FANCD2 Confers a Malignant Phenotype in Esophageal Squamous Cell Carcinoma by Regulating Cell Cycle Progression
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
2.2. RNA Sequence Analysis
2.3. Cell Lines
2.4. Plasmids and Lentivirus Preparation and Infection
2.5. Western Blot Analysis
2.6. In Vivo Tumorigenicity Assay
2.7. MTT Assay
2.8. Colony Formation Assay
2.9. Synchronization and Flow Cytometry Analysis
2.10. In Vitro Chemotherapy Treatment
2.11. Subcellular Fractionation
2.12. Statistical Analysis
3. Results
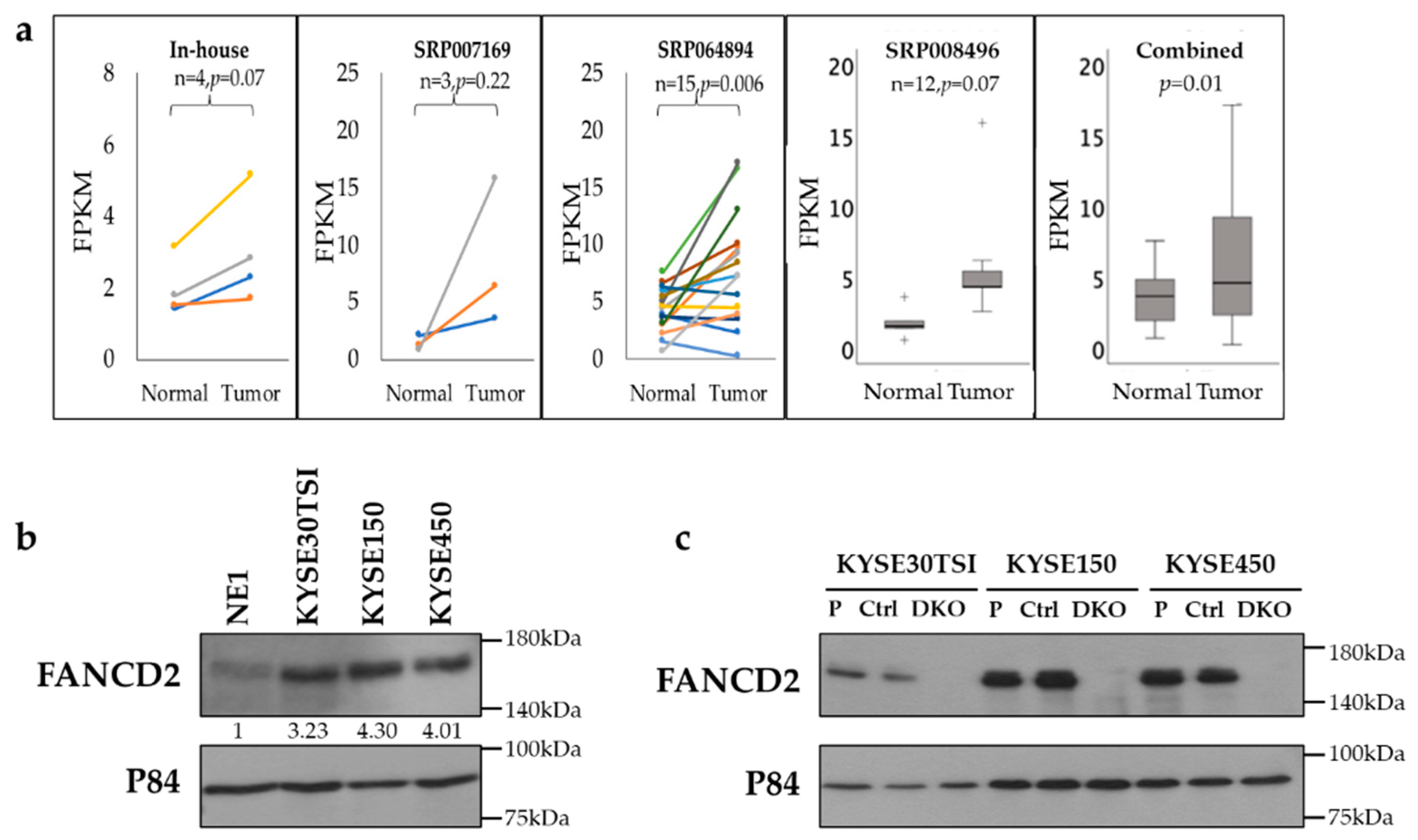
3.1. Upregulation of FANCD2 Gene Expression in ESCC Tissues and Cell Lines
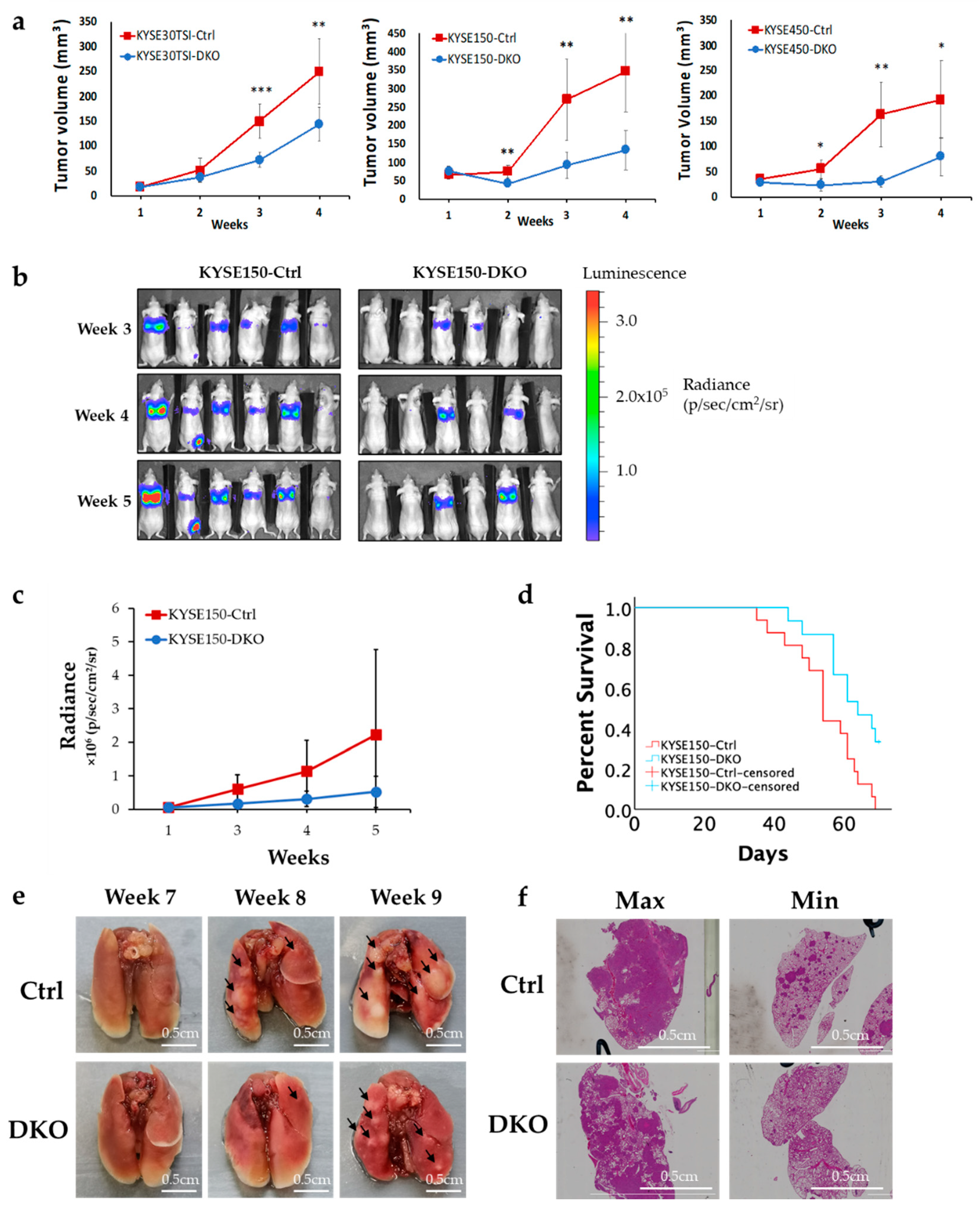
3.2. Depletion of FANCD2 Protein Expression Inhibits In Vivo Tumor Growth and Metastasis
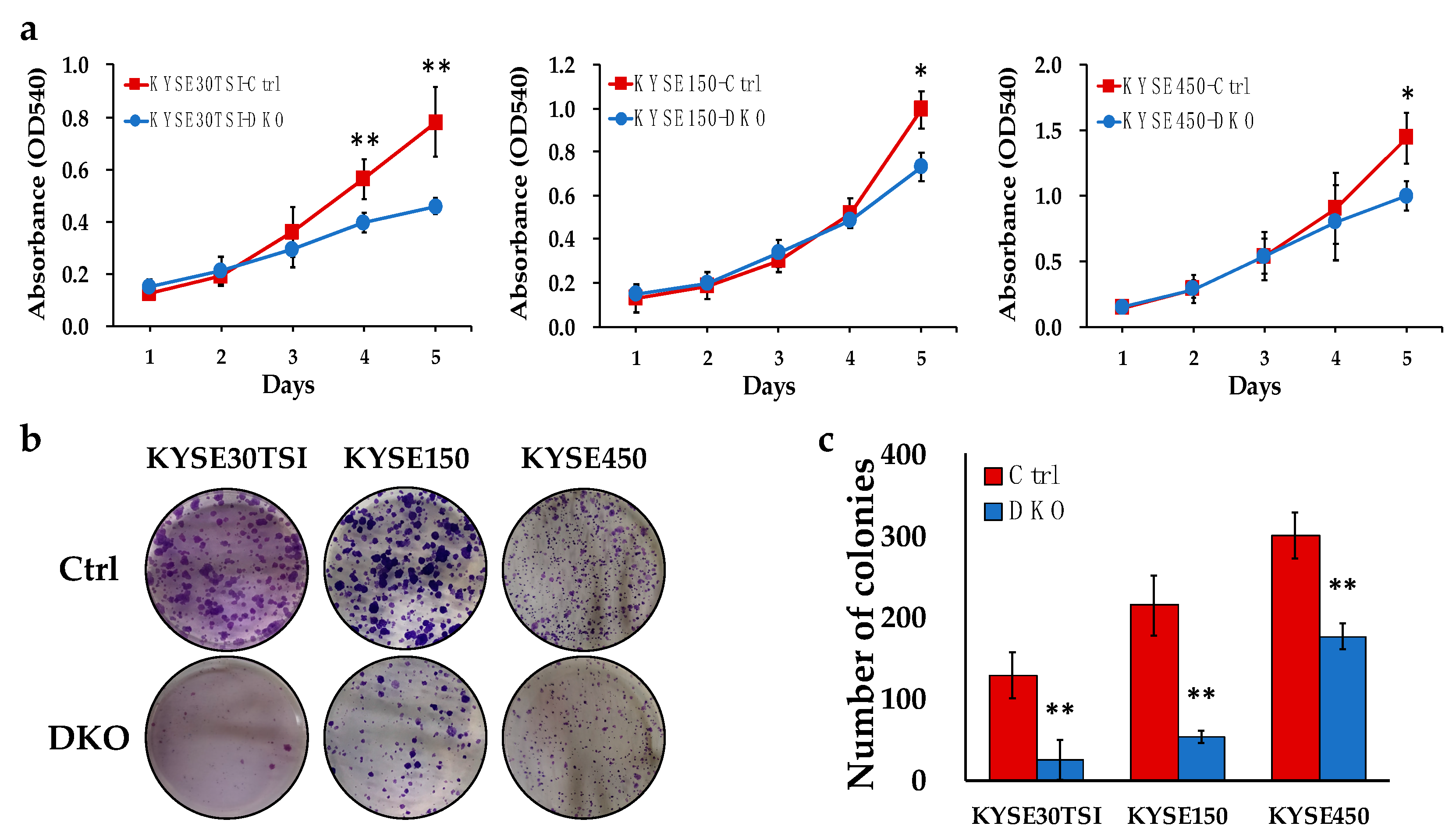
3.3. FANCD2-KO Suppresses In Vitro Cell Proliferation and Colony Formation in ESCC Cells
3.4. FANCD2-KO Suppresses Cell Cycle Progression in ESCC Cells
3.5. FANCD2-KO Affects G1/S Transition and Delays Entry to Mitosis in ESCC Cells
3.6. Ubiquitinated FANCD2 Localizes to the Nucleus to Regulate Cell Cycle Progression in ESCC Cells
3.7. FANCD2 Is Likely Dispensable for DNA Damage Repair in ESCC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conteduca, V.; Sansonno, D.E.; Ingravallo, G.; Marangi, S.; Russi, S.; Lauletta, G.; Dammacco, F. Barrett’s esophagus and esophageal cancer: An overview. Int. J. Oncol. 2012, 41, 414–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houghtaling, S.; Timmers, C.; Noll, M.; Finegold, M.J.; Jones, S.N.; Meyn, M.S.; Grompe, M. Epithelial cancer in Fanconi anemia knockout mice. Genes Dev. 2003, 2, 2021–2035. [Google Scholar] [CrossRef] [Green Version]
