The Diagnostic Accuracy of Mutant KRAS Detection from Pancreatic Secretions for the Diagnosis of Pancreatic Cancer: A Meta-Analysis
Abstract
1. Introduction
2. Results
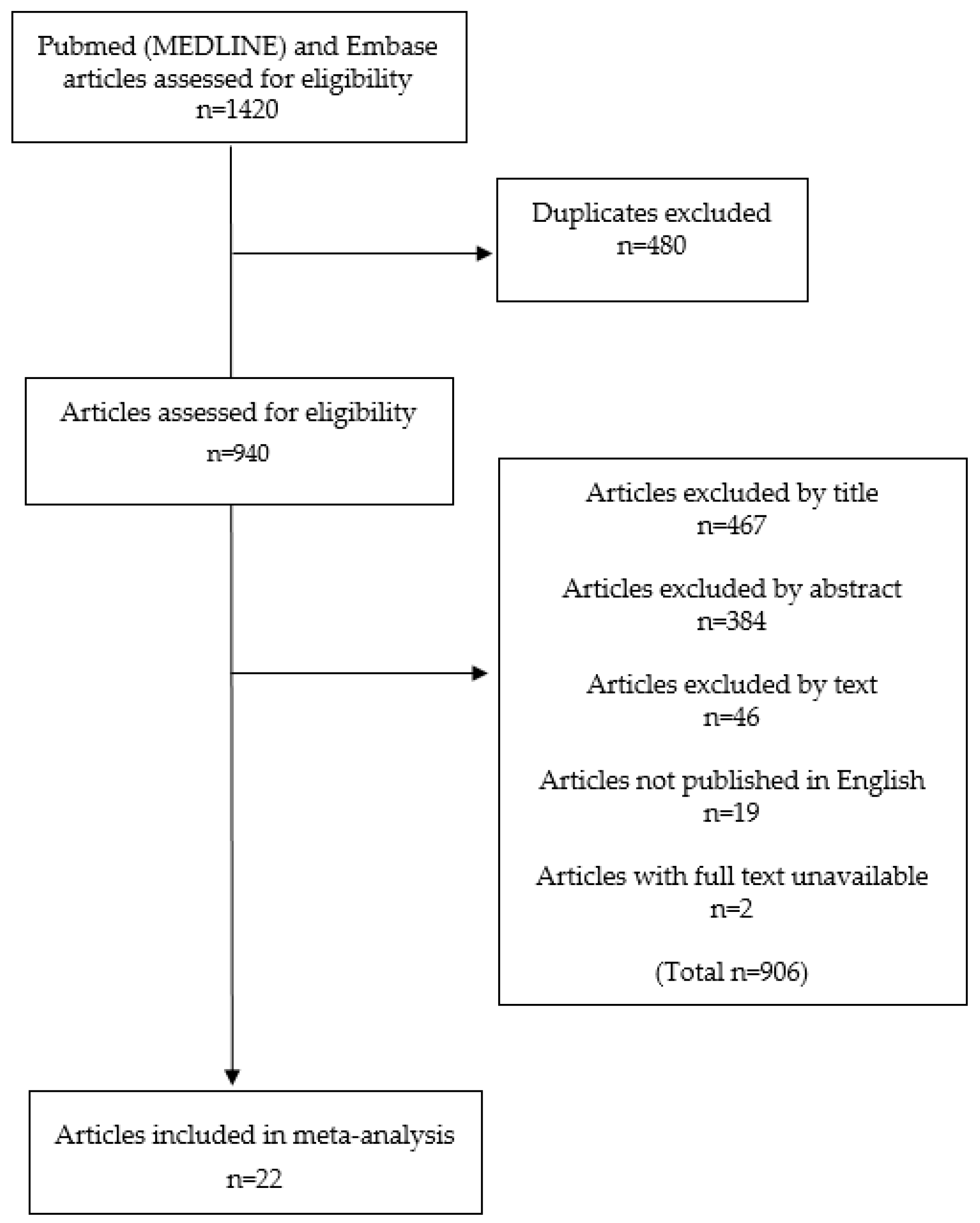
2.1. Study Inclusion
2.2. Study Demographics
2.3. Mutant KRAS Determination.
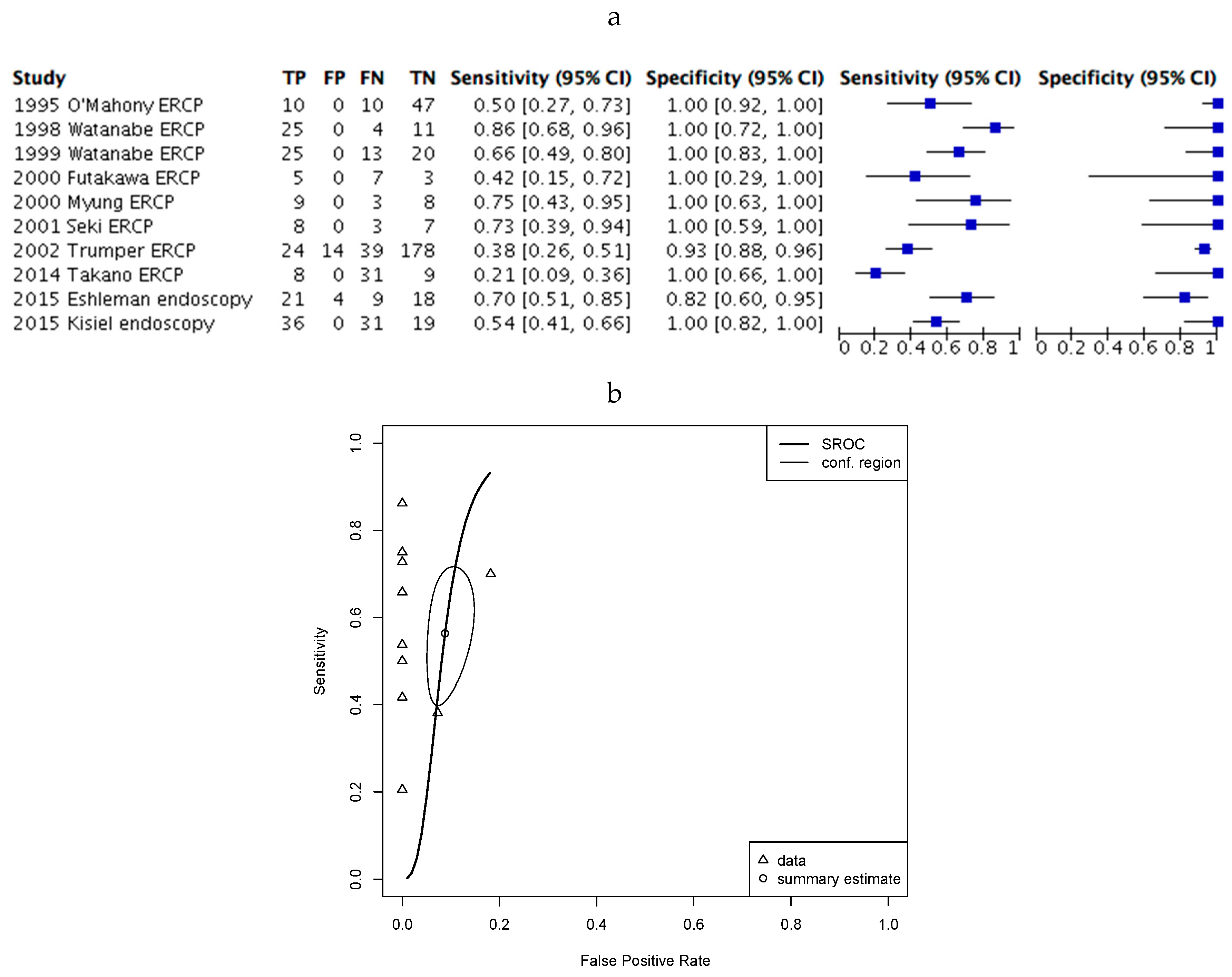
2.4. Assessment of Diagnostic Accuracy.
3. Discussion
4. Materials and Methods
4.1. Search Strategy
4.2. Inclusion/Exclusion Criteria
4.3. Data Extraction and Statistics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fokas, E.; O’Neill, E.; Gordon-Weeks, A.; Mukherjee, S.; McKenna, W.G.; Muschel, R.J. Pancreatic ductal adenocarcinoma: From genetics to biology to radiobiology to oncoimmunology and all the way back to the clinic. Biochim. Biophys. Acta 2015, 1855, 61–82. [Google Scholar] [CrossRef]
- Ghaneh, P.; Hanson, R.; Titman, A.; Lancaster, G.; Plumpton, C.; Lloyd-Williams, H.; Yeo, S.T.; Edwards, R.T.; Johnson, C.; Abu Hilal, M.; et al. PET-PANC: Multicentre prospective diagnostic accuracy and health economic analysis study of the impact of combined modality 18fluorine-2-fluoro-2-deoxy-d-glucose positron emission tomography with computed tomography scanning in the diagnosis and management of pancreatic cancer. Health Technol. Assess. 2018, 22, 1–114. [Google Scholar] [CrossRef] [PubMed]
- Klaiber, U.; Hackert, T.; Neoptolemos, J.P. Adjuvant treatment for pancreatic cancer. Transl. Gastroenterol. Hepatol. 2019, 4, 27. [Google Scholar] [CrossRef]
- Chawla, A.; Ferrone, C.R. Neoadjuvant Therapy for Resectable Pancreatic Cancer: An Evolving Paradigm Shift. Front. Oncol. 2019, 9, 1085. [Google Scholar] [CrossRef] [PubMed]
- Handgraaf, H.J.M.; Boonstra, M.C.; Van Erkel, A.R.; Bonsing, B.A.; Putter, H.; Van De Velde, C.J.; Vahrmeijer, A.L.; Mieog, J.S.D. Current and future intraoperative imaging strategies to increase radical resection rates in pancreatic cancer surgery. Biomed. Res. Int. 2014, 890230. [Google Scholar] [CrossRef] [PubMed]
- Saad, A.M.; Turk, T.; Al-Husseini, M.J.; Abdel-Rahman, O. Trends in pancreatic adenocarcinoma incidence and mortality in the United States in the last four decades; a SEER-based study. BMC Cancer 2018, 18, 688. [Google Scholar] [CrossRef]
- Ghaneh, P.; Kleeff, J.; Halloran, C.M.; Raraty, M.; Jackson, R.; Melling, J.; Jones, O.; Palmer, D.H.; Cox, T.F.; Smith, C.J.; et al. The Impact of Positive Resection Margins on Survival and Recurrence Following Resection and Adjuvant Chemotherapy for Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2019, 269, 520–529. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Stocken, D.D.; Dunn, J.A.; Almond, J.; Beger, H.G.; Pederzoli, P.; Bassi, C.; Dervenis, C.; Fernandez-Cruz, L.; Lacaine, F.; et al. Influence of resection margins on survival for patients with pancreatic cancer treated by adjuvant chemoradiation and/or chemotherapy in the ESPAC-1 randomized controlled trial. Ann. Surg. 2001, 234, 758–768. [Google Scholar] [CrossRef]
