Beta Human Papillomavirus 8E6 Attenuates Non-Homologous End Joining by Hindering DNA-PKcs Activity
Abstract
1. Introduction
2. Results
2.1. β-HPV 8E6 Decreases NHEJ Efficiency
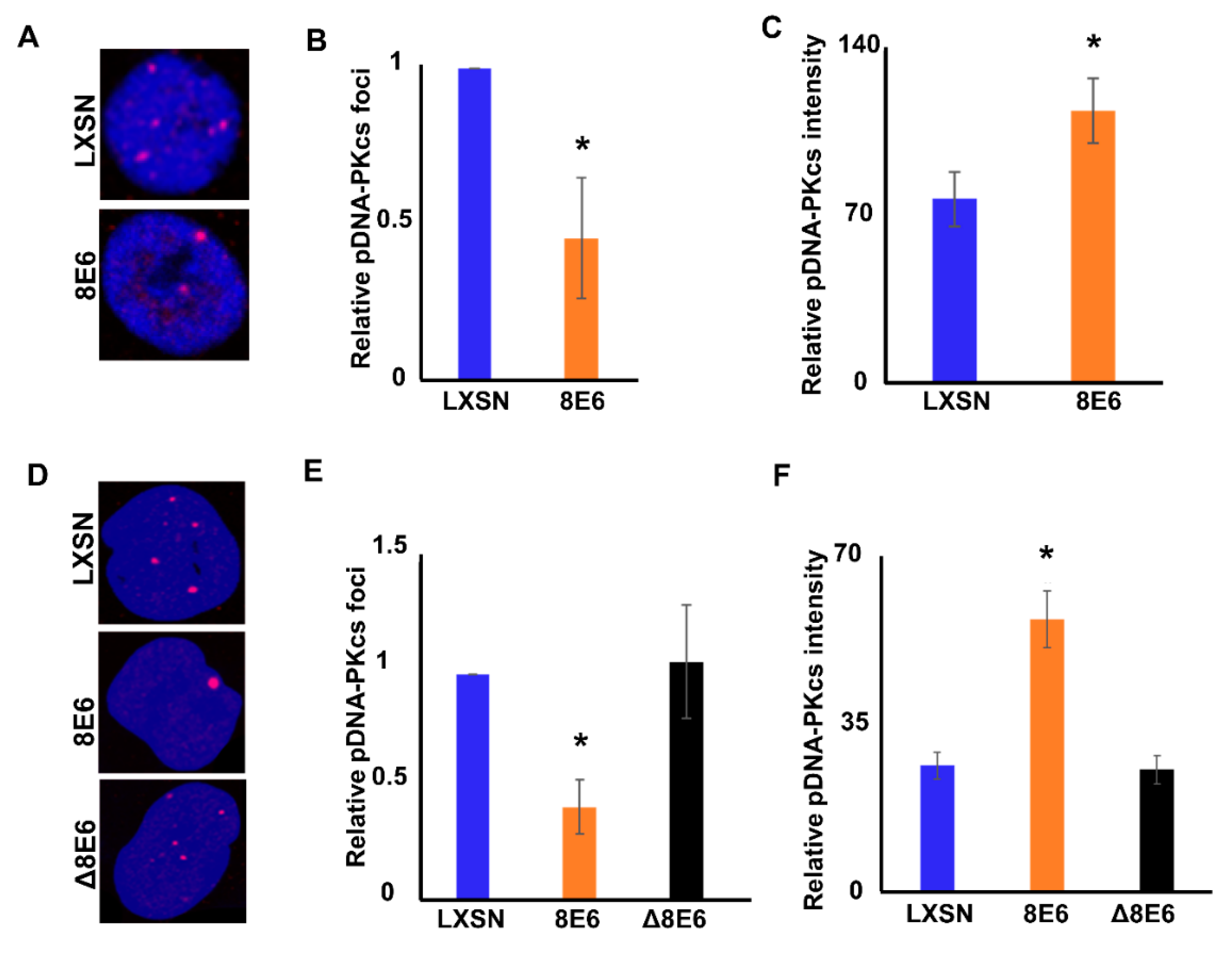
2.2. β-HPV 8E6 Attenuates DNA-PKcs Phosphorylation
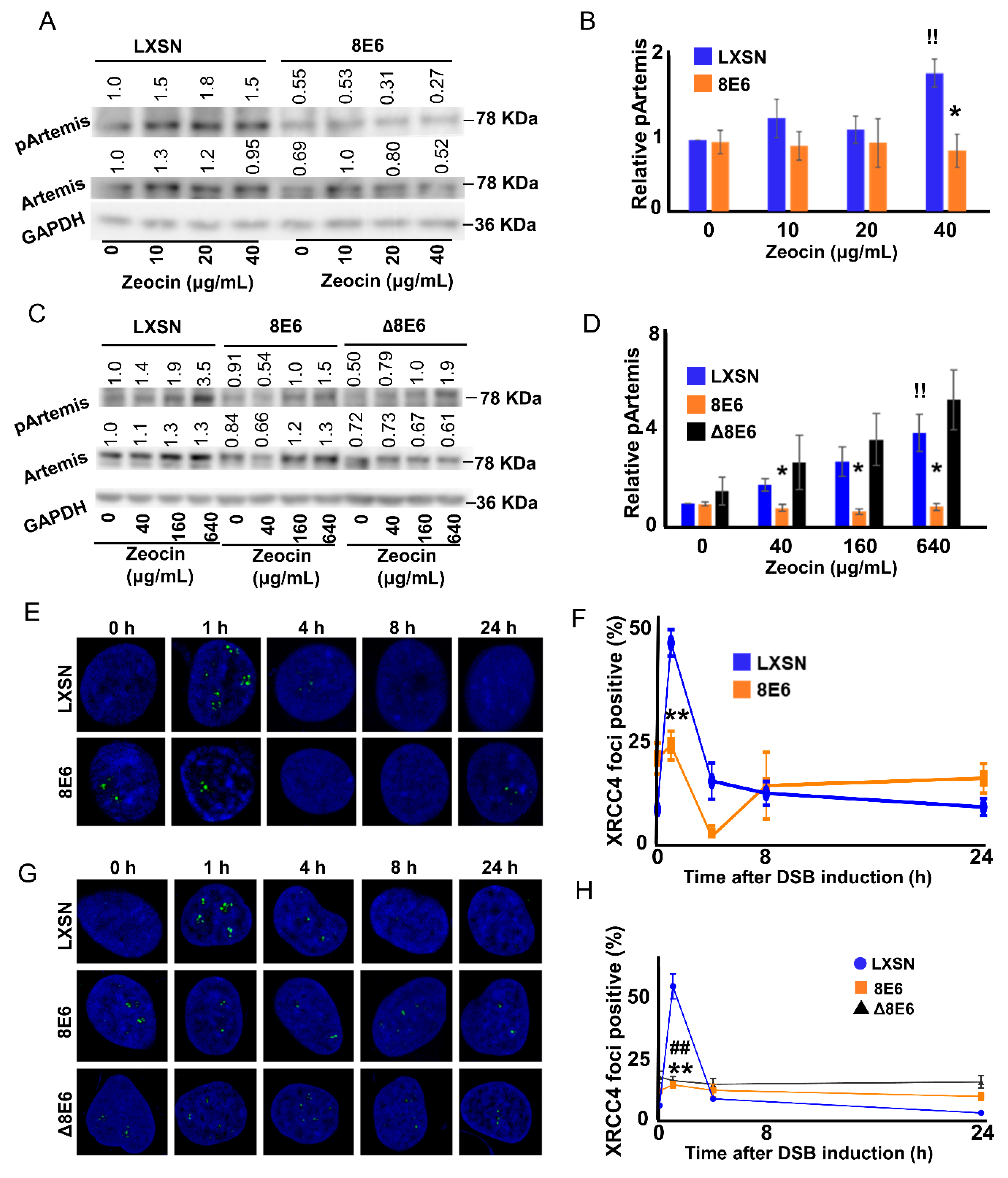
2.3. β-HPV 8E6 Attenuates DNA-PKcs-Dependent Signaling
2.4. p300 is Required for Robust DNA-PKcs Signaling
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Immunoblotting
4.3. Immunofluorescence Microscopy
4.4. End Joining Reporter Assay
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bzhalava, D.; Mühr, L.S.A.; Lagheden, C.; Ekström, J.; Forslund, O.; Dillner, J.; Hultin, E. Deep sequencing extends the diversity of human papillomaviruses in human skin. Sci. Rep. 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Cubie, H.A. Diseases associated with human papillomavirus infection. Virology 2013, 445, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Wendel, S.O.; Wallace, N.A. Loss of Genome Fidelity: Beta HPVs and the DNA Damage Response. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association: Human papillomavirus. Rev. Med. Virol. 2015, 25, 2–23. [Google Scholar] [CrossRef]
- Pfister, H.; Nürnberger, F.; Gissmann, L.; Hausen, H.Z. Characterization of a human papillomavirus from epidermodysplasia verruciformis lesions of a patient from upper-volta. Int. J. Cancer 1981, 27, 645–650. [Google Scholar] [CrossRef]
- Mudigonda, T.; Pearce, D.J.; Yentzer, B.A.; Williford, P.; Feldman, S.R. The Economic Impact of Non-Melanoma Skin Cancer: A review. J. Natl. Compr. Canc. Netw. 2010, 8, 888–896. [Google Scholar] [CrossRef]
- Kremsdorf, D.; Jablonska, S.; Favre, M.; Orth, G. Biochemical characterization of two types of human papillomaviruses associated with epidermodysplasia verruciformis. J. Virol. 1982, 43, 436–447. [Google Scholar] [CrossRef]