- Parmar, K.; D’Andrea, A.; Niedernhofer, L.J. Mouse models of Fanconi anemia. Mutat. Res. Mol. Mech. Mutagen. 2009, 668, 133–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barroso, E.; Milne, R.; Zamora, P.; Arias, J.; Fernández, L.; Benitez, J.; Ribas, G. FANCD2 associated with sporadic breast cancer risk. Carcinogenesis 2006, 27, 1930–1937. [Google Scholar] [CrossRef]
- Mantere, T.; Tervasmäki, A.; Nurmi, A.; Rapakko, K.; Kauppila, S.; Tang, J.; Schleutker, J.; Kallioniemi, A.; Hartikainen, J.M.; Mannermaa, A.; et al. Case-control analysis of truncating mutations in DNA damage response genes connects TEX15 and FANCD2 with hereditary breast cancer susceptibility. Sci. Rep. 2017, 7, 681. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekharappa, S.C.; Chinn, S.B.; Donovan, F.X.; Chowdhury, N.I.; Kamat, A.; Adeyemo, A.A.; Thomas, J.W.; Vemulapalli, M.; Hussey, C.S.; Reid, H.H.; et al. Assessing the spectrum of germline variation in Fanconi anemia genes among patients with head and neck carcinoma before age 50. Cancer 2017, 123, 3943–3954. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Jin, F. Expression and prognostic significance of Fanconi anemia group D2 protein and breast cancer type 1 susceptibility protein in familial and sporadic breast cancer. Oncol. Lett. 2019, 17, 3687–3700. [Google Scholar] [CrossRef] [Green Version]
- Fagerholm, R.; Sprott, K.; Heikkinen, T.; Bartkova, J.; Heikkilä, P.; Aittomäki, K.; Bartek, J.; Weaver, D.; Blomqvist, C.; Nevanlinna, H. Overabundant FANCD2, alone and combined with NQO1, is a sensitive marker of adverse prognosis in breast cancer. Ann. Oncol. 2013, 24, 2780–2785. [Google Scholar] [CrossRef]