- Paniccia, A.; Hosokawa, P.; Henderson, W.; Schulick, R.D.; Edil, B.H.; McCarter, M.D.; Gajdos, C. Characteristics of 10-Year Survivors of Pancreatic Ductal Adenocarcinoma. JAMA Surg. 2015, 150, 701–710. [Google Scholar] [CrossRef]
- Schnelldorfer, T.; Ware, A.L.; Sarr, M.G.; Smyrk, T.C.; Zhang, L.; Qin, R.; Gullerud, R.E.; Donohue, J.H.; Nagorney, D.M.; Farnell, M.B. Long-Term Survival After Pancreatoduodenectomy for Pancreatic Adenocarcinoma: Is Cure Possible? Ann. Surg. 2008, 247, 456–462. [Google Scholar] [CrossRef]
- Ying, H.; Dey, P.; Yao, W.; Kimmelman, A.C.; Draetta, G.F.; Maitra, A.; DePinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016, 30, 355–385. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Azim, S.; Zubair, H.; Bhardwaj, A.; Patel, G.K.; Khushman, M.; Singh, S.; Singh, A.P. Molecular Drivers of Pancreatic Cancer Pathogenesis: Looking Inward to Move Forward. Int. J. Mol. Sci. 2017, 18, 779. [Google Scholar] [CrossRef] [PubMed]
- Hruban, R.H.; Takaori, K.; Klimstra, D.S.; Adsay, N.V.; Albores-Saavedra, J.; Biankin, A.V.; Biankin, S.A.; Compton, C.; Fukushima, N.; Furukawa, T.; et al. An Illustrated Consensus on the Classification of Pancreatic Intraepithelial Neoplasia and Intraductal Papillary Mucinous Neoplasms. Am. J. Surg. Pathol. 2004, 28, 977–987. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203.e13. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Jones, S.; Bozic, I.; Antal, T.; Leary, R.; Fu, B.; Kamiyama, M.; Hruban, R.H.; Eshleman, J.R.; Nowak, M.A.; et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010, 467, 1114–1117. [Google Scholar] [CrossRef]
- Urban, T.; Ricci, S.; Grange, J.-D.; Lacave, R.; Boudghene, F.; Breittmayer, F.; Languille, O.; Roland, J.; Bernaudin, J.-F. Detection of c-Ki-ras Mutation by PCR/RFLP Analysis and Diagnosis of Pancreatic Adenocarcinomas. J. Natl. Cancer Inst. 1993, 85, 2008–2012. [Google Scholar] [CrossRef]
- Bournet, B.; Buscail, C.; Muscari, F.; Cordelier, P.; Buscail, L. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities. Eur. J. Cancer 2016, 54, 75–83. [Google Scholar] [CrossRef]
- Narayanan, V.; Konstantinov, S.R.; Peppelenbosch, M.P. Mutations in KRAS: Are they a valid biomarker for pancreatic ductal adenocarcinomas diagnosis? Transl. Cancer Res. 2017, 6, S72–S77. [Google Scholar] [CrossRef]
- Lang, A.H.; Drexel, H.; Geller-Rhomberg, S.; Stark, N.; Winder, T.; Geiger, K.; Muendlein, A. Optimized Allele-Specific Real-Time PCR Assays for the Detection of Common Mutations in KRAS and BRAF. J. Mol. Diagn. 2011, 13, 23–28. [Google Scholar] [CrossRef]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Sanjuanbenito, A.; Rodríguez-Garrote, M.; Lisa, E.; Mendía, E.; Lobo, E.; Malats, N.; et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef] [PubMed]