- Rollison, D.E.; Viarisio, D.; Amorrortu, R.P.; Gheit, T.; Tommasino, M. An Emerging Issue in Oncogenic Virology: The Role of Beta Human Papillomavirus Types in the Development of Cutaneous Squamous Cell Carcinoma. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- White, E.A.; Walther, J.; Javanbakht, H.; Howley, P.M. Genus Beta Human Papillomavirus E6 Proteins Vary in Their Effects on the Transactivation of p53 Target Genes. J. Virol. 2014, 88, 8201–8212. [Google Scholar] [CrossRef]
- Howley, P.M.; Pfister, H.J. Beta Genus Papillomaviruses and Skin Cancer. Virology 2015, 290–296. [Google Scholar] [CrossRef]
- Shanmugasundaram, S.; You, J. Targeting Persistent Human Papillomavirus Infection. Viruses 2017, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- 1Tommasino, M. HPV and skin carcinogenesis. Papillomavirus Res. 2019, 7, 129–131. [Google Scholar] [CrossRef] [PubMed]
- Viarisio, D.; Müller-Decker, K.; Accardi, R.; Robitaille, A.; Dürst, M.; Beer, K.; Jansen, L.; Flechtenmacher, C.; Bozza, M.; Harbottle, R.; et al. Beta HPV38 oncoproteins act with a hit-and-run mechanism in ultraviolet radiation-induced skin carcinogenesis in mice. PLoS Pathog. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Hufbauer, M.; Akgül, B. Molecular Mechanisms of Human Papillomavirus Induced Skin Carcinogenesis. Viruses 2017, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Strickley, J.D.; Messerschmidt, J.L.; Awad, M.E.; Li, T.; Hasegawa, T.; Ha, D.T.; Nabeta, H.W.; Bevins, P.A.; Ngo, K.H.; Asgari, M.M.; et al. Immunity to commensal papillomaviruses protects against skin cancer. Nature 2019, 575, 519–522. [Google Scholar] [CrossRef]
