Expression Profiling of Extracellular Matrix Genes Reveals Global and Entity-Specific Characteristics in Adenoid Cystic, Mucoepidermoid and Salivary Duct Carcinomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
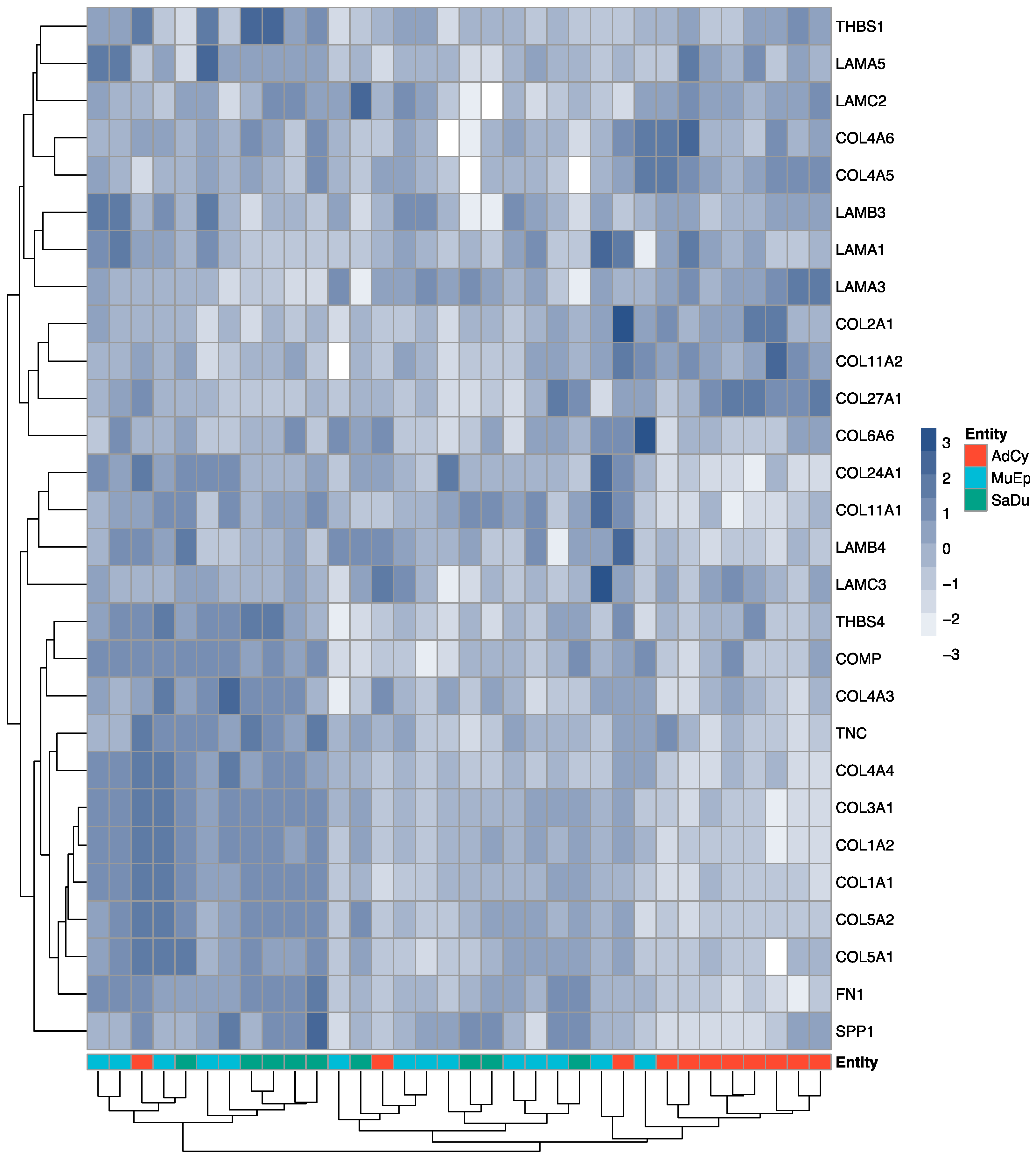
2.2. Exploratory Data Analysis Revealed a Distinct ECM Profile of Adenoid Cystic Carcinomas
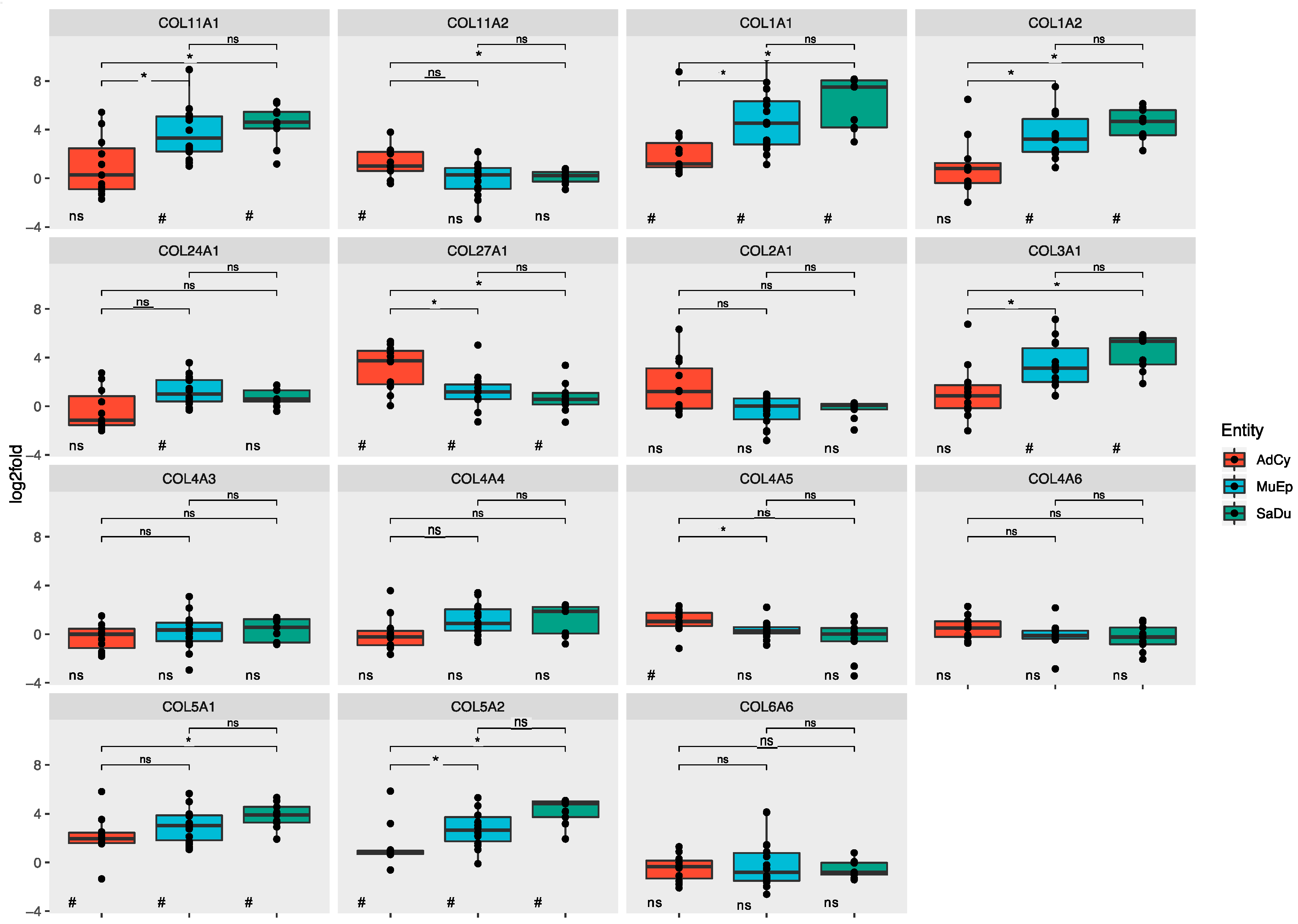
2.3. The ECM of Salivary Duct and Mucoepidermoid Carcinomas Shared a Set of Mutually Overexpressed Genes
2.3.1. Salivary Duct Carcinoma
2.3.2. Mucoepidermoid Carcinoma
2.3.3. Adenoid Cystic Carcinoma
2.4. In Situ Validations Revealed ECM Production by Adenoid Cystic Tumour Cells
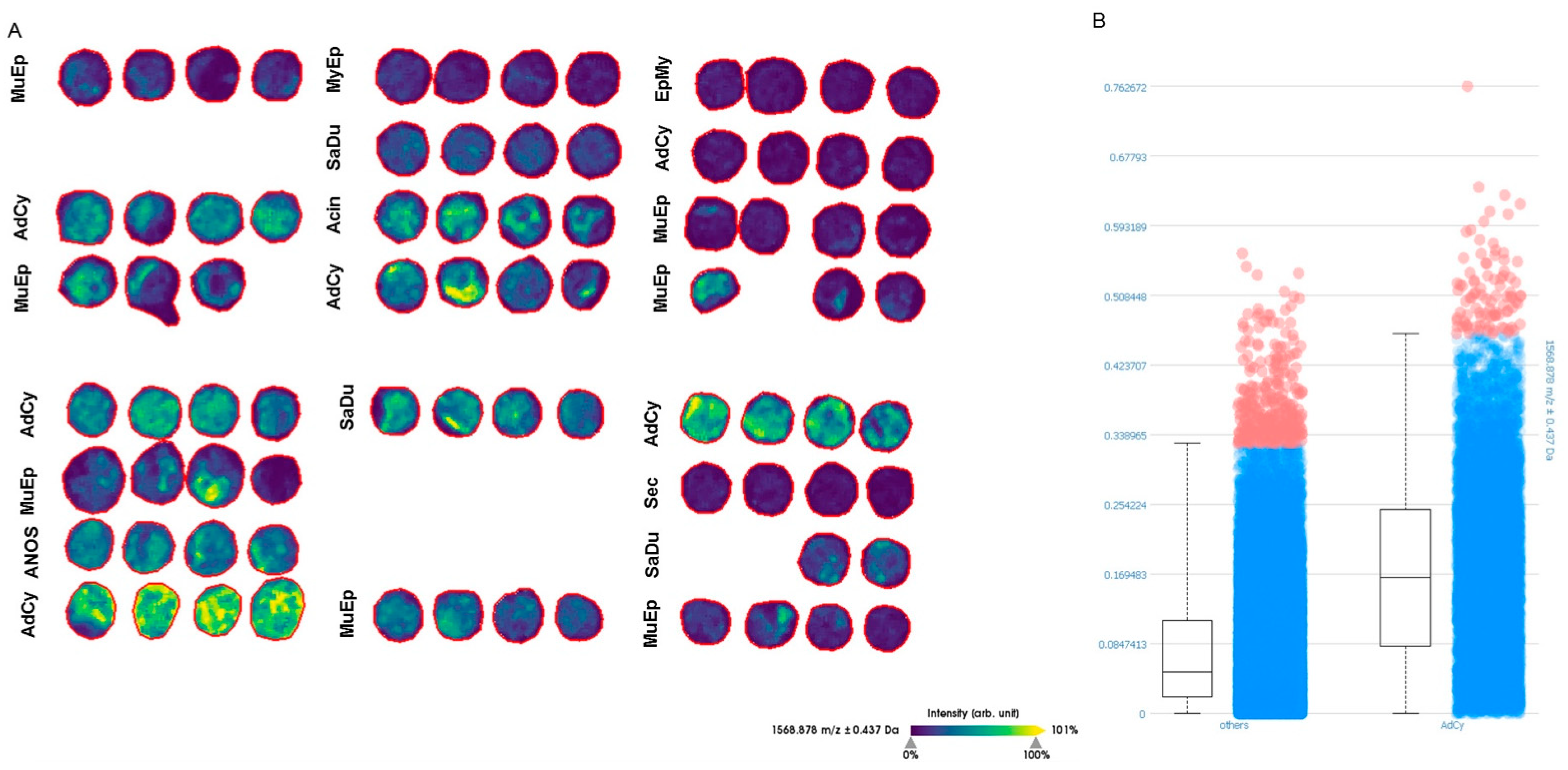
2.5. MALDI-TOF-MS-Imaging (MSI) Confirmed Specific Upregulation of COL27A1 in AdCy at the Protein Level
3. Discussion
4. Materials and Methods
4.1. Patient Characteristics
4.2. Immunohistochemistry
4.3. RNA In Situ Hybridisation (RNA-ISH)
4.4. RNA Processing
4.5. Data Processing
4.6. Statistical Analysis
4.7. MALDI-TOF-MS-Imaging (MSI)
4.8. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Congress Presentation
References
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerarduzzi, C.; Hartmann, U.; Leask, A.; Drobetsky, E. The Matrix Revolution: Matricellular Proteins and Restructuring of the Cancer Microenvironment. Cancer Res. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hynes, R.O.; Naba, A. Overview of the Matrisome—An Inventory of Extracellular Matrix Constituents and Functions. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafaeva, M.; Erler, J.T. Framing cancer progression: Influence of the organ- and tumour-specific matrisome. FEBS J. 2020, 287, 1454–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toss, M.S.; Miligy, I.M.; Gorringe, K.L.; Aleskandarany, M.A.; Alkawaz, A.; Mittal, K.; Aneja, R.; Ellis, I.O.; Green, A.R.; Rakha, E.A. Collagen (XI) alpha-1 chain is an independent prognostic factor in breast ductal carcinoma in situ. Mod. Pathol. 2019, 32, 1460–1472. [Google Scholar] [CrossRef]
- Leppänen, J.; Bogdanoff, S.; Lehenkari, P.P.; Saarnio, J.; Kauppila, J.H.; Karttunen, T.J.; Huhta, H.; Helminen, O. Tenascin-C and fibronectin in normal esophageal mucosa, Barrett’s esophagus, dysplasia and adenocarcinoma. Oncotarget 2017, 8, 66865–66877. [Google Scholar] [CrossRef] [Green Version]
- DeFilippis, R.A.; Chang, H.; Dumont, N.; Rabban, J.T.; Chen, Y.-Y.; Fontenay, G.V.; Berman, H.K.; Gauthier, M.L.; Zhao, J.; Hu, D.; et al. CD36 Repression Activates a Multicellular Stromal Program Shared by High Mammographic Density and Tumor Tissues. Cancer Discov. 2012, 2, 826–839. [Google Scholar] [CrossRef] [Green Version]
- Fordyce, C.A.; Patten, K.T.; Fessenden, T.B.; DeFilippis, R.; Hwang, E.S.; Zhao, J.; Tlsty, T.D. Cell-extrinsic consequences of epithelial stress: Activation of protumorigenic tissue phenotypes. Breast Cancer Res. BCR 2012, 14, R155. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Sun, B.; Li, X.; Zhang, L.; Niu, Y.; Xiao, C.; Ning, L.; Fang, Z.; Wang, Y.; Zhang, L.; et al. Differentially expressed genes between primary cancer and paired lymph node metastases predict clinical outcome of node-positive breast cancer patients. Breast Cancer Res. Treat. 2007, 103, 319–329. [Google Scholar] [CrossRef]