- Rudland, P.S.; Platt-Higgins, A.M.; Davies, L.M.; Rudland, S.D.S.; Wilson, J.B.; Aladwani, A.; Winstanley, J.H.; Barraclough, D.L.; Barraclough, R.; West, C.R.; et al. Significance of the Fanconi Anemia FANCD2 Protein in Sporadic and Metastatic Human Breast Cancer. Am. J. Pathol. 2010, 176, 2935–2947. [Google Scholar] [CrossRef]
- Wysham, W.Z.; Mhawech-Fauceglia, P.; Li, H.; Hays, L.; Syriac, S.; Skrepnik, T.; Wright, J.; Pande, N.; Hoatlin, M.; Pejovic, T. BRCAness profile of sporadic ovarian cancer predicts disease recurrence. PLoS ONE 2012, 7, e30042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pejovic, T.; Yates, J.E.; Liu, H.Y.; Hays, L.E.; Akkari, Y.; Torimaru, Y.; Keeble, W.; Rathbun, R.K.; Rodgers, W.H.; Bale, A.E.; et al. Cytogenetic instability in ovarian epithelial cells from women at risk of ovarian cancer. Cancer Res. 2006, 66, 9017–9025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Zhao, F.; Liang, Z.; Feng, H.; Bao, Y.; Xu, W.; Zhao, C.; Qin, G. Expression of FANCD2 is associated with progosis in patients with nasopharyngeal carcinoma. Int. J. Clin. Exp. Pathol. 2019, 12, 3465–3473. [Google Scholar] [PubMed]
- Patil, A.A.; Sayal, P.; Depondt, M.L.; Beveridge, R.D.; Roylance, A.; Kriplani, D.H.; Myers, K.N.; Cox, A.; Jellinek, D.; Fernando, M.; et al. FANCD2 re-expression is associated with glioma grade and chemical inhibition of the Fanconi Anaemia pathway sensitises gliomas to chemotherapeutic agents. Oncotarget 2014, 5, 6414–6424. [Google Scholar] [CrossRef] [Green Version]
- Mhawech-Fauceglia, P.; Wang, D.; Kim, G.; Sharifian, M.; Chen, X.; Liu, Q.; Lin, Y.G.; Liu, S.; Pejovic, T. Expression of DNA repair proteins in endometrial cancer predicts disease outcome. Gynecol. Oncol. 2014, 132, 593–598. [Google Scholar] [CrossRef]
