Cellular Mechanisms Accounting for the Refractoriness of Colorectal Carcinoma to Pharmacological Treatment
Abstract
Simple Summary
Abstract
1. Introduction
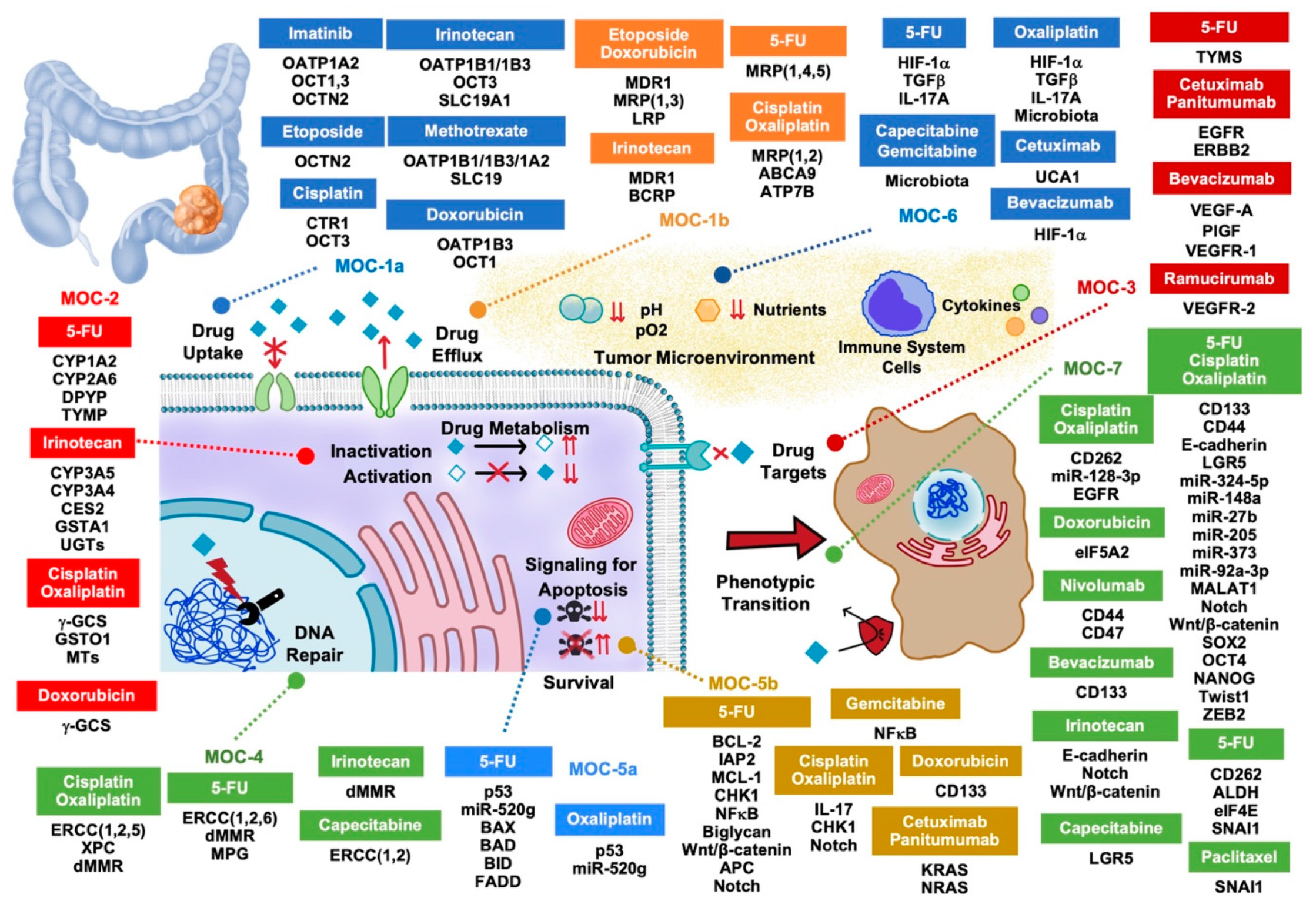
2. Drug Uptake and Export (MOC-1)
2.1. Drug Uptake Carriers (MOC-1a)
2.2. Drug Export Pumps (MOC-1b)
3. Drug Metabolism (MOC-2)
4. Changes in Drug Targets (MOC-3)
5. DNA Repairing (MOC-4)
6. Balance between Pro-Apoptotic and Pro-Survival Factors (MOC-5)
6.1. Pro-Apoptotic Factors (MOC-5a)
6.2. Survival Pathways (MOC-5b)
7. Adaptation to the Tumor Microenvironment (MOC-6)
8. Phenotype Transition (MOC-7)
9. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Sharma, R. An examination of colorectal cancer burden by socioeconomic status: Evidence from GLOBOCAN 2018. EPMA J. 2020, 11, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Cappell, M.S. Pathophysiology, clinical presentation, and management of colon cancer. Gastroenterol. Clin. N. Am. 2008, 37, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, M.G.; Tenesa, A.; Farrington, S.M.; Ballereau, S.; Brewster, D.H.; Koessler, T.; Pharoah, P.; Schafmayer, C.; Hampe, J.; Volzke, H.; et al. Cumulative impact of common genetic variants and other risk factors on colorectal cancer risk in 42,103 individuals. Gut 2013, 62, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.; Sanchez de Medina, F.; Castano, B.; Bujanda, L.; Romero, M.R.; Martinez-Augustin, O.; Moral-Avila, R.D.; Briz, O. Chemoprevention, chemotherapy, and chemoresistance in colorectal cancer. Drug Metab. Rev. 2012, 44, 148–172. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, R.; Gholghesaei, M.; Bonjer, H.J.; Meijer, D.W.; Buunen, M.; Jeekel, J.; Anderberg, B.; Cuesta, M.A.; Cuschierl, A.; Fingerhut, A.; et al. Laparoscopic resection of colon cancer: Consensus of the European Association of Endoscopic Surgery (EAES). Surg. Endosc. 2004, 18, 1163–1185. [Google Scholar] [CrossRef] [PubMed]
