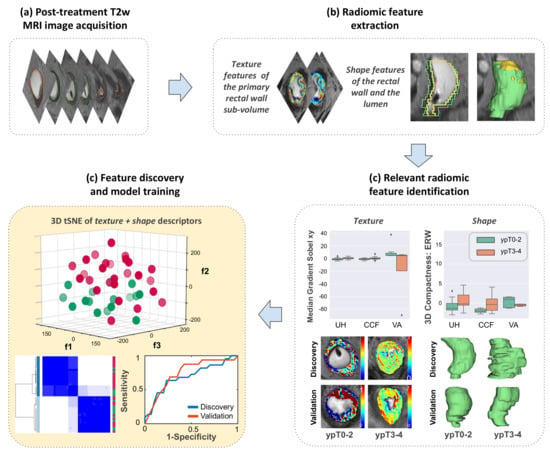
Radiomic Texture and Shape Descriptors of the Rectal Environment on Post-Chemoradiation T2-Weighted MRI are Associated with Pathologic Tumor Stage Regression in Rectal Cancers: A Retrospective, Multi-Institution Study
Abstract
:1. Introduction
2. Results
2.1. Data Description
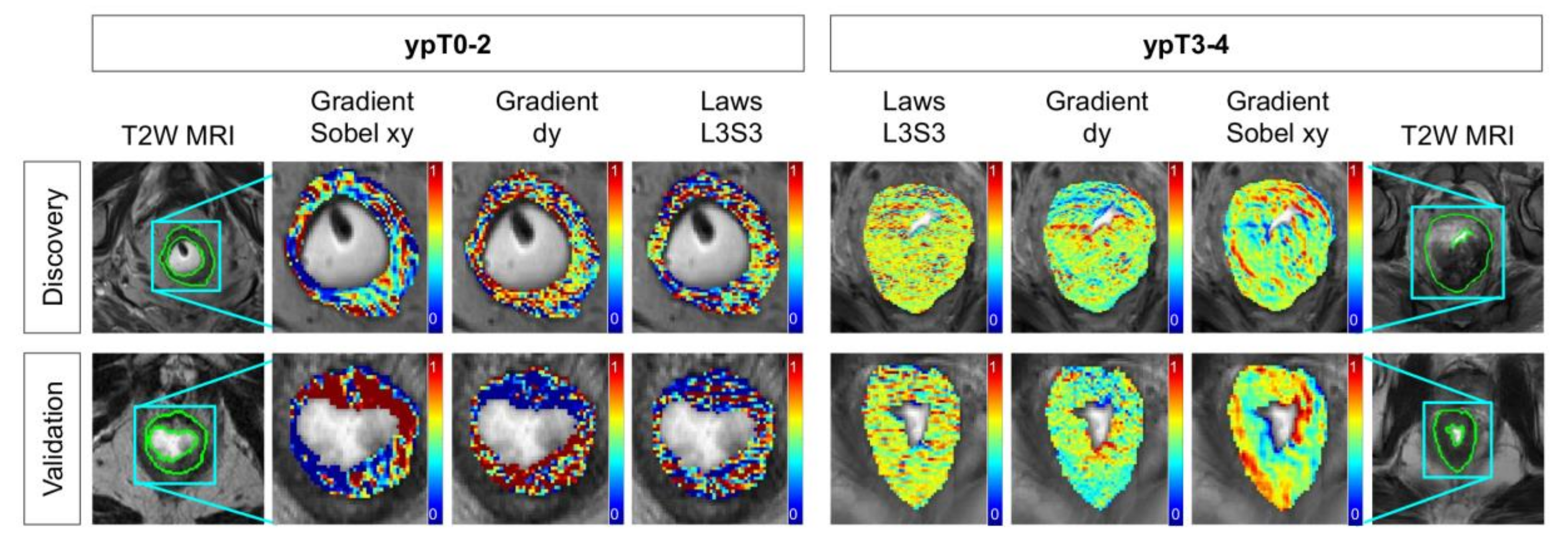
2.2. Identifying T2w Radiomic Texture Features Associated with Pathologic Tumor Down-Staging after Chemoradiation
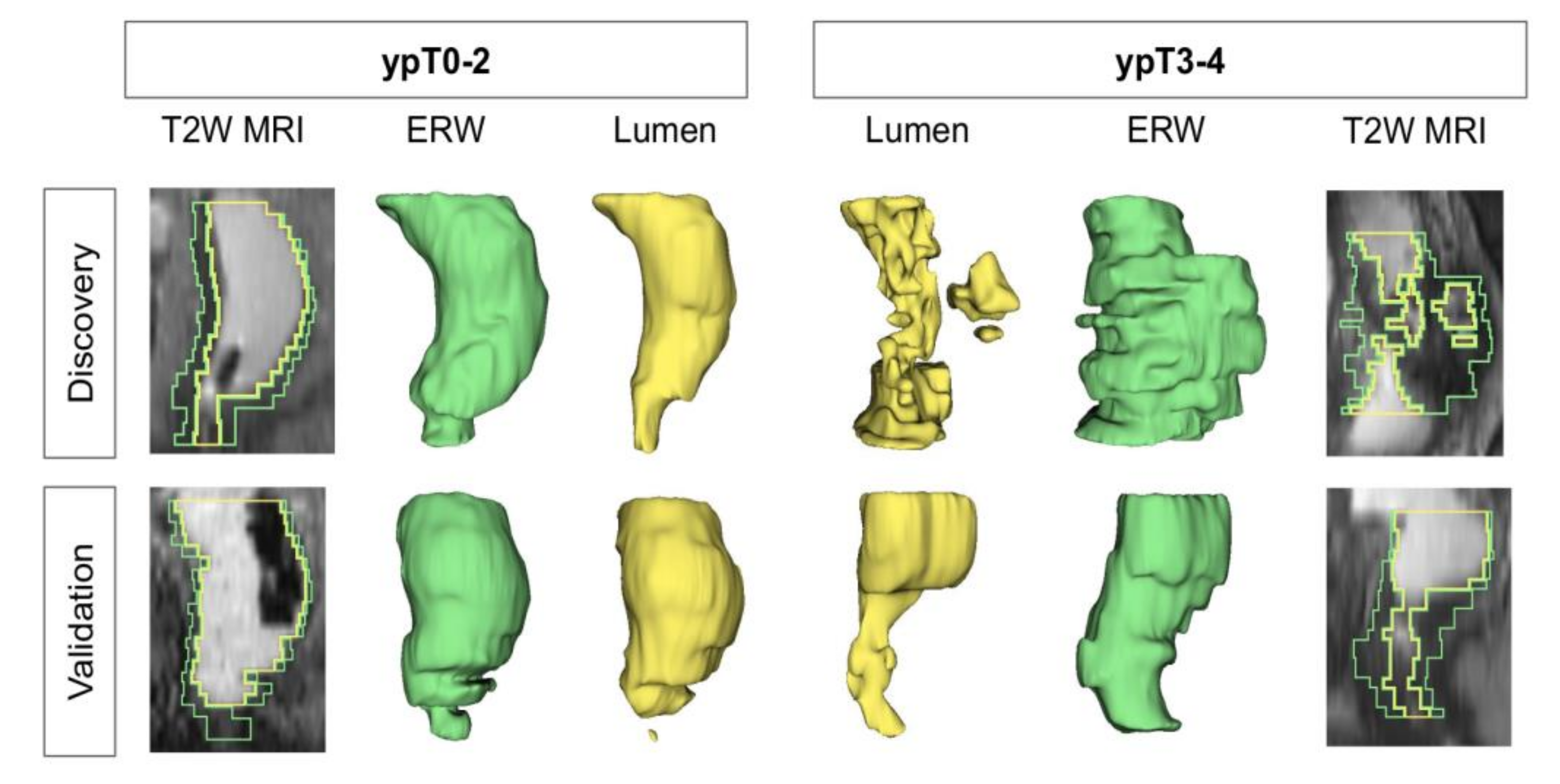
2.3. Identifying T2w Radiomic Shape Features Associated with Pathologic Tumor Down-Staging after Chemoradiation
2.4. Combining T2w Radiomic Texture and Shape Features Consistently Discriminates Pathologic Tumor Stage Groupings after Chemoradiation across Institutions and Magnetic Field Strengths
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Patient Selection
4.3. Neoadjuvant Treatment and Histopathologic Assessment
4.4. Annotation and ROI Identification on Post-nCRT T2w MRI Datasets
4.5. Radiomic Texture and Shape Feature Extraction
4.6. Identifying Relevant Radiomic Features Associated with Pathologic Stage after nCRT
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterol. Rev. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- National Comprehensive Cancer Network. NCCN Guidelines for Rectal Cancer Version 2.2019; National Comprehensive Cancer Network, Inc.: Plymouth Meeting, PA, USA, 2019. [Google Scholar]
- Deng, Y.; Chi, P.; Lan, P.; Wang, L.; Chen, W.; Cui, L.; Chen, D.; Cao, J.; Wei, H.; Peng, X.; et al. Modified FOLFOX6 with or without Radiation Versus Fluorouracil and Leucovorin with Radiation in Neoadjuvant Treatment of Locally Advanced Rectal Cancer: Initial Results of the Chinese FOWARC Multicenter, Open-Label, Randomized Three-Arm Phase III Tria. J. Clin. Oncol. 2016, 34, 3300–3307. [Google Scholar] [CrossRef]
- Smith, J.J.; Chow, O.S.; Gollub, M.J.; Nash, G.M.; Temple, L.K.; Weiser, M.R.; Guillem, J.G.; Paty, P.B.; Avila, K.; Garcia-Aguilar, J. Organ Preservation in Rectal Adenocarcinoma: A phase II randomized controlled trial evaluating 3-year disease-free survival in patients with locally advanced rectal cancer treated with chemoradiation plus induction or consolidation chemotherapy, and totalmesorectal excision or nonoperative management. BMC Cancer 2015, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renehan, A.G.; Malcomson, L.; Emsley, R.; Gollins, S.; Maw, A.; Myint, A.S.; Rooney, P.S.; Susnerwala, S.; Blower, A.; Saunders, M.P.; et al. Watch-and-wait approach versus surgical resection after chemoradiotherapy for patients with rectal cancer (the OnCoRe project): A propensity-score matched cohort analysis. Lancet Oncol. 2016, 17, 174–183. [Google Scholar] [CrossRef]
- Den Dulk, M.; Putter, H.; Collette, L.; Marijnen, C.A.M.; Folkesson, J.; Bosset, J.-F.; Rödel, C.; Bujko, K.; Påhlman, L.; van de Velde, C.J.H. The abdominoperineal resection itself is associated with an adverse outcome: The European experience based on a pooled analysis of five European randomised clinical trials on rectal cancer. Eur. J. Cancer 2009, 45, 1175–1183. [Google Scholar] [CrossRef]