- Leung, A.C.C.; Wong, V.C.L.; Li, C.Y.; Pui, L.C.; Daigo, Y.; Nakamura, Y.; Qi, R.Z.; Miller, L.D.; Liu, E.T.; Wang, L.D.; et al. Frequent decreased expression of candidate tumor suppressor gene, DEC1, and its anchorage-independent growth properties and impact on global gene expression in esophageal carcinoma. Int. J. Cancer 2008, 122, 587–594. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Cao, J.; Wu, W.; Zhu, Q.; Tang, Y.; Zhu, C.; Dai, J.; Li, Z.; Wang, J.; Xue, L.; et al. Genome-wide copy number variation analysis identified ANO1 as a novel oncogene and prognostic biomarker in esophageal squamous cell cancer. Carcinogenesis 2019, 20, 1–11. [Google Scholar] [CrossRef]
- Yu, V.Z.; Wong, V.C.-L.; Dai, W.; Ko, J.M.-Y.; Lam, A.K.-Y.; Chan, K.W.; Samant, R.S.; Lung, H.L.; Shuen, W.H.; Law, S.; et al. Nuclear localization of DNAJB6 is associated with survival of patients with esophageal cancer and reduces AKT signaling and proliferation of cancer cells. Gastroenterology 2015, 149, 1825–1836. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Tsao, S.W.; Chan, K.W.; Ludwig, D.L.; Novosyadlyy, R.; Li, Y.Y.; He, Q.Y.; Cheung, A.L. Id1-induced IGF-II and its autocrine/endocrine promotion of esophageal cancer progression and chemoresistance-implications for IGF-II and IGF-IR-targeted therapy. Clin. Cancer Res. 2014, 20, 2651–2662. [Google Scholar] [CrossRef] [Green Version]
- Song, I.Y.; Barkley, L.R.; Day, T.A.; Weiss, R.S.; Vaziri, C. A novel role for fanconi anemia (FA) pathway effector protein FANCD2 in cell cycle progression of untransformed primary human cells. Cell Cycle 2010, 9, 2375–2388. [Google Scholar] [CrossRef] [PubMed]