- Wallace, N.A.; Robinson, K.; Howie, H.L.; Galloway, D.A. β-HPV 5 and 8 E6 Disrupt Homology Dependent Double Strand Break Repair by Attenuating BRCA1 and BRCA2 Expression and Foci Formation. PLoS Pathog. 2015, 11, e1004687. [Google Scholar] [CrossRef]
- Wallace, N.A.; Gasior, S.L.; Faber, Z.J.; Howie, H.L.; Deininger, P.L.; Galloway, D.A. HPV 5 and 8 E6 expression reduces ATM protein levels and attenuates LINE-1 retrotransposition. Virology 2013, 443, 69–79. [Google Scholar] [CrossRef]
- Wallace, N.A.; Robinson, K.; Howie, H.L.; Galloway, D.A. HPV 5 and 8 E6 Abrogate ATR Activity Resulting in Increased Persistence of UVB Induced DNA Damage. PLoS Pathog. 2012, 8, e1002807. [Google Scholar] [CrossRef]
- Snow, J.A.; Murthy, V.; Dacus, D.; Hu, C.; Wallace, N.A. β-HPV 8E6 Attenuates ATM and ATR Signaling in Response to UV Damage. Pathogens 2019. [Google Scholar] [CrossRef]
- Howie, H.L.; Koop, J.I.; Weese, J.; Robinson, K.; Wipf, G.; Kim, L.; Galloway, D.A. Beta-HPV 5 and 8 E6 Promote p300 Degradation by Blocking AKT/p300 Association. PLoS Pathog. 2011, 7, e1002211. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Ahmed, K.M.; Pandita, R.K.; Singh, D.K.; Hunt, C.R.; Pandita, T.K. β1-Integrin Impacts Rad51 Stability and DNA Double-Strand Break Repair by Homologous Recombination. Mol. Cell Biol. 2018, 38. [Google Scholar] [CrossRef]
- Pierce, A.J.; Johnson, R.D.; Thompson, L.H.; Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999, 13, 2633–2638. [Google Scholar] [CrossRef]
- Bakr, A.; Oing, C.; Köcher, S.; Borgmann, K.; Dornreiter, I.; Petersen, C.; Dikomey, E.; Mansour, W.Y. Involvement of ATM in homologous recombination after end resection and RAD51 nucleofilament formation. Nucleic Acids Res. 2015, 43, 3154–3166. [Google Scholar] [CrossRef]
- Schwartz, M.; Zlotorynski, E.; Goldberg, M.; Ozeri, E.; Rahat, A.; le Sage, C.; Chen, B.P.C.; Chen, D.J.; Agami, R.; Kerem, B. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005, 19, 2715–2726. [Google Scholar] [CrossRef]