- Moriya, Y.; Akasu, T.; Fujita, S.; Yamamoto, S. Aggressive surgical treatment for patients with T4 rectal cancer. Color. Dis. 2003, 5, 427–431. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, H.; Du, X.L.; Wang, F.; Zhang, J.; Cui, X.; Li, E.; Yang, J.; Yi, M.; Zhang, Y. Impact of neoadjuvant and adjuvant radiotherapy on disease-specific survival in patients with stages II--IV rectal cancer. Oncotarget 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Jhaveri, K.S.; Hosseini-Nik, H. MRI of Rectal Cancer: An Overview and Update on Recent Advances. Am. J. Roentgenol. 2015, 205, W42–W55. [Google Scholar] [CrossRef]
- Dijkhoff, R.A.P.; Beets-Tan, R.G.H.; Lambregts, D.M.J.; Beets, G.L.; Maas, M. Value of DCE-MRI for staging and response evaluation in rectal cancer: A systematic review. Eur. J. Radiol. 2017, 95, 155–168. [Google Scholar] [CrossRef]
- Amodeo, S.; Rosman, A.S.; Desiato, V.; Hindman, N.M.; Newman, E.; Berman, R.; Pachter, H.L.; Melis, M. MRI-Based Apparent Diffusion Coefficient for Predicting Pathologic Response of Rectal Cancer after Neoadjuvant Therapy: Systematic Review and Meta-Analysis. Am. J. Roentgenol. 2018, 211, W205–W216. [Google Scholar] [CrossRef]
- Chen, C.-C.; Lee, R.-C.; Lin, J.-K.; Wang, L.-W.; Yang, S.-H. How Accurate is Magnetic Resonance Imaging in Restaging Rectal Cancer in Patients Receiving Preoperative Combined Chemoradiotherapy? Dis. Colon Rectum 2005, 48, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Van den Broek, J.J.; van der Wolf, F.S.W.; Lahaye, M.J.; Heijnen, L.A.; Meischl, C.; Heitbrink, M.A.; Schreurs, W.H. Accuracy of MRI in Restaging Locally Advanced Rectal Cancer after Preoperative Chemoradiation. Dis. Colon Rectum 2017, 60, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Patel, U.B.; Blomqvist, L.K.; Taylor, F.; George, C.; Guthrie, A.; Bees, N.; Brown, G. MRI after Treatment of Locally Advanced Rectal Cancer: How to Report Tumor Response-The MERCURY Experience. Am. J. Roentgenol. 2012, 199, W486–W495. [Google Scholar] [CrossRef] [PubMed]
- Taylor, F.G.M.; Swift, R.I.; Blomqvist, L.; Brown, G. A Systematic Approach to the Interpretation of Preoperative Staging MRI for Rectal Cancer. Am. J. Roentgenol. 2008, 191, 1827–1835. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.; Hill, V.; Statsevych, V.; Huang, R.; Prasanna, P.; Correa, R.; Singh, G.; Bera, K.; Beig, N.; Thawani, R.; et al. Shape Features of the Lesion Habitat to Differentiate Brain Tumor Progression from Pseudoprogression on Routine Multiparametric MRI: A Multisite Study. Am. J. Neuroradiol. 2018, 39, 2187–2193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reimer, R.P.; Reimer, P.; Mahnken, A.H. Assessment of Therapy Response to Transarterial Radioembolization for Liver Metastases by Means of Post-treatment MRI-Based Texture Analysis. Cardiovasc. Intervent. Radiol. 2018, 41, 1545–1556. [Google Scholar] [CrossRef]
- Bogowicz, M.; Leijenaar, R.T.H.; Tanadini-Lang, S.; Riesterer, O.; Pruschy, M.; Studer, G.; Unkelbach, J.; Guckenberger, M.; Konukoglu, E.; Lambin, P. Post-radiochemotherapy PET radiomics in head and neck cancer-The influence of radiomics implementation on the reproducibility of local control tumor models. Radiother. Oncol. 2017, 125, 385–391. [Google Scholar] [CrossRef] [Green Version]
- Shiradkar, R.; Podder, T.K.; Algohary, A.; Viswanath, S.; Ellis, R.J.; Madabhushi, A. Radiomics based targeted radiotherapy planning (Rad-TRaP): A computational framework for prostate cancer treatment planning with MRI. Radiat. Oncol. 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; He, Y.; Yuan, Z.; Benedict, S.; Valicenti, R.; Qiu, J.; Rong, Y. Radiomics for Response and Outcome Assessment for Non-Small Cell Lung Cancer. Technol. Cancer Res. Treat. 2018, 17, 153303381878278. [Google Scholar] [CrossRef]