- Brechbuhl, H.M.; Barrett, A.S.; Kopin, E.; Hagen, J.C.; Han, A.L.; Gillen, A.E.; Finlay-Schultz, J.; Cittelly, D.M.; Owens, P.; Horwitz, K.B.; et al. Fibroblast subtypes define a metastatic matrisome in breast cancer. JCI Insight 5 2020. [Google Scholar] [CrossRef] [Green Version]
- Oskarsson, T.; Acharyya, S.; Zhang, X.H.-F.; Vanharanta, S.; Tavazoie, S.F.; Morris, P.G.; Downey, R.J.; Manova-Todorova, K.; Brogi, E.; Massagué, J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 2011, 17, 867–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naba, A.; Clauser, K.R.; Lamar, J.M.; Carr, S.A.; Hynes, R.O. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, C.; Öhlund, D.; Rickelt, S.; Lidström, T.; Huang, Y.; Hao, L.; Zhao, R.T.; Franklin, O.; Bhatia, S.N.; Tuveson, D.A.; et al. Cancer Cell–Derived Matrisome Proteins Promote Metastasis in Pancreatic Ductal Adenocarcinoma. Cancer Res. 2020, 80, 1461–1474. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Liang, C.; Chen, M.; Su, W. Association between tumor-stroma ratio and prognosis in solid tumor patients: A systematic review and meta-analysis. Oncotarget 2016, 7, 68954–68965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiori, M.E.; Di Franco, S.; Villanova, L.; Bianca, P.; Stassi, G.; De Maria, R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol. Cancer 2019, 18, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, A.J.; Cortes, E.; Lachowski, D.; Cheung, B.C.H.; Karim, S.A.; Morton, J.P.; Del Río Hernández, A. Matrix stiffness induces epithelial-mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis 2017, 6, e352. [Google Scholar] [CrossRef] [Green Version]
- Diop-Frimpong, B.; Chauhan, V.P.; Krane, S.; Boucher, Y.; Jain, R.K. Losartan inhibits collagen I synthesis and improves the distribution and efficacy of nanotherapeutics in tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 2909–2914. [Google Scholar] [CrossRef] [Green Version]
- LIM, S.B.; TAN, S.J.; LIM, W.-T.; LIM, C.T. An extracellular matrix-related prognostic and predictive indicator for early-stage non-small cell lung cancer. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Lim, S.B.; Chua, M.L.K.; Yeong, J.P.S.; Tan, S.J.; Lim, W.-T.; Lim, C.T. Pan-cancer analysis connects tumor matrisome to immune response. NPJ Precis. Oncol. 2019, 3. [Google Scholar] [CrossRef] [Green Version]
- Pearce, O.M.T.; Delaine-Smith, R.; Maniati, E.; Nichols, S.; Wang, J.; Böhm, S.; Rajeeve, V.; Ullah, D.; Chakravarty, P.; Jones, R.R.; et al. Deconstruction of a metastatic tumor microenvironment reveals a common matrix response in human cancers. Cancer Discov. 2018, 8, 304–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuzhalin, A.E.; Urbonas, T.; Silva, M.A.; Muschel, R.J.; Gordon-Weeks, A.N. A core matrisome gene signature predicts cancer outcome. Br. J. Cancer 2018, 118, 435–440. [Google Scholar] [CrossRef] [Green Version]
- Luukkaa, H.; Klemi, P.; Hirsimäki, P.; Vahlberg, T.; Kivisaari, A.; Kähäri, V.-M.; Grénman, R. Matrix metalloproteinase (MMP)-7 in salivary gland cancer. Acta Oncol. 2010, 49, 85–90. [Google Scholar] [CrossRef]