- Lieber, M.R. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End Joining Pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Weterings, E.; van Gent, D.C. The mechanism of non-homologous end-joining: A synopsis of synapsis. DNA Repair 2004, 3, 1425–1435. [Google Scholar] [CrossRef]
- Bhargava, R.; Lopezcolorado, F.W.; Tsai, L.J.; Stark, J.M. The canonical non-homologous end joining factor XLF promotes chromosomal deletion rearrangements in human cells. J. Biol. Chem. 2020, 295, 125–137. [Google Scholar] [CrossRef]
- Zhang, Y.; Hefferin, M.L.; Chen, L.; Shim, E.Y.; Tseng, H.-M.; Kwon, Y.; Sung, P.; Lee, S.E.; Tomkinson, A.E. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 2007, 14, 639–646. [Google Scholar] [CrossRef]
- Patel, A.G.; Sarkaria, J.N.; Kaufmann, S.H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. PNAS 2011, 108, 3406–3411. [Google Scholar] [CrossRef]
- Gunn, A.; Bennardo, N.; Cheng, A.; Stark, J.M. Correct End Use during End Joining of Multiple Chromosomal Double Strand Breaks Is Influenced by Repair Protein RAD50, DNA-dependent Protein Kinase DNA-PKcs, and Transcription Context. J. Biol. Chem. 2011, 286, 42470–42482. [Google Scholar] [CrossRef]
- Gupta, A.; Hunt, C.R.; Chakraborty, S.; Pandita, R.K.; Yordy, J.; Ramnarain, D.B.; Horikoshi, N.; Pandita, T.K. Role of 53BP1 in the Regulation of DNA Double-Strand Break Repair Pathway Choice. Radiat. Res. 2014, 181, 1–8. [Google Scholar] [CrossRef]
- A Chromatin-Based Signalling Mechanism Directs the Switch from Mutagenic to Error-Free Repair of DNA Double Strand Breaks: Molecular & Cellular Oncology. Available online: https://www.tandfonline.com/doi/full/10.1080/23723556.2019.1605820 (accessed on 27 November 2019).