- Horvat, N.; Bates, D.D.B.; Petkovska, I. Novel imaging techniques of rectal cancer: What do radiomics and radiogenomics have to offer? A literature review. Abdom. Radiol. 2019, 44, 3764–3774. [Google Scholar] [CrossRef] [PubMed]
- Seo, N.; Kim, H.; Cho, M.S.; Lim, J.S. Response Assessment with MRI after Chemoradiotherapy in Rectal Cancer: Current Evidences. Korean J. Radiol. 2019, 20, 1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nardi, P. De How reliable is current imaging in restaging rectal cancer after neoadjuvant therapy? World J. Gastroenterol. 2013, 19, 5964. [Google Scholar] [CrossRef] [PubMed]
- Iafrate, F.; Laghi, A.; Paolantonio, P.; Rengo, M.; Mercantini, P.; Ferri, M.; Ziparo, V.; Passariello, R. Preoperative Staging of Rectal Cancer with MR Imaging: Correlation with Surgical and Histopathologic Findings. RadioGraphics 2006, 26, 701–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Dresen, R.C.; Kusters, M.; Daniels-Gooszen, A.W.; Cappendijk, V.C.; Nieuwenhuijzen, G.A.P.; Kessels, A.G.H.; de Bruïne, A.P.; Beets, G.L.; Rutten, H.J.T.; Beets-Tan, R.G.H. Absence of Tumor Invasion into Pelvic Structures in Locally Recurrent Rectal Cancer: Prediction with Preoperative MR Imaging. Radiology 2010, 256, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Bayanati, H.; Thornhill, R.E.; Souza, C.A.; Sethi-Virmani, V.; Gupta, A.; Maziak, D.; Amjadi, K.; Dennie, C. Quantitative CT texture and shape analysis: Can it differentiate benign and malignant mediastinal lymph nodes in patients with primary lung cancer? Eur. Radiol. 2014, 25, 480–487. [Google Scholar] [CrossRef]
- Dhara, A.K.; Mukhopadhyay, S.; Dutta, A.; Garg, M.; Khandelwal, N. A Combination of Shape and Texture Features for Classification of Pulmonary Nodules in Lung CT Images. J. Digit. Imaging 2016, 29, 466–475. [Google Scholar] [CrossRef] [Green Version]
- Newell, D.; Nie, K.; Chen, J.-H.; Hsu, C.-C.; Yu, H.J.; Nalcioglu, O.; Su, M.-Y. Selection of diagnostic features on breast MRI to differentiate between malignant and benign lesions using computer-aided diagnosis: Differences in lesions presenting as mass and non-mass-like enhancement. Eur. Radiol. 2009, 20, 771–781. [Google Scholar] [CrossRef] [Green Version]
- Klessen, C.; Rogalla, P.; Taupitz, M. Local staging of rectal cancer: The current role of MRI. Eur. Radiol. 2006, 17, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Bulens, P.; Couwenberg, A.; Intven, M.; Debucquoy, A.; Vandecaveye, V.; Van Cutsem, E.; D’Hoore, A.; Wolthuis, A.; Mukherjee, P.; Gevaert, O.; et al. Predicting the tumor response to chemoradiotherapy for rectal cancer: Model development and external validation using {MRI} radiomics. Radiother. Oncol. 2020, 142, 246–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Yang, X.; Shi, Z.; Yang, Z.; Du, X.; Zhao, Z.; Cheng, X. Radiomics analysis of multiparametric {MRI} for prediction of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Eur. Radiol. 