- Le Calvez-Kelm, F.; Foll, M.; Wozniak, M.B.; Delhomme, T.M.; Durand, G.; Chopard, P.; Pertesi, M.; Fabianova, E.; Adamcakova, Z.; Holcatova, I.; et al. KRAS mutations in blood circulating cell-free DNA: A pancreatic cancer case-control. Oncotarget 2016, 7, 78827–78840. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Ding, X.-Q.; Zhu, H.; Wang, R.-X.; Pan, X.-R.; Tong, J.-H. KRAS Mutant Allele Fraction in Circulating Cell-Free DNA Correlates with Clinical Stage in Pancreatic Cancer Patients. Front. Oncol. 2019, 9, 1295. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.L.; Zhao, W.; Project, C.G.; Leung, S.Y.; Yuen, S.T. BRAF and KRAS Mutations in Colorectal Hyperplastic Polyps and Serrated Adenomas. Cancer Res. 2003, 63, 4878–4881. [Google Scholar] [PubMed]
- Tímár, J. The clinical relevance of KRAS gene mutation in non-small-cell lung cancer. Curr. Opin. Oncol. 2014, 26, 138–144. [Google Scholar] [CrossRef]
- O’mahony, S.; Longfellow, M.; McMahon, M.J.; Axon, A.T.; Quirke, P. Detection of c-Ki-ras mutations in bile samples from patients with pancreatic and biliary cancers. Clin. Mol. Pathol. 1995, 48, M316–M318. [Google Scholar] [CrossRef]
- Iguchi, H.; Sugano, K.; Fukayama, N.; Ohkura, H.; Sadamoto, K.; Ohkoshi, K.; Seo, Y.; Tomoda, H.; Funakoshi, A.; Wakasugi, H. Analysis of Ki-ras codon 12 mutations in the duodenal juice of patients with pancreatic cancer. Gastroenterology 1996, 110, 221–226. [Google Scholar] [CrossRef]
- Uehara, H.; Nakaizumi, A.; Baba, M.; Iishi, H.; Tatsuta, M.; Kitamura, T.; Ohigashi, H.; Ishikawa, O.; Takenaka, A.; Ishiguro, S. Diagnosis of pancreatic cancer by K-ras point mutation and cytology of pancreatic juice. Am. J. Gastroenterol. 1996, 91, 1616–1621. [Google Scholar]
- Furuya, N.; Kawa, S.; Akamatsu, T.; Furihata, K. Long-term follow-up of patients with chronic pancreatitis and K-ras gene mutation detected in pancreatic juice. Gastroenterology 1997, 113, 593–598. [Google Scholar] [CrossRef]
- Fukushima, N.; Suzuki, M.; Fukayama, M. Analysis of K-ras oncogene mutation directly applied to atypical cell clusters on cytologic smear slides of bile and pancreatic juice. Pathol. Int. 1998, 48, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Yamaguchi, Y.; Ha, A.; Hu, Y.X.; Motoo, Y.; Okai, T.; Yoshimura, T.; Sawabu, N. Quantitative determination of K-ras mutations in pancreatic juice for diagnosis of pancreatic cancer using hybridization protection assay. Pancreas 1998, 17, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Ha, A.; Hu, Y.X.; Ohtsubo, K.; Yamaguchi, Y.; Motoo, Y.; Okai, T.; Toya, D.; Tanaka, N.; Sawabu, N. K-ras mutations in duodenal aspirate without secretin stimulation for screening of pancreatic and biliary tract carcinoma. Cancer 1999, 86, 1441–1448. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Watanabe, H.; Yrdiran, S.; Ohtsubo, K.; Motoo, Y.; Okai, T.; Sawabu, N. Detection of mutations of p53 tumor suppressor gene in pancreatic juice and its application to diagnosis of patients with pancreatic cancer: Comparison with K-ras mutation. Clin. Cancer Res. 1999, 5, 1147–1153. [Google Scholar] [PubMed]