- Souza Freitas, V.; Dos Santos, J.N.; de Andrade Santos, P.P.; Nonaka, C.F.W.; Pereira Pinto, L.; de Souza, L.B. Expression of matrix metalloproteinases (MMPs-2, -7, -9, and -26) and tissue inhibitors of metalloproteinases (TIMPs-1 and -2) in pleomorphic adenomas and adenoid cystic carcinomas. Eur. Arch. Oto-Rhino-Laryngol. 2018, 275, 3075–3082. [Google Scholar] [CrossRef] [PubMed]
- Raitz, R.; Martins, M.D.; Araújo, V.C. A study of the extracellular matrix in salivary gland tumors. J. Oral Pathol. Med. 2003, 32, 290–296. [Google Scholar] [CrossRef]
- El-Naggar, A.K.; Chan, J.K.C.; Grandis, J.R.; Takata, T.; Slootweg, P.J. WHO Classification of Head and Neck Tumours; WHO: Geneva, Switzerland, 2017; ISBN 978-92-832-2438-9. [Google Scholar]
- Birkeland, A.C.; Foltin, S.K.; Michmerhuizen, N.L.; Hoesli, R.C.; Rosko, A.J.; Byrd, S.; Yanik, M.; Nor, J.E.; Bradford, C.R.; Prince, M.E.; et al. Correlation of crtc1/3-maml2 fusion status, grade and survival in mucoepidermoid carcinoma. Oral Oncol. 2017, 68, 5–8. [Google Scholar] [CrossRef] [Green Version]
- Seethala, R.R. An Update on Grading of Salivary Gland Carcinomas. Head Neck Pathol. 2009, 3, 69–77. [Google Scholar] [CrossRef] [Green Version]
- Brayer, K.J.; Frerich, C.A.; Kang, H.; Ness, S.A. Recurrent Fusions in MYB and MYBL1 Define a Common, Transcription Factor-Driven Oncogenic Pathway in Salivary Gland Adenoid Cystic Carcinoma. Cancer Discov. 2016, 6, 176. [Google Scholar] [CrossRef] [Green Version]
- Fujii, K.; Murase, T.; Beppu, S.; Saida, K.; Takino, H.; Masaki, A.; Ijichi, K.; Kusafuka, K.; Iida, Y.; Onitsuka, T.; et al. MYB, MYBL1, MYBL2 and NFIB gene alterations and MYC overexpression in salivary gland adenoid cystic carcinoma. Histopathology 2017, 71, 823–834. [Google Scholar] [CrossRef]
- Togashi, Y.; Dobashi, A.; Sakata, S.; Sato, Y.; Baba, S.; Seto, A.; Mitani, H.; Kawabata, K.; Takeuchi, K. MYB and MYBL1 in adenoid cystic carcinoma: Diversity in the mode of genomic rearrangement and transcripts. Mod. Pathol. 2018, 31, 934–946. [Google Scholar] [CrossRef]
- Wang, K.; Russell, J.S.; McDermott, J.D.; Elvin, J.A.; Khaira, D.; Johnson, A.; Jennings, T.A.; Ali, S.M.; Murray, M.; Marshall, C.; et al. Profiling of 149 Salivary Duct Carcinomas, Carcinoma Ex Pleomorphic Adenomas, and Adenocarcinomas, Not Otherwise Specified Reveals Actionable Genomic Alterations. Clin. Cancer Res. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalin, M.G.; Desrichard, A.; Katabi, N.; Makarov, V.; Walsh, L.A.; Lee, K.-W.; Wang, Q.; Armenia, J.; West, L.; Dogan, S.; et al. Comprehensive Molecular Characterization of Salivary Duct Carcinoma Reveals Actionable Targets and Similarity to Apocrine Breast Cancer. Clin. Cancer Res. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alame, M.; Cornillot, E.; Cacheux, V.; Tosato, G.; Four, M.; Oliveira, L.D.; Gofflot, S.; Delvenne, P.; Turtoi, E.; Cabello-Aguilar, S.; et al. The molecular landscape and microenvironment of salivary duct carcinoma reveal new therapeutic opportunities. bioRxiv 2019, 810028. [Google Scholar] [CrossRef] [PubMed]
- Locati, L.D.; Perrone, F.; Cortelazzi, B.; Lo Vullo, S.; Bossi, P.; Dagrada, G.; Quattrone, P.; Bergamini, C.; Potepan, P.; Civelli, E.; et al. Clinical activity of androgen deprivation therapy in patients with metastatic/relapsed androgen receptor-positive salivary gland cancers. Head Neck 2016, 38, 724–731. [Google Scholar] [CrossRef]