- Sollazzo, A.; Brzozowska, B.; Cheng, L.; Lundholm, L.; Scherthan, H.; Wojcik, A. Live Dynamics of 53BP1 Foci Following Simultaneous Induction of Clustered and Dispersed DNA Damage in U2OS Cells. Int. J. Mol. Sci. 2018, 19, 519. [Google Scholar] [CrossRef]
- Daley, J.M.; Sung, P. 53BP1, BRCA1, and the Choice between Recombination and End Joining at DNA Double-Strand Breaks. Mol. Cell. Biol. 2014, 34, 1380–1388. [Google Scholar] [CrossRef]
- Dimitrova, N.; Chen, Y.-C.M.; Spector, D.L.; de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 2008, 456, 524–528. [Google Scholar] [CrossRef]
- Bunting, S.F.; Callén, E.; Wong, N.; Chen, H.-T.; Polato, F.; Gunn, A.; Bothmer, A.; Feldhahn, N.; Fernandez-Capetillo, O.; Cao, L.; et al. 53BP1 Inhibits Homologous Recombination in Brca1-Deficient Cells by Blocking Resection of DNA Breaks. Cell 2010, 141, 243–254. [Google Scholar] [CrossRef]
- Zhen, Y.; Li, S.; Zhu, Y.; Wang, X.; Zhou, X.; Zhu, L. Identification of DNA-PKcs as a primary resistance factor of salinomycin in osteosarcoma cells. Oncotarget 2016, 7, 79417–79427. [Google Scholar] [CrossRef]
- Jiang, W.; Crowe, J.L.; Liu, X.; Nakajima, S.; Wang, Y.; Li, C.; Lee, B.J.; Dubois, R.L.; Liu, C.; Yu, X.; et al. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during non-homologous end-joining. Mol. Cell 2015, 58, 172–185. [Google Scholar] [CrossRef]
- Uematsu, N.; Weterings, E.; Yano, K.; Morotomi-Yano, K.; Jakob, B.; Taucher-Scholz, G.; Mari, P.-O.; van Gent, D.C.; Chen, B.P.C.; Chen, D.J. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007, 177, 219–229. [Google Scholar] [CrossRef]
- Smith, G.C.M.; Jackson, S.P. The DNA-dependent protein kinase. Genes Dev. 1999, 13, 916–934. [Google Scholar] [CrossRef]
- Veuger, S.J.; Curtin, N.J.; Richardson, C.J.; Smith, G.C.M.; Durkacz, B.W. Radiosensitization and DNA Repair Inhibition by the Combined Use of Novel Inhibitors of DNA-dependent Protein Kinase and Poly(ADP-Ribose) Polymerase-1. Cancer Res. 2003, 63, 6008–6015. [Google Scholar]
- Zhao, Y.; Thomas, H.D.; Batey, M.A.; Cowell, I.G.; Richardson, C.J.; Griffin, R.J.; Calvert, A.H.; Newell, D.R.; Smith, G.C.M.; Curtin, N.J. Preclinical Evaluation of a Potent Novel DNA-Dependent Protein Kinase Inhibitor NU7441. Cancer Res. 2006, 66, 5354–5362. [Google Scholar] [CrossRef]
- Mamo, T.; Mladek, A.C.; Shogren, K.L.; Gustafson, C.; Gupta, S.K.; Riester, S.M.; Maran, A.; Galindo, M.; van Wijnen, A.J.; Sarkaria, J.N.; et al. Inhibiting DNA-PKCS Radiosensitizes Human Osteosarcoma Cells. Biochem. Biophys. Res. Commun. 2017, 486, 307–313. [Google Scholar] [CrossRef]
- Beucher, A.; Birraux, J.; Tchouandong, L.; Barton, O.; Shibata, A.; Conrad, S.; Goodarzi, A.A.; Krempler, A.; Jeggo, P.A.; Löbrich, M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009, 28, 3413–3427. [Google Scholar] [CrossRef]
- Soubeyrand, S.; Pope, L.; De Chasseval, R.; Gosselin, D.; Dong, F.; de Villartay, J.-P.; Haché, R.J.G. Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin. J. Mol. Biol. 2006, 358, 1200–1211. [Google Scholar] [CrossRef]
- Li, S.; Chang, H.H.; Niewolik, D.; Hedrick, M.P.; Pinkerton, A.B.; Hassig, C.A.; Schwarz, K.; Lieber, M.R. Evidence That the DNA Endonuclease ARTEMIS also Has Intrinsic 5′-Exonuclease Activity. J. Biol. Chem. 2014, 289, 7825–7834. [Google Scholar] [CrossRef]
- Mari, P.-O.; Florea, B.I.; Persengiev, S.P.; Verkaik, N.S.; Bruggenwirth, H.T.; Modesti, M.; Giglia-Mari, G.; Bezstarosti, K.; Demmers, J.A.A.; Luider, T.M.; et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA 2006, 103, 18597–18602. [Google Scholar] [CrossRef]
