NGS | next-generation sequencing |
| SNPs | single nucleotide polymorphisms |
References
- Gullo, I.; Carneiro, F.; Oliveira, C.; Almeida, G.M. Heterogeneity in Gastric Cancer: From Pure Morphology to Molecular Classifications. Pathobiology 2018, 85, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.P.; Xu, W.; Liu, W.T.; Yan, M.; Zhu, Z.G. Tumor heterogeneity of gastric cancer: From the perspective of tumor-initiating cell. World J. Gastroenterol. 2018, 24, 2567–2581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Zhang, P.Y. Gastric cancer: Somatic genetics as a guide to therapy. J. Med. Genet. 2017, 54, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Kokkola, A.; Sipponen, P. Gastric carcinoma in young adults. Hepato Gastroenterol. 2001, 48, 1552–1555. [Google Scholar]
- Milne, A.N.; Carvalho, R.; Morsink, F.M.; Musler, A.R.; de Leng, W.W.; Ristimäki, A.; Offerhaus, G.J. Early-onset gastric cancers have a different molecular expression profile than conventional gastric cancers. Mod. Pathol. 2006, 19, 564–572. [Google Scholar] [CrossRef]
- Pucułek, M.; Machlowska, J.; Wierzbicki, R.; Baj, J.; Maciejewski, R.; Sitarz, R. Helicobacter pylori associated factors in the development of gastric cancer with special reference to the early-onset subtype. Oncotarget 2018, 9, 31146–31162. [Google Scholar] [CrossRef]
- Rocco, A.; Nardone, G. Diet, H pylori infection and gastric cancer: Evidence and controversies. World J. Gastroenterol. 2007, 13, 2901–2912. [Google Scholar] [CrossRef]
- Baj, J.; Brzozowska, K.; Forma, A.; Maani, A.; Sitarz, E.; Portincasa, P. Immunological Aspects of the Tumor Microenvironment and Epithelial-Mesenchymal Transition in Gastric Carcinogenesis. Int. J. Mol. Sci. 2020, 21, 2544. [Google Scholar] [CrossRef]
- Baj, J.; Korona-Głowniak, I.; Forma, A.; Maani, A.; Sitarz, E.; Rahnama-Hezavah, M.; Radzikowska, E.; Portincasa, P. Mechanisms of the Epithelial–Mesenchymal Transition and Tumor Microenvironment in Helicobacter pylori-Induced Gastric Cancer. Cells 2020, 9, 1055. [Google Scholar] [CrossRef] [PubMed]
- Machlowska, J.; Maciejewski, R.; Sitarz, R. The Pattern of Signatures in Gastric Cancer Prognosis. Int. J. Mol. Sci. 2018, 19, 19. [Google Scholar] [CrossRef] [PubMed]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef] [PubMed]
- Rimini, M.; Casadei-Gardini, A.; Ravaioli, A.; Rovesti, G.; Conti, F.; Borghi, A.; Dall’Aglio, A.C.; Bedogni, G.; Domenicali, M.; Giacomoni, P.; et al. Could Inflammatory Indices and Metabolic Syndrome Predict the Risk of Cancer Development? Analysis from the Bagnacavallo Population Study. J. Clin. Med. 2020, 9, 1177. [Google Scholar] [CrossRef] [PubMed]
- Salati, M.; Pipitone, S.; Rimini, M.; Gelsomino, F.; Gardini, A.C.; Andrikou, K.; Schipilliti, F.; Cortesi, G.; Cassanelli, L.; Caffari, E.; et al. Immune-inflammatory and clinicopathologic prognostic factors in a Western cohort of resected gastric cancers (GCs). Ann. Oncol. 2019, 30, 75. [Google Scholar] [CrossRef]
- Laurén, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar]
- Bosman, F.T.; Carneiro, F.; Hruban, R.H.; Theise, N.D. WHO Classification of Tumours of the Digestive System, 4th ed.; International Agency for Research on Cancer (IARC): Lyon, France, 2010. [Google Scholar]
- Liang, H.; Kim, Y.H. Identifying molecular drivers of gastric cancer through next-generation sequencing. Cancer Lett. 2013, 340, 241–246. [Google Scholar] [CrossRef][Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Riquelme, I.; Saavedra, K.; Espinoza, J.A.; Weber, H.; García, P.; Nervi, B.; Garrido, M.; Corvalán, A.H.; Roa, J.C.; Bizma, C. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget 2015, 6, 24750–24779. [Google Scholar] [CrossRef]
- Sitarz, R.; Leguit, R.J.; de Leng, W.W.; Polak, M.; Morsink, F.M.; Bakker, O.; Maciejewski, R.; Offerhaus, G.J.; Milne, A.N. The COX-2 promoter polymorphism -765 G>C is associated with early-onset, conventional and stump gastric cancers. Mod. Pathol. 2008, 21, 685–690. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Narzisi, G.; O’Rawe, J.A.; Iossifov, I.; Fang, H.; Lee, Y.H.; Wang, Z.; Wu, Y.; Lyon, G.J.; Wigler, M.; Schatz, M.C. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat. Methods 2014, 11, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Patru, C.L.; Surlin, V.; Georgescu, I.; Patru, E. Current issues in gastric cancer epidemiology. Rev. Med. Chir. Soc. Med. Nat. Iasi 2013, 117, 199–204. [Google Scholar] [PubMed]