- Takahashi, H.; Tada, Y.; Saotome, T.; Akazawa, K.; Ojiri, H.; Fushimi, C.; Masubuchi, T.; Matsuki, T.; Tani, K.; Osamura, R.Y.; et al. Phase II Trial of Trastuzumab and Docetaxel in Patients With Human Epidermal Growth Factor Receptor 2-Positive Salivary Duct Carcinoma. J. Clin. Oncol. 2019, 37, 125–134. [Google Scholar] [CrossRef] [PubMed]
- García-Pravia, C.; Galván, J.A.; Gutiérrez-Corral, N.; Solar-García, L.; García-Pérez, E.; García-Ocaña, M.; Amo-Iribarren, J.D.; Menéndez-Rodríguez, P.; García-García, J.; de los Toyos, J.R.; et al. Overexpression of COL11A1 by Cancer-Associated Fibroblasts: Clinical Relevance of a Stromal Marker in Pancreatic Cancer. PLoS ONE 2013, 8, e78327. [Google Scholar] [CrossRef]
- Jia, D.; Liu, Z.; Deng, N.; Tan, T.Z.; Huang, R.Y.-J.; Taylor-Harding, B.; Cheon, D.-J.; Lawrenson, K.; Wiedemeyer, W.R.; Walts, A.E.; et al. A COL11A1-correlated pan-cancer gene signature of activated fibroblasts for the prioritization of therapeutic targets. Cancer Lett. 2016, 382, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Farmer, P.; Bonnefoi, H.; Anderle, P.; Cameron, D.; Wirapati, P.; Wirapati, P.; Becette, V.; André, S.; Piccart, M.; Campone, M.; et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat. Med. 2009, 15, 68–74. [Google Scholar] [CrossRef]
- Boguslawska, J.; Kedzierska, H.; Poplawski, P.; Rybicka, B.; Tanski, Z.; Piekielko-Witkowska, A. Expression of Genes Involved in Cellular Adhesion and Extracellular Matrix Remodeling Correlates with Poor Survival of Patients with Renal Cancer. J. Urol. 2016, 195, 1892–1902. [Google Scholar] [CrossRef]
- Cheon, D.-J.; Tong, Y.; Sim, M.-S.; Dering, J.; Berel, D.; Cui, X.; Lester, J.; Beach, J.A.; Tighiouart, M.; Walts, A.E.; et al. A collagen-remodeling gene signature regulated by TGF-β signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin. Cancer Res. 2014, 20, 711–723. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-H.; Chang, T.-H.; Huang, Y.-F.; Huang, H.-D.; Chou, C.-Y. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene 2014, 33, 3432–3440. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-H.; Chang, T.-H.; Huang, Y.-F.; Chen, C.-C.; Chou, C.-Y. COL11A1 confers chemoresistance on ovarian cancer cells through the activation of Akt/c/EBPβ pathway and PDK1 stabilization. Oncotarget 2015, 6, 23748–23763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Wu, J.; Zhang, J.; Yang, Z.; Jin, W.; Li, Y.; Jin, L.; Yin, L.; Liu, H.; Wang, Z. Integrated bioinformatics analysis of key genes involved in progress of colon cancer. Mol. Genet. Genom. Med. 2019, 7, e00588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Du, J.; Gu, J.; Jin, L.; Pu, Y.; Fei, B. A 65-gene signature for prognostic prediction in colon adenocarcinoma. Int. J. Mol. Med. 2018, 41, 2021–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raglow, Z.; Thomas, S.M. Tumor matrix protein collagen XIα1 in cancer. Cancer Lett. 2015, 357, 448–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilkes, D.M.; Semenza, G.L.; Wirtz, D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat. Rev. Cancer 2014, 14, 430–439. [Google Scholar] [CrossRef] [Green Version]