2018, 29, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, C.N.; Ganeshan, B.; Ciolina, M.; Rengo, M.; Meinel, F.G.; Musio, D.; De Felice, F.; Raffetto, N.; Tombolini, V.; Laghi, A. Texture Analysis as Imaging Biomarker of Tumoral Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer Patients Studied with 3-T Magnetic Resonance. Invest. Radiol. 2015, 50, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Zhang, C.; Zou, S.; Zhao, X.; Xu, K.; Zhang, H.; Zhou, C. MRI texture analysis in predicting treatment response to neoadjuvant chemoradiotherapy in rectal cancer. Oncotarget 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Shu, Z.; Fang, S.; Ye, Q.; Mao, D.; Cao, H.; Pang, P.; Gong, X. Prediction of efficacy of neoadjuvant chemoradiotherapy for rectal cancer: The value of texture analysis of magnetic resonance images. Abdom. Radiol. 2019, 44, 3775–3784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, H.; Yu, J.; Wei, Y.; Wu, J.; Xu, Q. Response to neoadjuvant chemoradiotherapy for locally advanced rectum cancer: Texture analysis of dynamic contrast-enhanced MRI. J. Magn. Reson. Imaging 2018, 49, 885–893. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, X.-Y.; Shi, Y.-J.; Wang, L.; Zhu, H.-T.; Tang, Z.; Wang, S.; Li, X.-T.; Tian, J.; Sun, Y.-S. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin. Cancer Res. 2017, 23, 7253–7262. [Google Scholar] [CrossRef] [Green Version]
- Chandramohan, A.; Siddiqi, U.M.; Mittal, R.; Eapen, A.; Jesudason, M.R.; Ram, T.S.; Singh, A.; Masih, D. Diffusion weighted imaging improves diagnostic ability of MRI for determining complete response to neoadjuvant therapy in locally advanced rectal cancer. Eur. J. Radiol. Open 2020, 7, 100223. [Google Scholar] [CrossRef]
- Horvat, N.; Veeraraghavan, H.; Khan, M.; Blazic, I.; Zheng, J.; Capanu, M.; Sala, E.; Garcia-Aguilar, J.; Gollub, M.J.; Petkovska, I. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology 2018, 287, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Nie, K.; Shi, L.; Chen, Q.; Hu, X.; Jabbour, S.K.; Yue, N.; Niu, T.; Sun, X. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clin. Cancer Res. 2016, 22, 5256–5264. [Google Scholar] [CrossRef] [Green Version]
- Iannicelli, E.; Di Renzo, S.; Ferri, M.; Pilozzi, E.; Di Girolamo, M.; Sapori, A.; Ziparo, V.; David, V. Accuracy of High-Resolution MRI with Lumen Distention in Rectal Cancer Staging and Circumferential Margin Involvement Prediction. Korean J. Radiol. 2014, 15, 37. [Google Scholar] [CrossRef] [Green Version]
- Dinapoli, N.; Barbaro, B.; Gatta, R.; Chiloiro, G.; Casà, C.; Masciocchi, C.; Damiani, A.; Boldrini, L.; Gambacorta, M.A.; Dezio, M.; et al. Magnetic Resonance, Vendor-independent, Intensity Histogram Analysis Predicting Pathologic Complete Response after Radiochemotherapy of Rectal Cancer. Int. J. Radiat. Oncol. 2018, 102, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, T.; Gollins, S.; Maw, A.; Hobson, P.; Byrne, R.; Widdowson, D. Magnetic resonance imaging in rectal cancer downstaged using neoadjuvant chemoradiation: Accuracy of prediction of tumour stage and circumferential resection margin status. Color. Dis. 2008, 10, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Torkzad, M.R.; Påhlman, L.; Glimelius, B. Magnetic resonance imaging (MRI) in rectal cancer: A comprehensive review. Insights Imaging 2010, 1, 245–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, F.L.; Page, D.L.; Fleming, I.D.; Fritz, A.G.; Balch, C.M.; Haller, D.G.; Morrow, M. AJCC Cancer Staging Manual 2002; Springer-Verlag: New York, NY, USA, 2002; Volume 6, pp. 32–34. [Google Scholar]