- Normanno, D.; Négrel, A.; de Melo, A.J.; Betzi, S.; Meek, K.; Modesti, M. Mutational phospho-mimicry reveals a regulatory role for the XRCC4 and XLF C-terminal tails in modulating DNA bridging during classical non-homologous end joining. ELife 2017, 6, e22900. [Google Scholar] [CrossRef]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2017, 19, 1–9. [Google Scholar] [CrossRef]
- Heyer, W.-D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef]
- Salmon, P.; Giovane, A.; Wasylyk, B.; Klatzmann, D. Characterization of the human CD4 gene promoter: Transcription from the CD4 gene core promoter is tissue-specific and is activated by Ets proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 7739–7743. [Google Scholar] [CrossRef] [PubMed]
- Underbrink, M.P.; Howie, H.L.; Bedard, K.M.; Koop, J.I.; Galloway, D.A. E6 proteins from multiple human betapapillomavirus types degrade Bak and protect keratinocytes from apoptosis after UVB irradiation. J. Virol. 2008, 82, 10408–10417. [Google Scholar] [CrossRef] [PubMed]
- Delacôte, F.; Deriano, L.; Lambert, S.; Bertrand, P.; Saintigny, Y.; Lopez, B.S. Chronic exposure to sublethal doses of radiation mimetic ZeocinTM selects for clones deficient in homologous recombination. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2007, 615, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Tsukuda, M.; Miyazaki, K. DNA fragmentation caused by an overdose of Zeocin. J. Biosci. Bioeng. 2013, 116, 644–646. [Google Scholar] [CrossRef]
- Davis, A.J.; Chi, L.; So, S.; Lee, K.-J.; Mori, E.; Fattah, K.; Yang, J.; Chen, D.J. BRCA1 modulates the autophosphorylation status of DNA-PKcs in S phase of the cell cycle. Nucleic Acids Res. 2014, 42, 11487–11501. [Google Scholar] [CrossRef]
- Greinert, R.; Volkmer, B.; Henning, S.; Breitbart, E.W.; Greulich, K.O.; Cardoso, M.C.; Rapp, A. UVA-induced DNA double-strand breaks result from the repair of clustered oxidative DNA damages. Nucleic Acids Res. 2012, 40, 10263–10273. [Google Scholar] [CrossRef]
- Lee, C.-S.; Lee, K.; Legube, G.; Haber, J.E. Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Nat. Struct. Mol. Biol. 2014, 21, 103–109. [Google Scholar] [CrossRef]
- Polo, S.E.; Jackson, S.P. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011, 25, 409–433. [Google Scholar] [CrossRef]
- Murthy, V.; Dacus, D.; Gamez, M.; Hu, C.; Wendel, S.O.; Snow, J.; Kahn, A.; Walterhouse, S.H.; Wallace, N.A. Characterizing DNA Repair Processes at Transient and Long-lasting Double-strand DNA Breaks by Immunofluorescence Microscopy. J. Vis. Exp. 2018, e57653. [Google Scholar] [CrossRef]
- Drouet, J.; Delteil, C.; Lefrançois, J.; Concannon, P.; Salles, B.; Calsou, P. DNA-dependent Protein Kinase and XRCC4-DNA Ligase IV Mobilization in the Cell in Response to DNA Double Strand Breaks. J. Biol. Chem. 2005, 280, 7060–7069. [Google Scholar] [CrossRef]
- Koike, M.; Yutoku, Y.; Koike, A. Cloning, localization and focus formation at DNA damage sites of canine XRCC4. J. Vet. Med. Sci. 2016, 78, 1865–1871. [Google Scholar] [CrossRef] [PubMed]
