PLEKHS1 Over-Expression is Associated with Metastases and Poor Outcomes in Papillary Thyroid Carcinoma
Abstract
1. Introduction
2. Results
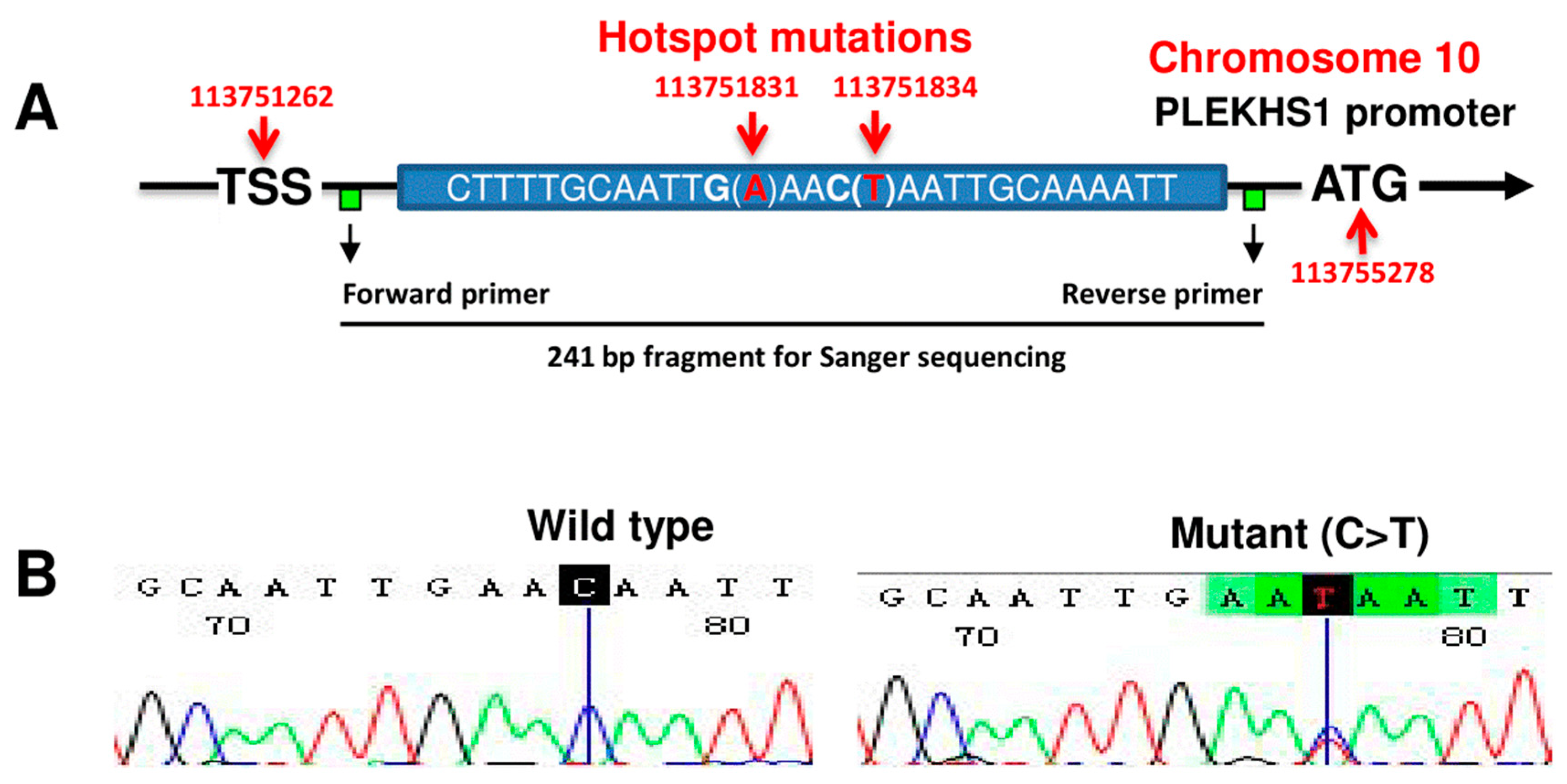
2.1. The Rarity of PLEKHS1 Hotspot Promoter Mutations in PTCs and ATCs
2.2. PLEKHS1 Expression in TC Cell Lines and Primary PTC and ATC Tumors
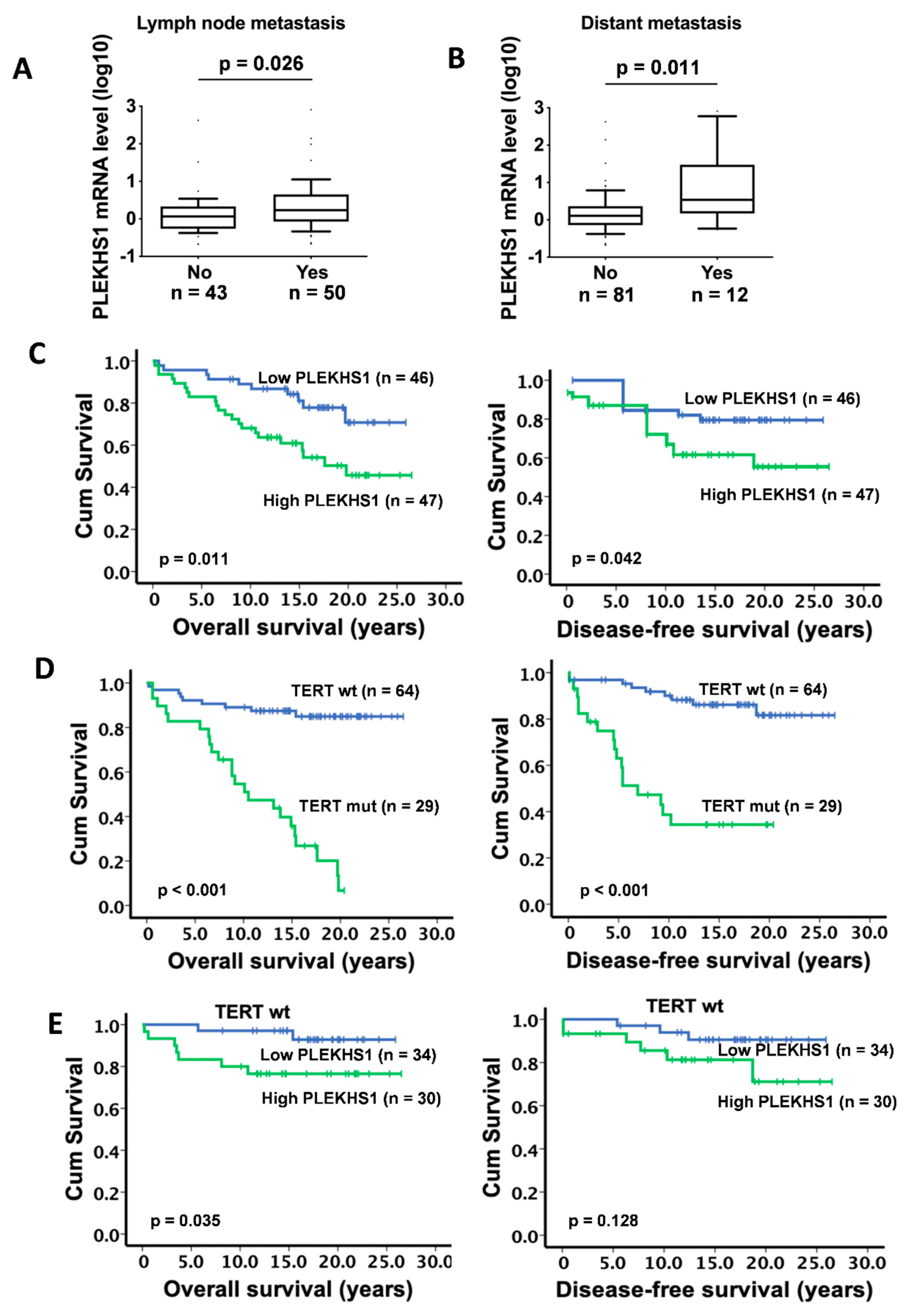
2.3. Association Between PLEKHS1 Over-Expression and Metastases and Shorter Survival in PTCs
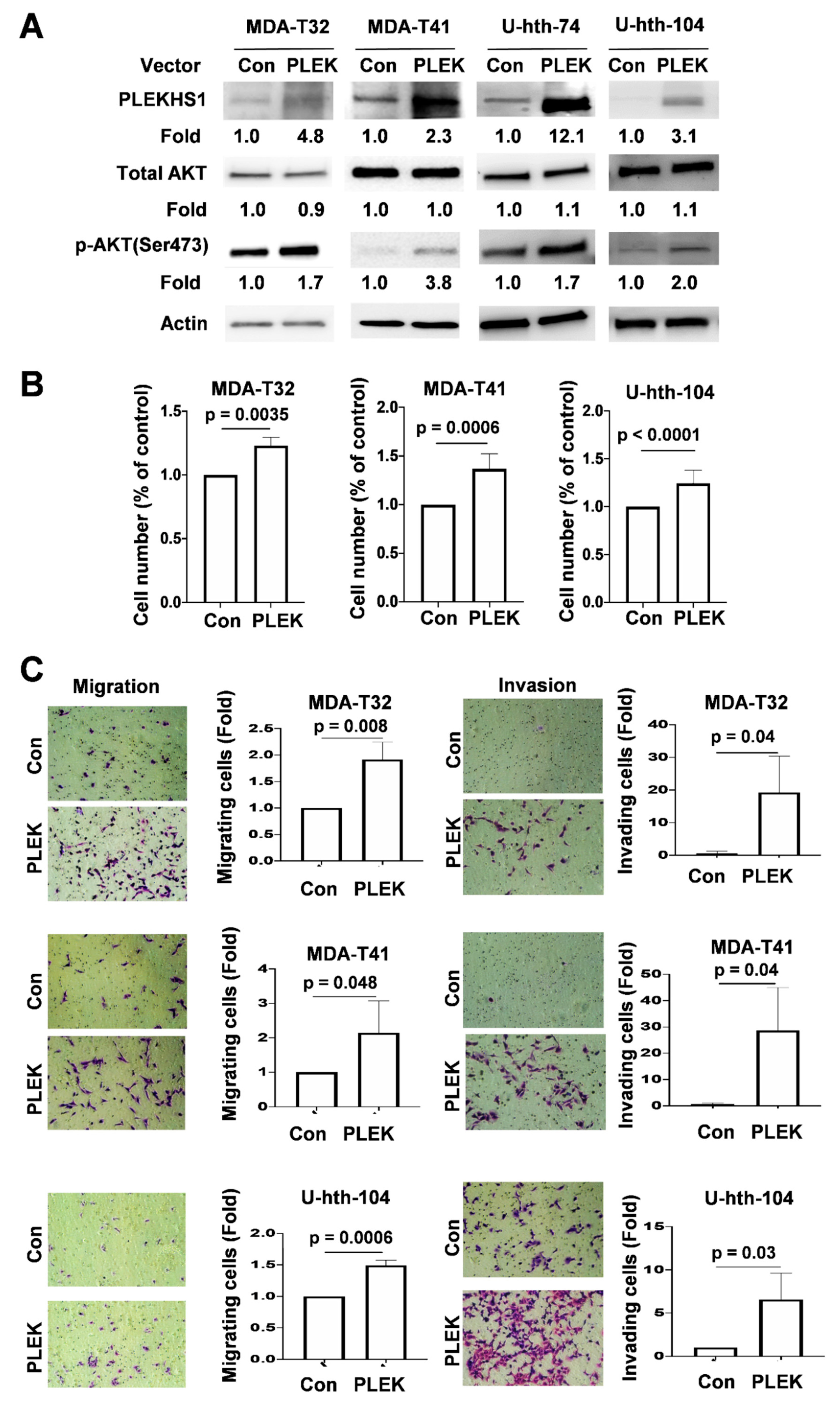
2.4. Enhanced AKT Phosphorylation, Proliferation and Invasiveness Mediated by PLEKHS1 Over-Expression in TC Cells
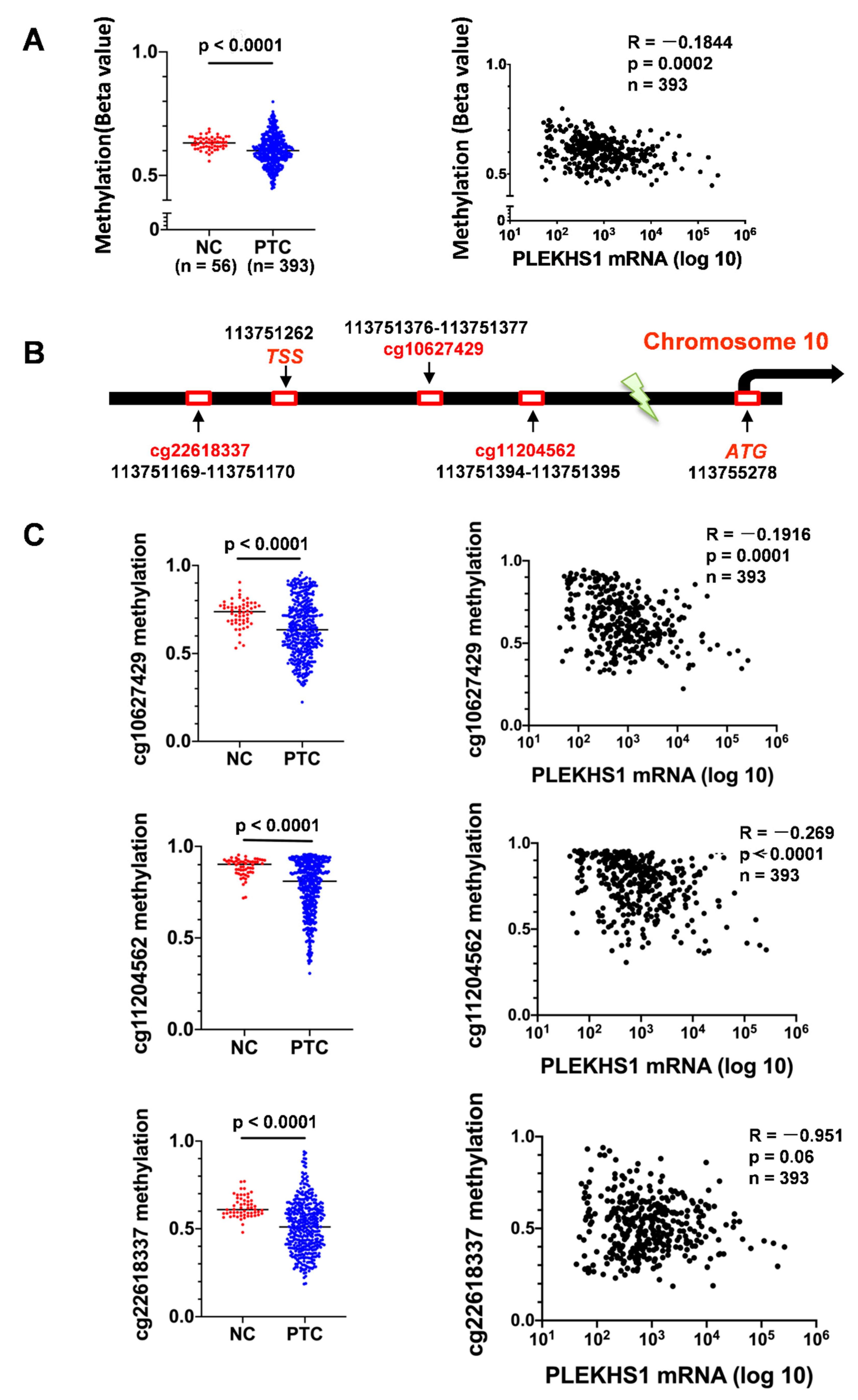
2.5. Association Between Demethylation and mRNA Expression of the PLEKHS1 Gene in PTCs
3. Discussion
4. Patients and Methods
4.1. Patients
4.2. Cell Lines and Cell Culture
4.3. DNA Extraction and Sanger Sequencing
4.4. RNA Extraction, Reverse Transcription and qPCR
4.5. Plasmid Transfection
4.6. Western Blot Analysis