- Pieper, S.; Halle, M.; Kikinis, R. 3D Slicer. In Proceedings of the 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821), Arlington, VA, USA, 18 April 2004; pp. 632–635. [Google Scholar]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Sobel, I.; Feldman, G. A 3x3 isotropic gradient operator for image processing, presented at a talk at the Stanford Artificial Project. Pattern Classif. Scene Anal. 1968, 271–272. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.S. Image representation using 2D Gabor wavelets. IEEE Trans. Pattern Anal. Mach. Intell. 1996, 18, 959–971. [Google Scholar] [CrossRef] [Green Version]
- Laws, K.I. Textured Image Segmentation; University of Southern California Los Angeles Image Processing INST: Los Angeles, CA, USA, 1980. [Google Scholar]
- Prasanna, P.; Tiwari, P.; Madabhushi, A. Co-occurrence of Local Anisotropic Gradient Orientations (CoLlAGe): A new radiomics descriptor. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Braman, N.; Prasanna, P.; Whitney, J.; Singh, S.; Beig, N.; Etesami, M.; Bates, D.D.B.; Gallagher, K.; Bloch, B.N.; Vulchi, M.; et al. Association of Peritumoral Radiomics with Tumor Biology and Pathologic Response to Preoperative Targeted Therapy for HER2 (ERBB2)-Positive Breast Cancer. JAMA Netw. Open 2019, 2, e192561. [Google Scholar] [CrossRef] [Green Version]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Foley, D. Considerations of sample and feature size. IEEE Trans. Inf. Theory 1972, 18, 618–626. [Google Scholar] [CrossRef]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Liang, C.; Huang, Y.; He, L.; Chen, X.; Ma, Z.; Dong, D.; Tian, J.; Liang, C.; Liu, Z. The development and validation of a {CT}-based radiomics signature for the preoperative discrimination of stage I-{II} and stage {III}-{IV} colorectal cancer. Oncotarget 2016, 7, 31401–31412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef] [Green Version]
- R.C. Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Copenhagen, Denmark, 2013. [Google Scholar]
- Antunes, J.T.; Ofshteyn, A.; Bera, K.; Wang, E.Y.; Brady, J.T.; Willis, J.E.; Friedman, K.A.; Marderstein, E.L.; Kalady, M.F.; Stein, S.L.; et al. Radiomic Features of Primary Rectal Cancers on Baseline T2 -Weighted MRI Are Associated with Pathologic Complete Response to Neoadjuvant Chemoradiation: A Multisite Study. J. Magn. Reson. Imaging 2020. [Google Scholar] [CrossRef]



| Imaging Parameter | Institution 1 UHCMC (n = 52) | Institution 2 CCF (n = 31) | Institution 3 VAMC (n = 11) |
|---|---|---|---|
| In-plane Resolution (mm) | 0.256–0.977 | 0.313–0.898 | 0.398–0.938 |
| Slice Thickness (mm) | 3.0–5.0 | 3.0–6.0 | 3.0–8.0 |
| Field of view (px) | 224–576 224–576 20–57 | 256–640 252–640 13–79 | 234–576 256–528 24–50 |
| Repetition Time (ms) | 3253–12690 | 3400–13333 | 3420–7200 |
| Echo Time (ms) | 67–110 | 84–166 | 80–100 |
| Sequence | TSE | TSE | FSE |
| Magnet Strength | |||
| 3 T | 51 | 3 | 8 |
| 1.5 T | 1 | 28 | 3 |
| Scanner | |||
| Siemens Symphony | 6 | ||
| Siemens Avanto | 14 | ||
| Siemens Espree | 3 | ||
| Siemens Aera | 4 | ||
| Siemens Skyra | 3 | ||
| Siemens Verio | 39 | ||
| Philips Achieva | 1 | 8 | |
| Philips Medical System Ingenuity | 5 | ||
| Philips Healthcare Ingenia | 8 | ||
| Toshiba Titan | 2 | ||