- Kamps, R.; Brandão, R.D.; Bosch, B.J.; Paulussen, A.D.; Xanthoulea, S.; Blok, M.J.; Romano, A. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. Int. J. Mol. Sci. 2017, 18, 308. [Google Scholar] [CrossRef] [PubMed]
- Shyr, D.; Liu, Q. Next generation sequencing in cancer research and clinical application. Biol. Proced. Online 2013, 15, 4. [Google Scholar] [CrossRef]
- Marini, F.; Falchetti, A.; Luzi, E.; Tonelli, F.; Maria Luisa, B. Multiple Endocrine Neoplasia Type 1 (MEN1) Syndrome. In Cancer Syndromes; Riegert-Johnson, D.L., Boardman, L.A., Hefferon, T., Roberts, M., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2009. [Google Scholar]
- Debelenko, L.V.; Emmert-Buck, M.R.; Zhuang, Z.; Epshteyn, E.; Moskaluk, C.A.; Jensen, R.T.; Liotta, L.A.; Lubensky, I.A. The multiple endocrine neoplasia type I gene locus is involved in the pathogenesis of type II gastric carcinoids. Gastroenterology 1997, 113, 773–781. [Google Scholar] [CrossRef]
- Capelli, L.; Petracci, E.; Quagliuolo, V.; Saragoni, L.; Colombo, P.; Morgagni, P.; Calistri, D.; Tomezzoli, A.; Di Cosmo, M.; Roviello, F.; et al. GISTs: Analysis of c-Kit, PDGFRA and BRAF mutations in relation to prognosis and clinical pathological characteristics of patients—A GIRCG study. Eur. J. Surg. Oncol. 2016, 42, 1206–1214. [Google Scholar] [CrossRef]
- Swift, M. Fanconi’s anaemia in the genetics of neoplasia. Nature 1971, 230, 370–373. [Google Scholar] [CrossRef]
- Goto, M.; Miller, R.W.; Ishikawa, Y.; Sugano, H. Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol. Prev. Biomark. 1996, 5, 239–246. [Google Scholar]
- Kastan, M.B. DNA damage responses: Mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial Award Lecture. Mol. Cancer Res. 2008, 6, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell. 2007, 28, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Arai, H.; Wada, R.; Ishino, K.; Kudo, M.; Uchida, E.; Naito, Z. Expression of DNA damage response proteins in gastric cancer: Comprehensive protein profiling and histological analysis. Int. J. Oncol. 2018, 52, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, L.; Melucci, E.; De Nicola, F.; Goeman, F.; Casini, B.; Sperati, F.; Pallocca, M.; Terrenato, I.; Pizzuti, L.; Vici, P.; et al. DNA damage repair and survival outcomes in advanced gastric cancer patients treated with first-line chemotherapy. Int. J. Cancer. 2017, 140, 2587–2595. [Google Scholar] [CrossRef]
- Lee, H.E.; Han, N.; Kim, M.A.; Lee, H.S.; Yang, H.K.; Lee, B.L.; Kim, W.H. DNA damage response-related proteins in gastric cancer: ATM, Chk2 and p53 expression and their prognostic value. Pathobiology 2014, 81, 25–35. [Google Scholar] [CrossRef]
- Wang, W. A major switch for the Fanconianemia DNA damage-response pathway. Nat. Struct. Mol. Biol. 2008, 15, 1128–1130. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Lasota, J. Succinate dehydrogenase deficient gastrointestinal stromal tumors (GISTs)—A review. Int. J. Biochem. Cell Biol. 2014, 53, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.C.; Luo, Y.H.; Yang, S.M.; Li, X.A.; Ling, X.L.; Fang, L. Mutation analysis of APC gene in gastric cancer with microsatellite instability. World J. Gastroenterol. 2002, 8, 787–791. [Google Scholar] [CrossRef]
- Wong, H.; Yau, T. Molecular targeted therapies in advanced gastric cancer: Does tumor histologymatter. Therap. Adv. Gastroenterol. 2013, 6, 15–31. [Google Scholar] [CrossRef]
- Ishimoto, T.; Miyake, K.; Nandi, T.; Yashiro, M.; Onishi, N.; Huang, K.K.; Lin, S.J.; Kalpana, R.; Tay, S.T.; Suzuki, Y.; et al. Activation of Transforming Growth Factor Beta 1 Signaling in Gastric Cancer-associated Fibroblasts Increases Their Motility, via Expression of Rhomboid 5 Homolog 2, and Ability to Induce Invasiveness of Gastric Cancer Cells. Gastroenterology 2017, 153, 191–204. [Google Scholar] [CrossRef]
- Fenoglio-Preiser, C.M.; Wang, J.; Stemmermann, G.N.; Noffsinger, A. TP53 and gastric carcinoma: A review. Hum. Mutat. 2003, 21, 258–270. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).