- Spivak-Kroizman, T.R.; Hostetter, G.; Posner, R.; Aziz, M.; Hu, C.; Demeure, M.J.; Von Hoff, D.; Hingorani, S.R.; Palculict, T.B.; Izzo, J.; et al. Hypoxia triggers Hedgehog-mediated tumor stromal interactions in pancreatic cancer. Cancer Res. 2013, 73, 3235–3247. [Google Scholar] [CrossRef] [Green Version]
- Lampi, M.C.; Reinhart-King, C.A. Targeting extracellular matrix stiffness to attenuate disease: From molecular mechanisms to clinical trials. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, H.; Miyagi, Y.; Kikkawa, Y.; Yamanaka, N.; Yasumitsu, H.; Misugi, K.; Miyazaki, K. Differential Expression of Laminin-5/Ladsin Subunits in Human Tissues and Cancer Cell Lines and Their Induction by Tumor Promoter and Growth Factors. J. Biochem. (Tokyo) 1996, 120, 1196–1202. [Google Scholar] [CrossRef]
- Waterman, E.A.; Sakai, N.; Nguyen, N.T.; Horst, B.A.J.; Veitch, D.P.; Dey, C.N.; Ortiz-Urda, S.; Khavari, P.A.; Marinkovich, M.P. A Laminin-Collagen Complex Drives Human Epidermal Carcinogenesis through Phosphoinositol-3-Kinase Activation. Cancer Res. 2007, 67, 4264–4270. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, A.; Sawai, H.; Takahashi, H.; Ochi, N.; Matsuo, Y.; Funahashi, H.; Sato, M.; Okada, Y.; Takeyama, H.; Manabe, T. Stem Cell Factor/c-kit Receptor Signaling Enhances the Proliferation and Invasion of Colorectal Cancer Cells Through the PI3K/Akt Pathway. Dig. Dis. Sci. 2007, 52, 2292–2300. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Iyama, K.I.; Hirashima, K.; Nagai, Y.; Yoshida, N.; Hayashi, N.; Miyanari, N.; Baba, H. Laminin-332 promotes the invasion of oesophageal squamous cell carcinoma via PI3K activation. Br. J. Cancer 2008, 98, 974–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, P.M.; Ziogas, A.; Markham, E.M.; Cantillep, A.S.; Yan, R.; Anton-Culver, H. Laminin 332 expression and prognosis in breast cancer. Hum. Pathol. 2018, 82, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, W.; Wei, J.; Zhou, D.; Zhao, X.; Song, W.; Sun, Q.; Huang, P.; Zheng, S. Overexpression of β3 Chains of Laminin-332 is Associated With Clinicopathologic Features and Decreased Survival in Patients With Pancreatic Adenocarcinoma. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 516–521. [Google Scholar] [CrossRef] [Green Version]
- Giannelli, G.; Fransvea, E.; Bergamini, C.; Marinosci, F.; Antonaci, S. Laminin-5 Chains Are Expressed Differentially in Metastatic and Nonmetastatic Hepatocellular Carcinoma. Clin. Cancer Res. 2003, 9, 3684–3691. [Google Scholar]
- Takahashi, S.; Hasebe, T.; Oda, T.; Sasaki, S.; Kinoshita, T.; Konishi, M.; Ochiai, T.; Ochiai, A. Cytoplasmic expression of laminin γ2 chain correlates with postoperative hepatic metastasis and poor prognosis in patients with pancreatic ductal adenocarcinoma. Cancer 2002, 94, 1894–1901. [Google Scholar] [CrossRef]
- Plumb, D.A.; Dhir, V.; Mironov, A.; Poulsom, R.; Kadler, K.E.; Thornton, D.J.; Briggs, M.D.; Boot-Handford, R.P. Collagen XXVII is developmentally-regulated and forms thin fibrillar structures distinct from those of classical vertebrate fibrillar collagens. J. Biol. Chem. 2007, 282, 12791–12795. [Google Scholar] [CrossRef] [Green Version]