- Futakawa, N.; Kimura, W.; Yamagata, S.; Zhao, B.; Ilsoo, H.; Inoue, T.; Sata, N.; Kawaguchi, Y.; Kubota, Y.; Muto, T. Significance of K-ras mutation and CEA level in pancreatic juice in the diagnosis of pancreatic cancer. J. Hepato-Biliary-Pancreat. Surg. 2000, 7, 63–71. [Google Scholar] [CrossRef]
- Myung, S.J.; Kim, M.H.; Kim, Y.S.; Kim, H.J.; Park, E.T.; Yoo, K.S.; Lim, B.C.; Wan Seo, D.; Lee, S.K.; Min, Y.I.; et al. Telomerase activity in pure pancreatic juice for the diagnosis of pancreatic cancer may be complementary to K-ras mutation. Gastrointest. Endosc. 2000, 51, 708–713. [Google Scholar] [CrossRef]
- Boadas, J.; Mora, J.; Urgell, E.; Puig, P.; Roca, M.; Cussó, X.; Capellà, G.; Lluís, F.; Farré, A. Clinical usefulness of K-ras gene mutation detection and cytology in pancreatic juice in the diagnosis and screening of pancreatic cancer. Eur. J. Gastroenterol. Hepatol. 2001, 13, 1153–1159. [Google Scholar] [CrossRef]
- Ha, A.; Watanabe, H.; Yamaguchi, Y.; Ohtsubo, K.; Wang, Y.; Motoo, Y.; Okai, T.; Wakabayahi, T.; Sawabu, N. Usefulness of supernatant of pancreatic juice for genetic analysis of K-ras in diagnosis of pancreatic carcinoma. Pancreas 2001, 23, 356–363. [Google Scholar] [CrossRef]
- Pugliese, V.; Pujic, N.; Saccomanno, S.; Gatteschi, B.; Pera, C.; Aste, H.; Ferrara, G.B.; Nicolò, G. Pancreatic intraductal sampling during ERCP in patients with chronic pancreatitis and pancreatic cancer: Cytologic studies and k-ras-2 codon 12 molecular analysis in 47 cases. Gastrointest. Endosc. 2001, 54, 595–599. [Google Scholar] [CrossRef]
- Seki, K.; Suda, T.; Aoyagi, Y.; Sugawara, S.; Natsui, M.; Motoyama, H.; Shirai, Y.; Sekine, T.; Kawai, H.; Mita, Y.; et al. Diagnosis of pancreatic adenocarcinoma by detection of human telomerase reverse transcriptase messenger RNA in pancreatic juice with sample qualification. Clin. Cancer Res. 2001, 7, 1976–1981. [Google Scholar]
- Costentin, L.; Pagès, P.; Bouisson, M.; Berthelémy, P.; Buscail, L.; Escourrou, J.; Pradayrol, L.; Vaysse, N. Frequent deletions of tumor suppressor genes in pure pancreatic juice from patients with tumoral or nontumoral pancreatic diseases. Pancreatology 2002, 2, 17–25. [Google Scholar] [CrossRef]
- Wang, Y.; Yamaguchi, Y.; Watanabe, H.; Ohtsubo, K.; Motoo, Y.; Sawabu, N. Detection of p53 gene mutations in the supernatant of pancreatic juice and plasma from patients with pancreatic carcinomas. Pancreas 2004, 28, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Trümper, L.; Menges, M.; Daus, H.; Köhler, D.; Reinhard, J.-O.; Sackmann, M.; Moser, C.; Sek, A.; Jacobs, G.; Zeitz, M.; et al. Low sensitivity of the ki-ras polymerase chain reaction for diagnosing pancreatic cancer from pancreatic juice and bile: A multicenter prospective trial. J. Clin. Oncol. 2002, 20, 4331–4337. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Fukushima, N.; Abe, T.; Bian, Y.; Hua, L.; Wendelburg, B.J.; Yeo, C.J.; Hruban, R.H.; Goggins, M.G.; Eshleman, J.R. Sensitive and quantitative detection of KRAS2 gene mutations in pancreatic duct juice differentiates patients with pancreatic cancer from chronic pancreatitis, potential for early detection. Cancer Biol. 2008, 7, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Takano, S.; Fukasawa, M.; Maekawa, S.; Kadokura, M.; Miura, M.; Shindo, H.; Takahashi, E.; Sato, T.; Enomoto, N. Deep sequencing of cancer-related genes revealed GNAS mutations to be associated with intraductal papillary mucinous neoplasms and its main pancreatic duct dilation. PLoS ONE 2014, 9, e98718. [Google Scholar] [CrossRef]
- Eshleman, J.R.; Norris, A.L.; Sadakari, Y.; Debeljak, M.; Borges, M.; Harrington, C.; Lin, E.; Brant, A.; Barkley, T.; Almario, J.A.; et al. KRAS and guanine nucleotide-binding protein mutations in pancreatic juice collected from the duodenum of patients at high risk for neoplasia undergoing endoscopic ultrasound. Clin. Gastroenterol. Hepatol. 2015, 13, 963–969.e4. [Google Scholar] [CrossRef]
- Kisiel, J.B.; Raimondo, M.; Taylor, W.R.; Yab, T.C.; Mahoney, D.W.; Sun, Z.; Middha, S.; Baheti, S.; Zou, H.; Smyrk, T.C.; et al. New DNA Methylation Markers for Pancreatic Cancer: Discovery, Tissue Validation, and Pilot Testing in Pancreatic Juice. Clin. Cancer Res. 2015, 21, 4473–4481. [Google Scholar] [CrossRef]
- Ginesta, M.M.; Diaz-Riascos, Z.V.; Busquets, J.; Pelaez, N.; Serrano, T.; Peinado, M.À.; Jorba, R.; García-Borobia, F.J.; Capella, G.; Fabregat, J. APC promoter is frequently methylated in pancreatic juice of patients with pancreatic carcinomas or periampullary tumors. Oncol. Lett. 2016, 12, 2210–2216. [Google Scholar] [CrossRef]
- Canto, M.I.; Harinck, F.; Hruban, R.H.; Offerhaus, G.J.; Poley, J.-W.; Kamel, I.; Nio, Y.; Schulick, R.S.; Bassi, C.; Kluijt, I.; et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut 2013, 62, 339–347. [Google Scholar] [CrossRef]
- Li, W.-M.; Hu, T.-T.; Zhou, L.-L.; Feng, Y.-M.; Wang, Y.-Y.; Fang, J. Highly sensitive detection of the PIK3CAH1047R mutation in colorectal cancer using a novel PCR-RFLP method. BMC Cancer 2016, 16, 454. [Google Scholar] [CrossRef]
- Rutjes, A.W.S.; Reitsma, J.B.; Di Nisio, M.; Smidt, N.; van Rijn, J.C.; Bossuyt, P.M.M. Evidence of bias and variation in diagnostic accuracy studies. CMAJ 2006, 174, 469–476. [Google Scholar] [CrossRef]
- Lijmer, J.G.; Mol, B.W.; Heisterkamp, S.; Bonsel, G.J.; Prins, M.H.; van der Meulen, J.H.; Bossuyt, P.M. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA 1999, 282, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Pourshams, A.; Sepanlou, S.G.; Ikuta, K.S.; Bisignano, C.; Safiri, S.; Roshandel, G.; Sharif, M.; Khatibian, M.; Fitzmaurice, C.; Nixon, M.R.; et al. The global, regional, and national burden of pancreatic cancer and its attributable risk factors in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2019, 4, 934–947. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Chen, F.; Zhong, Z.; Qi, L. Detection of K-ras gene mutations in feces by magnetic nanoprobe in patients with pancreatic cancer: A preliminary study. Exp. Med. 2018, 15, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Kisiel, J.B.; Yab, T.C.; Taylor, W.R.; Chari, S.T.; Petersen, G.M.; Mahoney, D.W.; Ahlquist, D.A. Stool DNA testing for the detection of pancreatic cancer: Assessment of methylation marker candidates. Cancer 2012, 118, 2623–2631. [Google Scholar] [CrossRef]
- Choi, M.H.; Mejlænder-Andersen, E.; Manueldas, S.; El Jellas, K.; Steine, S.J.; Tjensvoll, K.; Sætran, H.A.; Knappskog, S.; Hoem, D.; Nordgård, O.; et al. Mutation analysis by deep sequencing of pancreatic juice from patients with pancreatic ductal adenocarcinoma. BMC Cancer 2019, 19, 11. [Google Scholar] [CrossRef]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Reid, M.D.; Choi, H.; Balci, S.; Akkas, G.; Adsay, V. Serous cystic neoplasms of the pancreas: Clinicopathologic and molecular characteristics. Semin. Diagn. Pathol. 2014, 31, 475–483. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Khalid, A.; Fasanella, K.E.; McGrath, K.M.; Brand, R.E.; Chennat, J.S.; Slivka, A.; Zeh, H.J.; Zureikat, A.H.; Krasinskas, A.M.; et al. Integration of KRAS testing in the diagnosis of pancreatic cystic lesions: A clinical experience of 618 pancreatic cysts. Mod. Pathol. 2013, 26, 1478–1487. [Google Scholar] [CrossRef]
- Singhi, A.D.; Nikiforova, M.N.; Fasanella, K.E.; McGrath, K.M.; Pai, R.K.; Ohori, N.P.; Bartholow, T.L.; Brand, R.E.; Chennat, J.S.; Lu, X.; et al. Preoperative GNAS and KRAS testing in the diagnosis of pancreatic mucinous cysts. Clin. Cancer Res. 2014, 20, 4381–4389. [Google Scholar] [CrossRef]
- Singhi, A.D.; McGrath, K.; Brand, R.E.; Khalid, A.; Zeh, H.J.; Chennat, J.S.; Fasanella, K.E.; Papachristou, G.I.; Slivka, A.; Bartlett, D.L.; et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut 2018, 67, 2131–2141. [Google Scholar] [CrossRef]
- van Enst, W.A.; Ochodo, E.; Scholten, R.J.; Hooft, L.; Leeflang, M.M. Investigation of publication bias in meta-analyses of diagnostic test accuracy: A meta-epidemiological study. BMC Med. Res. Methodol. 2014, 14, 70. [Google Scholar] [CrossRef] [PubMed]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B.; et al. Meta-analysis of Observational Studies in Epidemiology: A Proposal for Reporting. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- McInnes, M.D.F.; Moher, D.; Thombs, B.D.; McGrath, T.A.; Bossuyt, P.M.; Clifford, T.; Cohen, J.F.; Deeks, J.J.; Gatsonis, C.; Hooft, L.; et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018, 319, 388–396. [Google Scholar] [CrossRef] [PubMed]
- R-Forge: Meta-Analysis of Diagnostic Accuracy: Project Home. Available online: http://r-forge.r-project.org/projects/mada/ (accessed on 13 August 2020).