| Unknown | 1 | ||
| Imaging Plane Axial Through Tumor | |||
| Transverse | 43 | 28 | 10 |
| Coronal | 9 | 3 | 1 |
| Gel use | Yes | Yes | No |
| Rank | FT | FS | FT+S |
|---|---|---|---|
| 1 | Median Gradient Sobel xy (p = 0.0002) | 3D Compactness Entire Rectal Wall (p = 0.003) | Median Gradient Sobel xy (p = 0.0002) |
| 2 | Skewness Gradient dy (p = 0.0007) | Skewness - 2D Eccentricity Lumen (p = 0.004) | 3D Compactness Entire Rectal Wall (p = 0.003) |
| 3 | Median Laws L3S3 (p = 0.0009) | Variance - 2D Convexity Entire Rectal Wall (p = 0.0009) | Variance - 2D Convexity Entire Rectal Wall (p = 0.0009) |
| 4 | Median CoLlAGe sum-av ws = 5 (p = 0.002) | Mean - 2D Compactness Entire Rectal Wall (p = 0.002) | Median Laws L3S3 (p = 0.0009) |
| 5 | Median Haralick sum-av ws = 3 (p = 0.006) | Variance - 2D Minor Axis Length Lumen (p = 0.0009) | Skewness Gradient dy (p = 0.0007) |
| 6 | Variance Haralick sum-av ws = 3 (p = 0.006) | Kurtosis - 2D Major Axis Length Lumen (p = 0.02) | Median CoLlAGe sum-av ws = 5 (p = 0.002) |
| Ranked Feature | 1.5 T Median (IQR) | 3 T Median (IQR) | Unadjusted p-Value |
|---|---|---|---|
| Median Gradient Sobel xy | 0.64 (−0.05–1.40) | 0.31 (−1.34–1.12) | 0.127 |
| 3D Compactness: ERW | −0.43 (−2.09–1.77) | −0.64 (−1.42–0.17) | 0.921 |
| Variance—2D Convexity ERW | −2.26 (−2.72–−0.91) | −1.37 (−2.31–0.34) | 0.015 |
| Median Laws L3S3 | 0.79 (−0.80–1.57) | 0.60 (−1.04–1.65) | 0.814 |
| Skewness Gradient dy | 0.26 (−0.77–0.96) | 0.30 (-1.12–0.88) | 0.423 |
| Median CoLIAGe sum-av ws = 5 | −0.21 (−1.07–0.84) | −0.50 (−1.31–1.03) | 0.789 |
| Median Haralick sum-av ws = 3 | −0.97 (−1.82–0.29) | −0.03 (−0.9–1.21) | 0.014 |
| Variance Haralick sum-av ws = 3 | 0.05 (−0.95–0.79) | −0.54 (−1.34–0.71) | 0.369 |
| Skewness—2D Eccentricity Lumen | 0.11 (−1.54–1.04) | 0.06 (-0.67–0.99) | 0.510 |
| Mean—2D Compactness ERW | −0.2 (−0.98–0.58) | 0.37 (−0.97–1.17) | 0.166 |
| Variance—2D Minor Axis Length Lumen | −0.03 (−0.80–1.82) | −0.45 (−1.49–0.43) | 0.030 |
| Kurtosis—2D Major Axis Length Lumen | −1.33 (−1.81–0.09) | −0.60 (−1.64–1.05) | 0.423 |
| Radiomic Feature | Unadjusted p-Value | ICC |
|---|---|---|
| Median Gradient Sobel xy | 0.457 | 0.549 |
| 3D Compactness: ERW | 0.776 | 0.923 |
| Variance—2D Convexity ERW | 0.441 | 0.375 |
| Median Laws L3S3 | 0.693 | 0.908 |
| Skewness Gradient dy | 0.962 | 0.484 |
| Median CoLIAGe sum-av ws = 5 | 0.602 | 0.829 |
| Median Haralick sum-av ws = 3 | 0.912 | 0.947 |
| Variance Haralick sum-av ws = 3 | 0.079 | 0.745 |
| Skewness - 2D Eccentricity Lumen | 0.925 | 0.473 |
| Mean—2D Compactness ERW | 0.903 | 0.842 |
| Variance—2D Minor Axis Length Lumen | 0.903 | 0.831 |
| Kurtosis—2D Major Axis Length Lumen | 0.285 | 0.695 |
| Clinical Variable | Inst. 1 UHCMC (n = 52) | Inst. 2 CCF (n = 31) | Inst. 3 VAMC (n = 11) |
|---|---|---|---|
| Gender | |||
| Male | 30 | 20 | 11 |
| Female | 22 | 11 | 0 |
| Age at diagnosis (yrs) | 62.8 ± 13.6 | 58.2 ± 11.4 | 65.8 ± 12.0 |
| Rectal wall volume (cm3) | 43.1 ± 33.6 | 62.4 ± 66.1 | 35.9 ± 17.6 |
| Lumen wall volume (cm3) | 40.1 ± 31.1 | 69.5 ± 43.4 | 21.8 ± 8.8 |
| Pathologic Staging | |||
| ypN0M0 | |||
| ypT0–2 | 18 | 7 | 5 |
| ypT3–4 | 15 | 9 | 2 |
| ypN+ or ypM+ | |||
| ypT0–2 | 4 | 2 | 2 |
| ypT3–4 | 15 | 13 | 2 |
| Feature Group | Quantity | Description & Rationale |
|---|---|---|