- Hjorten, R.; Hansen, U.; Underwood, R.A.; Telfer, H.E.; Fernandes, R.J.; Krakow, D.; Sebald, E.; Wachsmann-Hogiu, S.; Bruckner, P.; Jacquet, R.; et al. Type XXVII collagen at the transition of cartilage to bone during skeletogenesis. Bone 2007, 41, 535–542. [Google Scholar] [CrossRef] [Green Version]
- Gariballa, N.; Ben-Mahmoud, A.; Komara, M.; Al-Shamsi, A.M.; John, A.; Ali, B.R.; Al-Gazali, L. A novel aberrant splice site mutation in COL27A1 is responsible for Steel syndrome and extension of the phenotype to include hearing loss. Am. J. Med. Genet. A 2017, 173, 1257–1263. [Google Scholar] [CrossRef]
- Gonzaga-Jauregui, C.; Gamble, C.N.; Yuan, B.; Penney, S.; Jhangiani, S.; Muzny, D.M.; Gibbs, R.A.; Lupski, J.R.; Hecht, J.T. Mutations in COL27A1 cause Steel syndrome and suggest a founder mutation effect in the Puerto Rican population. Eur. J. Hum. Genet. 2015, 23, 342–346. [Google Scholar] [CrossRef] [Green Version]
- Vargas, A.C.; McCart Reed, A.E.; Waddell, N.; Lane, A.; Reid, L.E.; Smart, C.E.; Cocciardi, S.; da Silva, L.; Song, S.; Chenevix-Trench, G.; et al. Gene expression profiling of tumour epithelial and stromal compartments during breast cancer progression. Breast Cancer Res. Treat. 2012, 135, 153–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirasuna, K.; Saka, M.; Hayashido, Y.; Yoshioka, H.; Sugiura, T.; Matsuya, T. Extracellular Matrix Production and Degradation by Adenoid Cystic Carcinoma Cells: Participation of Plasminogen Activator and Its Inhibitor in Matrix Degradation. Cancer Res. 1993, 53, 147–152. [Google Scholar] [PubMed]
- Sternlicht, M.D.; Safarians, S.; Rivera, S.P.; Barsky, S.H. Characterizations of the extracellular matrix and proteinase inhibitor content of human myoepithelial tumors. Lab. Investig. J. Tech. Methods Pathol. 1996, 74, 781–796. [Google Scholar]
- Weide, B.; Eigentler, T.; Catania, C.; Ascierto, P.A.; Cascinu, S.; Becker, J.C.; Hauschild, A.; Romanini, A.; Danielli, R.; Dummer, R.; et al. A phase II study of the L19IL2 immunocytokine in combination with dacarbazine in advanced metastatic melanoma patients. Cancer Immunol. Immunother. CII 2019, 68, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Lingasamy, P.; Tobi, A.; Kurm, K.; Kopanchuk, S.; Sudakov, A.; Salumäe, M.; Rätsep, T.; Asser, T.; Bjerkvig, R.; Teesalu, T. Tumor-penetrating peptide for systemic targeting of Tenascin-C. Sci. Rep. 2020, 10, 5809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daubon, T.; Léon, C.; Clarke, K.; Andrique, L.; Salabert, L.; Darbo, E.; Pineau, R.; Guérit, S.; Maitre, M.; Dedieu, S.; et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat. Commun. 2019, 10, 1146. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Wong, J.P.C.; Kwok, H.F. Osteopontin—a promising biomarker for cancer therapy. J. Cancer 2017, 8, 2173–2183. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zhou, L.; Li, D.; Andl, T.; Zhang, Y. Cancer-Associated Fibroblasts Build and Secure the Tumor Microenvironment. Front. Cell Dev. Biol. 2019, 7, 60. [Google Scholar] [CrossRef] [Green Version]
- Johannsen, M.; Spitaleri, G.; Curigliano, G.; Roigas, J.; Weikert, S.; Kempkensteffen, C.; Roemer, A.; Kloeters, C.; Rogalla, P.; Pecher, G.; et al. The tumour-targeting human L19-IL2 immunocytokine: Preclinical safety studies, phase I clinical trial in patients with solid tumours and expansion into patients with advanced renal cell carcinoma. Eur. J. Cancer 2010, 46, 2926–2935. [Google Scholar] [CrossRef]