4.7. Pyrosequencing for DNA Methylation Analyses
4.8. Transwell Assays for Cell Migration and Invasion
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lloyd, R.V. WHO Classification of Tumours of Endocrine Organs. In WHO/IARC Classification of Tumours, 4th ed.; WHO: Geneva, Switzerland, 2017; Volume 10. [Google Scholar]
- Kitahara, C.M.; Sosa, J.A. The changing incidence of thyroid cancer. Nat. Rev. Endocrinol. 2016, 12, 646–653. [Google Scholar] [CrossRef]
- Cabanillas, M.E.; McFadden, D.G.; Durante, C. Thyroid cancer. Lancet 2016, 388, 2783–2795. [Google Scholar] [CrossRef]
- Simoes-Pereira, J.; Capitao, R.; Limbert, E.; Leite, V. Anaplastic Thyroid Cancer: Clinical Picture of the Last Two Decades at a Single Oncology Referral Centre and Novel Therapeutic Options. Cancers 2019, 11, 1188. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar]
- Ravi, N.; Yang, M.; Gretarsson, S.; Jansson, C.; Mylona, N.; Sydow, S.R.; Woodward, E.L.; Ekblad, L.; Wennerberg, J.; Paulsson, K. Identification of Targetable Lesions in Anaplastic Thyroid Cancer by Genome Profiling. Cancers 2019, 11, 402. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Kim, Y.; Jeong, Y.M.; Bae, J.S.; Jung, C.K. CCND1 Splice Variant as A Novel Diagnostic and Predictive Biomarker for Thyroid Cancer. Cancers 2018, 10, 437. [Google Scholar] [CrossRef]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Mu, N.; Wang, N.; Straat, K.; Sofiadis, A.; Guo, Y.; Stenman, A.; Li, K.; Cheng, G.; Zhang, L.; et al. GABPA inhibits invasion/metastasis in papillary thyroid carcinoma by regulating DICER1 expression. Oncogene 2019, 38, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Dai, M.; Xu, D. TERT promoter mutations and GABP transcription factors in carcinogenesis: More foes than friends. Cancer Lett. 2020. [Google Scholar] [CrossRef]
- Liu, T.; Wang, N.; Cao, J.; Sofiadis, A.; Dinets, A.; Zedenius, J.; Larsson, C.; Xu, D. The age- and shorter telomere-dependent TERT promoter mutation in follicular thyroid cell-derived carcinomas. Oncogene 2014, 33, 4978–4984. [Google Scholar] [CrossRef]