| Histogram measures | 21 | First-order statistics of the original image signal intensity within local pixel neighborhoods, capturing basic variations in signal intensities due to intermixed tissue types (fibrosis, ulceration, mucosa) after nCRT |
| Gradient operators [49] | 10 | Identification of leading gradients and edges in the local signal within small neighborhoods of pixels, likely occurring due to impact of nCRT within the rectal wall |
| Haralick measures [50] | 65 | Quantify heterogeneity and entropy of local intensity texture as represented by the gray-level co-occurrence matrix pixel neighborhoods, widely shown to be related to underlying tissue heterogeneity as a result of intermixed treatment effects, residual disease, and irradiated tissue |
| Gabor operators [51] | 35 | Responses to Gabor wavelets which are defined at specific unit-length scales (λ = 0.765, 0.128, 1.786, 2.296, and 2.806; corresponding to window sizes 3, 5, 7, 9 or 11 pixels) and orientations (θ = ), thus capturing multi-scale and multi-oriented variations within the rectal wall |
| Laws operators [52] | 34 | Responses to local filters targeting combinations of specific textural patterns in the x- and y-directions. Descriptors include all combinations of 1D filters: level (L), edge (E), spot (S), wave (W), and ripple (R), which have been related to underlying abnormal structures or enhancement patterns |
| CoLlAGe [53] | 26 | Captures and exploits local anisotropic differences in voxel-level gradient orientations by assigning every image voxel an entropy value associated with the co-occurrence matrix of gradient orientations, which have been related to reflecting subtle local differences in tissue microarchitecture |
| Feature Name | Description | 2D | 3D |
|---|---|---|---|
| Contour-Based | |||
| Axis length | Length of a line drawn through the center of an ellipse (2D) or sphere (3D) that has the same normalized second central moments as the object | x | x |
| Convexity | Ratio between the convex perimeter and the perimeter of the original object | x | |
| Convex perimeter | Length of the outline of the convex object (smallest convex polygon that can contain the object) | x | |
| Eccentricity | Ratio of the distance between the foci of the ellipse (2D) or sphere (3D) and its major axis length, measuring how much a conic section deviates from being circular | x | x |
| Elongation | Ratio between the minor and the major axis, measuring the aspect ratio of the object | x | x |
| Equivalent diameter | Diameter of a circle that has the same area as the object | x | |
| Equivalent ellipsoid diameter | Diameter of an ellipse that has the same second-moments as the object | x | |
| Equivalent spherical radius | Radius of a sphere that has the same second-moments as the object | x | |
| Equivalent spherical perimeter | Perimeter of a sphere that has the same second-moments as the object | x | |
| Flatness | Measure that describe if the surface of the object is flat or if it has raised areas or indentations | x | |
| Orientation | Angle between the x-axis and the major axis of the ellipse (2D) or sphere (3D) that has the same second-moments as the object | x | x |
| Perimeter | Length of the outline of the object | x | |
| Region-based | |||
| Area | Measure of the number of pixels in a 2D object | x | |