| Study | Risk of Bias | Concerns Regarding Applicability | |||||
|---|---|---|---|---|---|---|---|
| Patient Selection | Index Tests | Reference Standard | Flow and Timing | Patient Selection | Index Tests | Reference Standard | |
| O’Mahony [26] | Low | Low | Unclear | Low | Low | High | Low |
| Iguchi [27] | Unclear | Low | Low | High | Low | Low | Low |
| Uehara [28] | Low | Unclear | High | High | Low | Unclear | Low |
| Furuya [29] | Low | Low | Low | Unclear | Low | Unclear | Low |
| Fukushima [30] | High | High | Low | Low | Low | High | Low |
| Watanabe (1998) [31] | Low | Unclear | Low | Unclear | Low | Unclear | Unclear |
| Watanabe (1999) [32] | Low | Unclear | High | Low | Low | Low | Low |
| Yamaguchi [33] | High | Low | Low | Unclear | Low | Unclear | Low |
| Futakawa [34] | Low | High | High | Unclear | Low | Low | Low |
| Myung [35] | Low | High | Unclear | Low | Low | High | Unclear |
| Boadas [36] | High | Unclear | Unclear | High | Low | Unclear | Unclear |
| Ha [37] | High | Unclear | Low | High | Low | Low | High |
| Pugliese [38] | High | High | High | High | Low | High | Unclear |
| Seki [39] | Unclear | Unclear | Unclear | Unclear | Low | Unclear | Low |
| Costentin [40] | High | High | Unclear | High | Low | Low | Unclear |
| Wang [41] | High | Low | High | High | Low | Unclear | Low |
| Trumper [42] | Unclear | Low | High | High | Low | Low | Low |
| Shi [43] | Unclear | Low | Low | Unclear | Low | Low | Low |
| Takano [44] | High | High | Unclear | Low | Low | Low | Low |
| Eshleman [45] | High | Unclear | Low | Low | Low | Unclear | Low |
| Kisiel [46] | High | Low | Low | Unclear | Low | Low | Low |
| Ginesta [47] | High | Low | Unclear | Unclear | Low | Low | Low |
| Author | Published | Country | Design | Number of Patients (% PDAC Prevalence) | Patient Population | Age | Male (%) |
|---|---|---|---|---|---|---|---|
| O’Mahony [26] | 1995 | UK | * | 67 (30) | PDAC, H | * | * |
| Iguchi [27] | 1996 | Japan | * | 60 (32) | PDAC, Pc, B | * | * |
| Uehara [28] | 1996 | Japan | Ret-Co | 47 (30) | PDAC, Pc, B, H | * | * |
| Furuya [29] | 1997 | Japan | Pro-Co | 70 (8.6) | PDAC, Pc, B | * | * |
| Fukushima [30] | 1998 | Japan | Ret-Co | 29 (72) | PDAC, Pc | * | * |
| Watanabe (1998) [31] | 1998 | Japan | Ret-Co | 66 (44) | PDAC, Pc, H | * (39–83) | 55 |
| Watanabe (1999) [32] | 1999 | Japan | Ret-Co | 140 (43) | PDAC, Pc, B, H | 61 (28–84) § | * |
| Yamaguchi [33] | 1999 | Japan | Ret-Co | 46(57) | PDAC, Pc, B | * | * |
| Futakawa [34] | 2000 | Japan | Pro-Co | 52 (23) | PDAC, Pc, B, H | * | * |
| Myung [35] | 2000 | Korea | Ret-Co | 31 (39) | PDAC, Pc, H | 63 (46–77) ± | 61 |
| Boadas [36] | 2001 | Spain | Pro-Co | 90 (20) | PDAC, Pc | * | * |
| Ha [37] | 2001 | Japan | Ret-Co | 44 (43) | PDAC, Pc | * (17–81) | * |
| Pugliese [38] | 2001 | Italy | Pro-Co | 45 (76) | PDAC, Pc | 66 (44–88) § | 56 |
| Seki [39] | 2001 | Japan | Ret-Co | 36 (47) | PDAC, Pc, H | * | 72 |
| Costentin [40] | 2002 | France | Ret-Co | 57 (32) | PDAC, Pc, B | * | * |
| Wang [41] | 2002 | Germany | Pro-Co | 358 (33) | PDAC, Pc, B, H | * | * |
| Trumper [42] | 2004 | Japan | Ret-Co | 46 (46) | PDAC, Pc | * | * |
| Shi [43] | 2008 | USA | Ret-Co | 36 (75) | PDAC, Pc | * | * |
| Takano [44] | 2014 | Japan | Ret-Co | 152(26) | PDAC, Pc, B, H | 65 (35–85) § | 58 |
| Eshleman [45] | 2015 | USA | Cas-Co | 272 (11) | PDAC, B, H | 57± | 48 |
| Kisiel [46] | 2015 | USA | Cas-Co | 102 (60) | PDAC, Pc, H | 64 (49–76) § | 52 |
| Ginesta [47] | 2016 | Spain | Cas-Co | 135 (82) | PDAC, Pc, B | 68 (40–79) ± | 60 |