- Xing, M.; Liu, R.; Liu, X.; Murugan, A.K.; Zhu, G.; Zeiger, M.A.; Pai, S.; Bishop, J. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol. 2014, 32, 2718–2726. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.; da Rocha, A.G.; Vinagre, J.; Batista, R.; Peixoto, J.; Tavares, C.; Celestino, R.; Almeida, A.; Salgado, C.; Eloy, C.; et al. TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J. Clin. Endocrinol. Metab. 2014, 99, E754–E765. [Google Scholar] [CrossRef] [PubMed]
- Hysek, M.; Paulsson, J.O.; Jatta, K.; Shabo, I.; Stenman, A.; Hoog, A.; Larsson, C.; Zedenius, J.; Juhlin, C.C. Clinical Routine TERT Promoter Mutational Screening of Follicular Thyroid Tumors of Uncertain Malignant Potential (FT-UMPs): A Useful Predictor of Metastatic Disease. Cancers 2019, 11, 1443. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, T.; Sofiadis, A.; Juhlin, C.C.; Zedenius, J.; Hoog, A.; Larsson, C.; Xu, D. TERT promoter mutation as an early genetic event activating telomerase in follicular thyroid adenoma (FTA) and atypical FTA. Cancer 2014, 120, 2965–2979. [Google Scholar] [CrossRef]
- Liu, T.; Yuan, X.; Xu, D. Cancer-Specific Telomerase Reverse Transcriptase (TERT) Promoter Mutations: Biological and Clinical Implications. Genes 2016, 7, 38. [Google Scholar] [CrossRef]
- Weinhold, N.; Jacobsen, A.; Schultz, N.; Sander, C.; Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014, 46, 1160–1165. [Google Scholar] [CrossRef]
- Pignot, G.; Le Goux, C.; Vacher, S.; Schnitzler, A.; Radvanyi, F.; Allory, Y.; Lallemand, F.; Delongchamps, N.B.; Zerbib, M.; Terris, B.; et al. PLEKHS1: A new molecular marker predicting risk of progression of non-muscle-invasive bladder cancer. Oncol. Lett. 2019, 18, 3471–3480. [Google Scholar] [CrossRef]
- GeneCards. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PLEKHS1 (accessed on 5 January 2019).