- Schlacher, K.; Wu, H.; Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012, 22, 106–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreassen, P.R.; D’Andrea, A.D.; Taniguchi, T. ATR couples FANCD2 monoubiquitination to the DNA-damage responsey. Genes Dev. 2004, 18, 1958–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guervilly, J.-H.; Macé-Aimé, G.; Rosselli, F. Loss of CHK1 function impedes DNA damage-induced FANCD2 monoubiquitination but normalizes the abnormal G2 arrest in Fanconi anemia. Hum. Mol. Genet. 2007, 17, 679–689. [Google Scholar] [CrossRef]
- Shen, C.; Oswald, D.; Phelps, D.; Çam, H.; Pelloski, C.E.; Pang, Q.; Houghton, P.J. Abstract 4437: Regulation of FANCD2 by the mTOR pathway contributes to the resistance of cancer cells to DNA double strand breaks. Exper. Mol. Ther. 2013, 73. [Google Scholar] [CrossRef]
- Nijman, S.M.; Huang, T.T.; Dirac, A.M.; Brummelkamp, T.R.; Kerkhoven, R.M.; D’Andrea, A.; Bernards, R. The Deubiquitinating Enzyme USP1 Regulates the Fanconi Anemia Pathway. Mol. Cell 2005, 17, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.-L.; Palmai-Pallag, T.; Ying, S.; Hickson, I.D. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature 2009, 11, 753–760. [Google Scholar] [CrossRef]
- Nalepa, G.; Enzor, R.; Sun, Z.; Marchal, C.; Park, S.-J.; Yang, Y.; Tedeschi, L.; Kelich, S.; Hanenberg, H.; Clapp, D.W. Fanconi anemia signaling network regulates the spindle assembly checkpoint. J. Clin. Investig. 2013, 123, 3839–3847. [Google Scholar] [CrossRef] [Green Version]
- Knipscheer, P.; Räschle, M.; Smogorzewska, A.; Enoiu, M.; Ho, T.V.; Schärer, O.D.; Elledge, S.J.; Walter, J.C. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science 2009, 326, 1698–1701. [Google Scholar] [CrossRef] [Green Version]