- Ross, J.S.; Gay, L.M.; Wang, K.; Vergilio, J.-A.; Suh, J.; Ramkissoon, S.; Somerset, H.; Johnson, J.M.; Russell, J.; Ali, S.; et al. Comprehensive genomic profiles of metastatic and relapsed salivary gland carcinomas are associated with tumor type and reveal new routes to targeted therapies. Ann. Oncol. 2017, 28, 2539–2546. [Google Scholar] [CrossRef]
- Senthebane, D.A.; Rowe, A.; Thomford, N.E.; Shipanga, H.; Munro, D.; Al Mazeedi, M.A.M.; Almazyadi, H.A.M.; Kallmeyer, K.; Dandara, C.; Pepper, M.S.; et al. The Role of Tumor Microenvironment in Chemoresistance: To Survive, Keep Your Enemies Closer. Int. J. Mol. Sci. 2017, 18, 1586. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.E.; Wo, J.Y.; Ryan, D.P.; Clark, J.W.; Jiang, W.; Yeap, B.Y.; Drapek, L.C.; Ly, L.; Baglini, C.V.; Blaszkowsky, L.S.; et al. Total Neoadjuvant Therapy With FOLFIRINOX in Combination With Losartan Followed by Chemoradiotherapy for Locally Advanced Pancreatic Cancer: A Phase 2 Clinical Trial. JAMA Oncol. 2019, 5, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, X.; Lu, J.; Salfenmoser, M.; Wirsik, N.M.; Schleussner, N.; Imle, A.; Valls, A.F.; Radhakrishnan, P.; Liang, J.; et al. Reduction of Liver Metastasis Stiffness Improves Response to Bevacizumab in Metastatic Colorectal Cancer. Cancer Cell 2020, 37, 800–817.e7. [Google Scholar] [CrossRef] [PubMed]
- Nallanthighal, S.; Rada, M.; Heiserman, J.P.; Cha, J.; Sage, J.; Zhou, B.; Yang, W.; Hu, Y.; Korgaonkar, C.; Hanos, C.T.; et al. Inhibition of collagen XI alpha 1-induced fatty acid oxidation triggers apoptotic cell death in cisplatin-resistant ovarian cancer. Cell Death Dis. 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Midwood, K.S.; Chiquet, M.; Tucker, R.P.; Orend, G. Tenascin-C at a glance. J. Cell Sci. 2016, 129, 4321–4327. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Velázquez-Quesada, I.; Murdamoothoo, D.; Ahowesso, C.; Yilmaz, A.; Spenlé, C.; Averous, G.; Erne, W.; Oberndorfer, F.; Oszwald, A.; et al. Tenascin-C increases lung metastasis by impacting blood vessel invasions. Matrix Biol. J. Int. Soc. Matrix Biol. 2019, 83, 26–47. [Google Scholar] [CrossRef]
- Gopal, S.; Veracini, L.; Grall, D.; Butori, C.; Schaub, S.; Audebert, S.; Camoin, L.; Baudelet, E.; Radwanska, A.; Beghelli-de la Forest Divonne, S.; et al. Fibronectin-guided migration of carcinoma collectives. Nat. Commun. 2017, 8, 14105. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Q.; Alam, A.; Cui, J.; Suen, K.C.; Soo, A.P.; Eguchi, S.; Gu, J.; Ma, D. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, C.; Feng, Y.; Quan, M.; Cui, Y.; Xuan, Y. Tenascin-C predicts poor outcomes for patients with colorectal cancer and drives cancer stemness via Hedgehog signaling pathway. Cancer Cell Int. 2020, 20, 122. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z.; Chen, M.; Ren, Q.; Yue, X.; Dai, Z. Fibronectin-targeted dual-acting micelles for combination therapy of metastatic breast cancer. Signal Transduct. Target. Ther. 2020, 5, 1–11. [Google Scholar] [CrossRef]