- The Cancer Genome Atlas (TCGA). Available online: http://cancergenome.nih.gov/ (accessed on 5 January 2019).
- Kotoh, J.; Sasaki, D.; Matsumoto, K.; Maeda, A. Plekhs1 and Prdx3 are candidate genes responsible for mild hyperglycemia associated with obesity in a new animal model of F344-fa-nidd6 rat. J. Vet. Med. Sci. 2016, 78, 1683–1691. [Google Scholar] [CrossRef]
- Tavares, C.; Eloy, C.; Melo, M.; Gaspar da Rocha, A.; Pestana, A.; Batista, R.; Bueno Ferreira, L.; Rios, E.; Sobrinho Simoes, M.; Soares, P. mTOR Pathway in Papillary Thyroid Carcinoma: Different Contributions of mTORC1 and mTORC2 Complexes for Tumor Behavior and SLC5A5 mRNA Expression. Int. J. Mol. Sci. 2018, 19, 1448. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar]
- cBioPortal for Cancer Genomics. Available online: https://www.cbioportal.org (accessed on 10 November 2019).
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef] [PubMed]
- DeLellis R: Pathology and Genetics of Tumours of Endocrine Organs. In WHO Classification of Tumours, 3rd ed.; WHO: Geneva, Switzerland, 2004.
- Andreasson, A.; Kiss, N.B.; Juhlin, C.C.; Hoog, A. Long-term storage of endocrine tissues at—80 degrees C does not adversely affect RNA quality or overall histomorphology. Biopreservation Biobanking 2013, 11, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Lee, J.J.; Foukakis, T.; Hashemi, J.; Grimelius, L.; Heldin, N.E.; Wallin, G.; Rudduck, C.; Lui, W.O.; Höög, A.; Larsson, C. Molecular cytogenetic profiles of novel and established human anaplastic thyroid carcinoma models. Thyroid 2007, 17, 289–301. [Google Scholar] [CrossRef]
- Guo, Y.; Yuan, X.; Li, K.; Dai, M.; Zhang, L.; Wu, Y.; Sun, C.; Chen, Y.; Cheng, G.; Liu, C.; et al. GABPA is a master regulator of luminal identity and restrains the aggressive disease in bladder cancer. Cell Death Differ. 2020, 27, 1862–1877. [Google Scholar] [CrossRef]


| Parameters n = (No of Informative Cases) | Observations | PLEKHS1 mRNA | p-Value |
|---|---|---|---|
| Median (Min–Max) | |||
| Age at diagnosis (n = 93) | 0.365 | ||
| Median (Min–Max) years | 51 (15–97) | ||
| <55 | n = 55 | 1.43 (0.21–139.30) | |
| ≥55 | n = 38 | 1.67 (0.37–811.88) | |
| Gender (n = 93) | 0.980 | ||
| Female | n = 67 | 1.43 (0.21–811.88) | |
| Male | n = 26 | 1.77 (0.21–421.76) | |
| Tumor size (n = 88) | 0.244 | ||
| ≤2cm (n = 40) | n = 40 | 1.06 (0.21–11.30) | |
| 2–4cm (n = 29) | n = 29 | 2.09 (0.21–811.88) | |
| >4cm (n = 19) | n = 19 | 1.67 (0.45–139.30) | |
| Lymph node metastases (n = 93) | 0.026 | ||
| No | n = 43 | 1.17 (0.21–421.76) | |
| Yes | n = 50 | 1.70 (0.21–811.88) | |
| Distant metastases (n = 93) | 0.011 | ||
| No | n = 81 | 1.28 (0.21–421.76) | |
| Yes | n = 12 | 3.43 (0.54–811.88) | |
| Extrathyroidal extension (n = 81) | 0.833 | ||
| No | n = 47 | 1.26 (0.21–97.07) | |
| Yes | n = 34 | 1.36 (0.22–811.88) | |
| TERT hotspot promoter mutation (n = 93) | 0.281 | ||