- Chirnomas, D.; Taniguchi, T.; De La Vega, M.; Vaidya, A.P.; Vasserman, M.; Hartman, A.-R.; Kennedy, R.; Foster, R.; Mahoney, J.; Seiden, M.V.; et al. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol. Cancer Ther. 2006, 5, 952–961. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Higuera, I.; Taniguchi, T.; Ganesan, S.; Meyn, M.; Timmers, C.; Hejna, J.; Grompe, M.; D’Andrea, A.D. Interaction of the Fanconi Anemia Proteins and BRCA1 in a Common Pathway. Mol. Cell 2001, 7, 249–262. [Google Scholar] [CrossRef]
- Pace, P.; Johnson, M.; Tan, W.M.; Mosedale, G.; Sng, C.; Hoatlin, M.; De Winter, J.; Joenje, H.; Gergely, F.; Patel, K.J. FANCE: The link between Fanconi anaemia complex assembly and activity. EMBO J. 2002, 21, 3414–3423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naim, V.; Rosselli, F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nature 2009, 11, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Du, W.; Wilson, A.F.; Namekawa, S.H.; Andreassen, P.R.; Meetei, A.R.; Pang, Q. Fancd2 in vivo interaction network reveals a non-canonical role in mitochondrial function. Sci. Rep. 2017, 7, 45626. [Google Scholar] [CrossRef] [Green Version]
- Jayabal, P.; Ma, C.; Nepal, M.; Shen, Y.; Che, R.; Turkson, J.; Fei, P. Involvement of FANCD2 in Energy Metabolism via ATP5α. Sci. Rep. 2017, 7, 4921. [Google Scholar] [CrossRef]
- Marcussen, M.; Larsen, P.J. Cell cycle-dependent regulation of cellular ATP concentration, and depolymerization of the interphase microtubular network induced by elevated cellular ATP concentration in whole fibroblasts. Cell Motil. Cytoskelet. 1996, 35, 94–99. [Google Scholar] [CrossRef]



| Antibody | Company | Catalog Number | Dilution | Antibody | Company | Catalog Number | Dilution |
|---|---|---|---|---|---|---|---|
| FANCD2 | Santa Cruz | sc20022 | 1:1000 | CDK4 | Cell Signaling | 12,790 | 1:1000 |
| p84 | GeneTex | GTX70220 | 1:2000 | CDK7 | Cell Signaling | 2090 | 1:1000 |