- Henke, E.; Nandigama, R.; Ergün, S. Extracellular Matrix in the Tumor Microenvironment and Its Impact on Cancer Therapy. Front. Mol. Biosci. 2020, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.-C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.-T.; Ma, X.-J.; Luo, Y. RNAscope. J. Mol. Diagn. JMD 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, F.; Umbreit, C.; Krüger, T.; Pelzel, D.; Ernst, G.; Kniemeyer, O.; Guntinas-Lichius, O.; Berndt, A.; von Eggeling, F. Identification of Proteomic Markers in Head and Neck Cancer Using MALDI-MS Imaging, LC-MS/MS, and Immunohistochemistry. Proteom. Clin. Appl. 2019, 13, e1700173. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Clinicopathological Parameter | AdCy (n = 11) | MuEp (n = 14) | SaDu (n = 9) | Total (n = 34) | |
|---|---|---|---|---|---|
| Grade | |||||
| 1 | 1 (9) | 8 (57) | 5 (56) | 14 (41) | |
| 2 | 5 (46) | 2 (14) | 4 (44) | 11 (32) | |
| 3 | 3 (27) | 4 (29) | 0 (0) | 7 (21) | |
| NA | 2 (18) | 0 (0) | 0 (0) | 2 (6) | |
| pT | |||||
| 1 | 2 (18) | 2 (15) | 0 (0) | 4 (12) | |
| 2 | 1 (9) | 6 (46) | 1 (11) | 8 (24) | |
| 3 | 2 (18) | 2 (15) | 4 (44) | 8 (24) | |
| 4a | 3 (27) | 1 (8) | 3 (33) | 7 (21) | |
| 4b | 1 (9) | 2 (15) | 1 (11) | 4 (12) | |
| NA | 2 (18) | 0 (0) | 0 (0) | 2 (6) | |
| pN | |||||
| 0 | 6 (55) | 9 (69) | 1 (11) | 16 (49) | |
| 1 | 3 (27) | 0 (0) | 1 (11) | 4 (12) | |
| 2 | 1 (9) | 4 (31) | 7 (78) | 12 (36) | |
| NA | 1 (9) | 0 (0) | 0 (0) | 1 (3) | |
| L | |||||
| L0 | 7 (70) | 9 (82) | 6 (67) | 22 (73) | |
| L1 | 0 (0) | 2 (18) | 3 (33) | 5 (17) | |
| NA | 3 (30) | 0 (0) | 0 (0) | 3 (10) | |
| V | |||||
| V0 | 7 (70) | 8 (73) | 8 (89) | 23 (77) | |
| V1 | 0 (0) | 3 (27) | 1 (11) | 4 (13) | |
| NA | 3 (30) | 0 (0) | 0 (0) | 3 (10) | |
| Pn | |||||
| Pn0 | 5 (46) | 6 (50) | 4 (44) | 15 (47) | |
| Pn1 | 3 (27) | 6 (50) | 5 (56) | 14 (44) | |
| NA | 3 (27) | 0 (0) | 0 (0) | 3 (9) | |
| Sex | |||||
| f | 8 (73) | 10 (71) | 4 (44) | 22 (65) | |
| m | 3 (27) | 4 (29) | 5 (56) | 12 (35) | |
| Age, years | |||||
| Median (range) | 42 (30–73) | 40 (16–68) | 63 (37–78) | 46 (16–78) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arolt, C.; Meyer, M.; Hoffmann, F.; Wagener-Ryczek, S.; Schwarz, D.; Nachtsheim, L.; Beutner, D.; Odenthal, M.; Guntinas-Lichius, O.; Buettner, R.; et al. Expression Profiling of Extracellular Matrix Genes Reveals Global and Entity-Specific Characteristics in Adenoid Cystic, Mucoepidermoid and Salivary Duct Carcinomas. Cancers 2020, 12, 2466. https://doi.org/10.3390/cancers12092466
Arolt C, Meyer M, Hoffmann F, Wagener-Ryczek S, Schwarz D, Nachtsheim L, Beutner D, Odenthal M, Guntinas-Lichius O, Buettner R, et al. Expression Profiling of Extracellular Matrix Genes Reveals Global and Entity-Specific Characteristics in Adenoid Cystic, Mucoepidermoid and Salivary Duct Carcinomas. Cancers. 2020; 12(9):2466. https://doi.org/10.3390/cancers12092466
Chicago/Turabian StyleArolt, Christoph, Moritz Meyer, Franziska Hoffmann, Svenja Wagener-Ryczek, David Schwarz, Lisa Nachtsheim, Dirk Beutner, Margarete Odenthal, Orlando Guntinas-Lichius, Reinhard Buettner, and et al. 2020. "Expression Profiling of Extracellular Matrix Genes Reveals Global and Entity-Specific Characteristics in Adenoid Cystic, Mucoepidermoid and Salivary Duct Carcinomas" Cancers 12, no. 9: 2466. https://doi.org/10.3390/cancers12092466