| Area of bounding box | Measure of the number of pixels in the bounding box (smallest rectangle containing the region) | x | |
| Compactness | Ratio between the area (2D) of the object and the area of a circle with the same perimeter | x | x |
| Convex area | Measure of the number of pixel in the convex hull (the smallest convex polygon that can contain the region) | x | |
| Elongation of the bounding box | Ratio between the minor and the major axis of the bounding box (smallest rectangle containing the region), measuring the aspect ratio of the object | x | |
| Elongation shape factor | Square root of the ratio of the two second moments of the object around its principal axes | x | |
| Extent | Ratio between pixels in the original object and pixels in the bounding box (smallest rectangle containing the region) | x | |
| Filled area | Number of pixels in the filled object (original object with all the holes filled) | x | |
| Principal moments | Measures that describe the moments of inertia at center of mass | x | |
| Roundness | Ratio between the area (2D) or volume (3D) of the object and the area of a circle (2D) or sphere (3D) with the same convex perimeter | x | x |
| Solidity | Density of the object measured as proportion of the pixels in the convex object (smallest convex polygon that can contain the object) that are also in the original object | x | |
| Volume | Measure of the number of pixels in a 3D object | x |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Jimenez, C.; Antunes, J.T.; Talasila, N.; Bera, K.; Brady, J.T.; Gollamudi, J.; Marderstein, E.; Kalady, M.F.; Purysko, A.; Willis, J.E.; et al. Radiomic Texture and Shape Descriptors of the Rectal Environment on Post-Chemoradiation T2-Weighted MRI are Associated with Pathologic Tumor Stage Regression in Rectal Cancers: A Retrospective, Multi-Institution Study. Cancers 2020, 12, 2027. https://doi.org/10.3390/cancers12082027
Alvarez-Jimenez C, Antunes JT, Talasila N, Bera K, Brady JT, Gollamudi J, Marderstein E, Kalady MF, Purysko A, Willis JE, et al. Radiomic Texture and Shape Descriptors of the Rectal Environment on Post-Chemoradiation T2-Weighted MRI are Associated with Pathologic Tumor Stage Regression in Rectal Cancers: A Retrospective, Multi-Institution Study. Cancers. 2020; 12(8):2027. https://doi.org/10.3390/cancers12082027
Chicago/Turabian StyleAlvarez-Jimenez, Charlems, Jacob T. Antunes, Nitya Talasila, Kaustav Bera, Justin T. Brady, Jayakrishna Gollamudi, Eric Marderstein, Matthew F. Kalady, Andrei Purysko, Joseph E. Willis, and et al. 2020. "Radiomic Texture and Shape Descriptors of the Rectal Environment on Post-Chemoradiation T2-Weighted MRI are Associated with Pathologic Tumor Stage Regression in Rectal Cancers: A Retrospective, Multi-Institution Study" Cancers 12, no. 8: 2027. https://doi.org/10.3390/cancers12082027
APA StyleAlvarez-Jimenez, C., Antunes, J. T., Talasila, N., Bera, K., Brady, J. T., Gollamudi, J., Marderstein, E., Kalady, M. F., Purysko, A., Willis, J. E., Stein, S., Friedman, K., Paspulati, R., Delaney, C. P., Romero, E., Madabhushi, A., & Viswanath, S. E. (2020). Radiomic Texture and Shape Descriptors of the Rectal Environment on Post-Chemoradiation T2-Weighted MRI are Associated with Pathologic Tumor Stage Regression in Rectal Cancers: A Retrospective, Multi-Institution Study. Cancers, 12(8), 2027. https://doi.org/10.3390/cancers12082027