| Cyclin A2 | Cell Signaling | 4656 | 1:1000 | CDK9 | Cell Signaling | 2316 | 1:1000 |
| Cyclin B1 | Cell Signaling | 4138 | 1:1000 | p-ATR | Cell Signaling | 2853 | 1:1000 |
| Cyclin D1 | Cell Signaling | 2978 | 1:1000 | p-ATM | Cell Signaling | 13,050 | 1:1000 |
| Cyclin D2 | Cell Signaling | 3741 | 1:1000 | p-Chk1 | Cell Signaling | 12,302 | 1:1000 |
| Cyclin E2 | Cell Signaling | 4132 | 1:1000 | p-Chk2 | Cell Signaling | 2661 | 1:1000 |
| Chk1 | Cell Signaling | 2345 | 1:500 | p-BRCA1 | Cell Signaling | 9009 | 1:1000 |
| Chk2 | Cell Signaling | 3440 | 1:1000 | Vinculin | Cell Signaling | 13,901 | 1:2000 |
| cdc2 | Cell Signaling | 28,439 | 1:1000 | ⍺-Tubulin | Cell Signaling | 2125 | 1:2000 |
| CDK2 | Cell Signaling | 2546 | 1:1000 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, L.C.; Yu, V.Z.; Ko, J.M.Y.; Ning, L.; Lung, M.L. FANCD2 Confers a Malignant Phenotype in Esophageal Squamous Cell Carcinoma by Regulating Cell Cycle Progression. Cancers 2020, 12, 2545. https://doi.org/10.3390/cancers12092545
Lei LC, Yu VZ, Ko JMY, Ning L, Lung ML. FANCD2 Confers a Malignant Phenotype in Esophageal Squamous Cell Carcinoma by Regulating Cell Cycle Progression. Cancers. 2020; 12(9):2545. https://doi.org/10.3390/cancers12092545
Chicago/Turabian StyleLei, Lisa Chan, Valen Zhuoyou Yu, Josephine Mun Yee Ko, Lvwen Ning, and Maria Li Lung. 2020. "FANCD2 Confers a Malignant Phenotype in Esophageal Squamous Cell Carcinoma by Regulating Cell Cycle Progression" Cancers 12, no. 9: 2545. https://doi.org/10.3390/cancers12092545
APA StyleLei, L. C., Yu, V. Z., Ko, J. M. Y., Ning, L., & Lung, M. L. (2020). FANCD2 Confers a Malignant Phenotype in Esophageal Squamous Cell Carcinoma by Regulating Cell Cycle Progression. Cancers, 12(9), 2545. https://doi.org/10.3390/cancers12092545