| Wild-type | n = 64 | 1.35 (0.21–421.76) | |
| Mutation C228T/C250T | n = 24/5 | 1.86 (0.37–811.88) | |
| TERT mRNA expression (n = 93) | 0.603 | ||
| No TERT mRNA | n = 40 | 1.46 (0.21–139.30) | |
| TERT mRNA expressed | n = 53 | 1.59 (0.37–811.88) | |
| Overall survival (n = 93) | <0.001 | ||
| Alive | n = 32 | 1.17 (0.21–139.30) | 95% CI = 1.002–1.007 |
| Dead | n = 61 | 2.17 (0.37–811.88) | HR = 1.005 |
| Follow-up:median (min–max) years | 14.8 (0.2–26.5) | ||
| Disease-free survival (n = 93) | 0.004 | ||
| No evidence of disease | n = 67 | 1.26 (0.21–421.76) | 95% CI = 1.001–1.007 |
| Relapsed/progression | n = 26 | 2.09 (0.34–811.88) | HR = 1.004 |
| Follow-up: Median (Min–Max) years | 13.5 (0.1–26.5) |
| Parameters (Number of Informative Cases) | Observations | PLEKHS1 mRNA | p-Value |
|---|---|---|---|
| Median (Min–Max) | |||
| Age at diagnosis (n = 18) | |||
| Median (Min–Max) years | 77.5 (54–91) | 0.101 | |
| <55 years | n = 1 | 1.03 (1.03–1.03) | |
| ≥55 years | n = 17 | 4.00 (1.80–85.57) | |
| Gender (n = 18) | 0.214 | ||
| Female | n = 10 | 4.68 (2.92–85.57) | |
| Male | n = 8 | 2.76 (1.03–18.91) | |
| Tumor size (n = 18) | 0.251 | ||
| ≤2 cm | n = 1 | 1.03 (1.03–1.03) | |
| 2–4 cm | n = 2 | 44.04 (2.52–85.57) | |
| >4 cm | n = 15 | 4.00 (1.80–18.91) | |
| TERT promoter mutation (n = 18) | 0.722 | ||
| Wild-type | n = 8 | 4.26 (1.03–85.57) | |
| Mutation (C228T/C250T) | n = 7/3 | 3.67 (1.80–18.91) | |
| TERT mRNA expression (n = 18) | 0.888 | ||
| No TERT mRNA | n = 2 | 10.02 (2.33–17.71) | |
| TERT mRNA expressed | n = 16 | 3.67 (1.03–85.57) | |
| Survival (n = 17) | 0.54 | ||
| Alive | n = 1 | 7.91 (7.91–7.91) | |
| Dead | n = 16 | 3.25 (1.03–85.57) | |
| Follow-up: Median (Min–Max) months | 3 (0–190) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, X.; Mu, N.; Yuan, X.; Wang, N.; Juhlin, C.C.; Strååt, K.; Larsson, C.; Xu, D. PLEKHS1 Over-Expression is Associated with Metastases and Poor Outcomes in Papillary Thyroid Carcinoma. Cancers 2020, 12, 2133. https://doi.org/10.3390/cancers12082133
Xing X, Mu N, Yuan X, Wang N, Juhlin CC, Strååt K, Larsson C, Xu D. PLEKHS1 Over-Expression is Associated with Metastases and Poor Outcomes in Papillary Thyroid Carcinoma. Cancers. 2020; 12(8):2133. https://doi.org/10.3390/cancers12082133
Chicago/Turabian StyleXing, Xiangling, Ninni Mu, Xiaotian Yuan, Na Wang, C. Christofer Juhlin, Klas Strååt, Catharina Larsson, and Dawei Xu. 2020. "PLEKHS1 Over-Expression is Associated with Metastases and Poor Outcomes in Papillary Thyroid Carcinoma" Cancers 12, no. 8: 2133. https://doi.org/10.3390/cancers12082133
APA StyleXing, X., Mu, N., Yuan, X., Wang, N., Juhlin, C. C., Strååt, K., Larsson, C., & Xu, D. (2020). PLEKHS1 Over-Expression is Associated with Metastases and Poor Outcomes in Papillary Thyroid Carcinoma. Cancers, 12(8), 2133. https://doi.org/10.3390/cancers12082133