- Collis, S.J.; DeWeese, T.L.; Jeggo, P.A.; Parker, A.R. The life and death of DNA-PK. Oncogene 2005, 24, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.J.; Chen, B.P.C.; Chen, D.J. DNA-PK: A dynamic enzyme in a versatile DSB repair pathway. DNA Repair 2014, 17, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Ghezraoui, H.; Piganeau, M.; Renouf, B.; Renaud, J.-B.; Sallmyr, A.; Ruis, B.; Oh, S.; Tomkinson, A.; Hendrickson, E.A.; Giovannangeli, C.; et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 2014, 55, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Iyer, N.G.; Chin, S.-F.; Ozdag, H.; Daigo, Y.; Hu, D.-E.; Cariati, M.; Brindle, K.; Aparicio, S.; Caldas, C. p300 regulates p53-dependent apoptosis after DNA damage in colorectal cancer cells by modulation of PUMA/p21 levels. Proc. Natl. Acad. Sci. USA 2004, 101, 7386–7391. [Google Scholar] [CrossRef] [PubMed]
- Francis, D.B.; Kozlov, M.; Chavez, J.; Chu, J.; Malu, S.; Hanna, M.; Cortes, P. DNA Ligase IV regulates XRCC4 nuclear localization. DNA Repair 2014, 21, 36–42. [Google Scholar] [CrossRef]
- Goodman, R.H.; Smolik, S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000, 14, 1553–1577. [Google Scholar] [CrossRef]
- Chen, J.; Halappanavar, S.S.; St-Germain, J.R.; Tsang, B.K.; Li, Q. Role of Akt/protein kinase B in the activity of transcriptional coactivator p300. Cell. Mol. Life Sci. 2004, 61, 1675–1683. [Google Scholar] [CrossRef]
- Ogiwara, H.; Ui, A.; Otsuka, A.; Satoh, H.; Yokomi, I.; Nakajima, S.; Yasui, A.; Yokota, J.; Kohno, T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 2011, 30, 2135–2146. [Google Scholar] [CrossRef]
- Mori, E.; Davis, A.J.; Hasegawa, M.; Chen, D.J. Lysines 3241 and 3260 of DNA-PKcs are important for genomic stability and radioresistance. Biochem. Biophys. Res. Commun. 2016, 477, 235–240. [Google Scholar] [CrossRef]
- Jette, N.; Lees-Miller, S.P. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 2015, 117, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Eccles, L.J.; Bell, A.C.; Powell, S.N. Inhibition of non-homologous end joining in Fanconi Anemia cells results in rescue of survival after interstrand crosslinks but sensitization to replication associated double-strand breaks. DNA Repair 2018, 64, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zhang, T.; Ren, Y.; Wang, Z.; Ling, C.C.; He, F.; Li, G.C.; Wang, C.; Wen, B. Inhibiting DNA-PKcs in a non-homologous end-joining pathway in response to DNA double-strand breaks. Oncotarget 2017, 8, 22662–22673. [Google Scholar] [CrossRef] [PubMed]
- Meyers, J.M.; Uberoi, A.; Grace, M.; Lambert, P.F.; Munger, K. Cutaneous HPV8 and MmuPV1 E6 Proteins Target the NOTCH and TGF-β Tumor Suppressors to Inhibit Differentiation and Sustain Keratinocyte Proliferation. PLoS Pathog. 2017, 13, e1006171. [Google Scholar] [CrossRef] [PubMed]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, C.; Bugbee, T.; Gamez, M.; Wallace, N.A. Beta Human Papillomavirus 8E6 Attenuates Non-Homologous End Joining by Hindering DNA-PKcs Activity. Cancers 2020, 12, 2356. https://doi.org/10.3390/cancers12092356
Hu C, Bugbee T, Gamez M, Wallace NA. Beta Human Papillomavirus 8E6 Attenuates Non-Homologous End Joining by Hindering DNA-PKcs Activity. Cancers. 2020; 12(9):2356. https://doi.org/10.3390/cancers12092356
Chicago/Turabian StyleHu, Changkun, Taylor Bugbee, Monica Gamez, and Nicholas A. Wallace. 2020. "Beta Human Papillomavirus 8E6 Attenuates Non-Homologous End Joining by Hindering DNA-PKcs Activity" Cancers 12, no. 9: 2356. https://doi.org/10.3390/cancers12092356
APA StyleHu, C., Bugbee, T., Gamez, M., & Wallace, N. A. (2020). Beta Human Papillomavirus 8E6 Attenuates Non-Homologous End Joining by Hindering DNA-PKcs Activity. Cancers, 12(9), 2356. https://doi.org/10.3390/cancers12092356