| Author | Modality | Site | Secretin Stimulation | Mucus Volume (mls) | DNA Extraction Method | Amplification Method | Mut-KRAS Detection Method | Codon/Mutant Tested | Gold Standard Test |
|---|---|---|---|---|---|---|---|---|---|
| O’Mahony [26] | ERCP | Bile duct | n | * | Pheno-chlor | PCR | RFLP | 12 | Histology |
| Iguchi [27] | Endoscopy | Duodenum | y | (30–40) | Pheno-chlor | PCR | RFLP, sequencing | 12 | Histology |
| Uehara [28] | ERCP | Pancreas | n | * | * | PCR | Slot-blot | 12 | * |
| Furuya [29] | Endoscopy | Duodenum | Y | 0.5 ± | Acet-chlor | PCR | RFLP | 12 | Histology |
| Fukushima [30] | ERCP | Pancreas, bile duct | n | * | Pheno-chlor | PCR | RFLP | 12 | Histology |
| Watanabe (1998) [31] | ERCP | Pancreas | n | * | * | PCR | Hybridization probe | 12 | Histology |
| Watanabe (1999) [32] | ERCP | Pancreas | n | (2–3) | Pheno-chlor, Prot-K | PCR with A-sA | 12 | Histology | |
| Yamaguchi [33] | ERCP | Pancreas | y | * | Pheno-chlor | PCR | SSCP | 12 | Histology |
| Futakawa [34] | ERCP | Pancreas | n | 0.1 | Pheno-chlor | PCR | RFLP | 12 | * |
| Myung [35] | ERCP | Pancreas | y | 0.1 | * | PCR | RFLP | 12 | Histology |
| Boadas [36] | ERCP | Pancreas | y | 4.6 ± | * | PCR | RFLP | 12 | Histology |
| Ha [37] | Endoscopy | Duodenum | y | * | Pheno-chlor | PCR with A-sA | RFLP | 12 | Histology |
| Pugliese [38] | ERCP | Pancreas | n | * | * | PCR | RFLP, sequencing | 12 | Histology |
| Seki [39] | ERCP | Pancreas | y | 2 ± | * | PCR | SSCP | 12 | * |
| Costentin [40] | ERCP | Pancreas | n | * | * | PCR | RFLP | 12 | * |
| Wang [41] | ERCP | Pancreas | n | (1–2) | * | RFLP-Targeted enrichment | RFLP, sequencing | 12 | * |
| Trumper [42] | Endoscopy | Duodenum | y | (10–15) | Pheno-chlor | PCR with A-sA | RFLP | 12 | Histology |
| Shi [43] | Surgery | Pancreas | n | * | * | PCR | qPCR-primer based | G12V, G12D, G12R | Histology |
| Takano [44] | ERCP | Pancreas | n | * | Proprietary (QiAMP®Kit) | PCR | DNA sequencing | G12D, G12R, G12V, Q61H | Histology |
| Eshleman [45] | Endoscopy | Pancreas | y | (5–10) | * | * | HRMA | * | |
| Kisiel [46] | Endoscopy | Duodenum | y | 2 ± | * | * | QuARTS | G12D | * |
| Ginesta [47] | Surgery | Pancreas | n | * | Pheno-chlor | PCR | qPCR primer-based | G12C, G12V, G12D, G12A, G12s, G12R, G13D | Histology |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, N.; Petrinic, T.; Silva, M.; Soonawalla, Z.; Reddy, S.; Gordon-Weeks, A. The Diagnostic Accuracy of Mutant KRAS Detection from Pancreatic Secretions for the Diagnosis of Pancreatic Cancer: A Meta-Analysis. Cancers 2020, 12, 2353. https://doi.org/10.3390/cancers12092353
Patel N, Petrinic T, Silva M, Soonawalla Z, Reddy S, Gordon-Weeks A. The Diagnostic Accuracy of Mutant KRAS Detection from Pancreatic Secretions for the Diagnosis of Pancreatic Cancer: A Meta-Analysis. Cancers. 2020; 12(9):2353. https://doi.org/10.3390/cancers12092353
Chicago/Turabian StylePatel, Nikhil, Tatjana Petrinic, Michael Silva, Zahir Soonawalla, Srikanth Reddy, and Alex Gordon-Weeks. 2020. "The Diagnostic Accuracy of Mutant KRAS Detection from Pancreatic Secretions for the Diagnosis of Pancreatic Cancer: A Meta-Analysis" Cancers 12, no. 9: 2353. https://doi.org/10.3390/cancers12092353
APA StylePatel, N., Petrinic, T., Silva, M., Soonawalla, Z., Reddy, S., & Gordon-Weeks, A. (2020). The Diagnostic Accuracy of Mutant KRAS Detection from Pancreatic Secretions for the Diagnosis of Pancreatic Cancer: A Meta-Analysis. Cancers, 12(9), 2353. https://doi.org/10.3390/cancers12092353