- Bujanda, L.; Sarasqueta, C.; Hijona, E.; Hijona, L.; Cosme, A.; Gil, I.; Elorza, J.L.; Asensio, J.I.; Larburu, S.; Enriquez-Navascues, J.M.; et al. Colorectal cancer prognosis twenty years later. World J. Gastroenterol. 2010, 16, 862–867. [Google Scholar] [CrossRef]
- Jacome, A.A.; Eng, C. Role of immune checkpoint inhibitors in the treatment of colorectal cancer: Focus on nivolumab. Expert Opin. Biol. Ther. 2019, 19, 1247–1263. [Google Scholar] [CrossRef]
- Marin, J.J.G.; Macias, R.I.R.; Monte, M.J.; Romero, M.R.; Asensio, M.; Sanchez-Martin, A.; Cives-Losada, C.; Temprano, A.; Espinosa-Escudero, R.; Reviejo, M.; et al. Molecular bases of drug resistance in hepatocellular carcinoma. Cancers 2020, 12, 1663. [Google Scholar] [CrossRef]
- Marin, J.J.G.; Lozano, E.; Herraez, E.; Asensio, M.; Di Giacomo, S.; Romero, M.R.; Briz, O.; Serrano, M.A.; Efferth, T.; Macias, R.I.R. Chemoresistance and chemosensitization in cholangiocarcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1444–1453. [Google Scholar] [CrossRef]
- Marin, J.J.G.; Cives-Losada, C.; Asensio, M.; Lozano, E.; Briz, O.; Macias, R.I.R. Mechanisms of anticancer drug resistance in hepatoblastoma. Cancers 2019, 11, 407. [Google Scholar] [CrossRef]
- Marin, J.J.; Romero, M.R.; Martinez-Becerra, P.; Herraez, E.; Briz, O. Overview of the molecular bases of resistance to chemotherapy in liver and gastrointestinal tumours. Curr. Mol. Med. 2009, 9, 1108–1129. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.G.; Perez-Silva, L.; Macias, R.I.R.; Asensio, M.; Peleteiro-Vigil, A.; Sanchez-Martin, A.; Cives-Losada, C.; Sanchon-Sanchez, P.; Sanchez De Blas, B.; Herraez, E.; et al. Molecular bases of mechanisms accounting for drug resistance in gastric adenocarcinoma. Cancers 2020, 12, 2116. [Google Scholar] [CrossRef] [PubMed]
- Okabe, M.; Szakacs, G.; Reimers, M.A.; Suzuki, T.; Hall, M.D.; Abe, T.; Weinstein, J.N.; Gottesman, M.M. Profiling SLCO and SLC22 genes in the NCI-60 cancer cell lines to identify drug uptake transporters. Mol. Cancer Ther. 2008, 7, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Ballestero, M.R.; Monte, M.J.; Briz, O.; Jimenez, F.; Gonzalez-San Martin, F.; Marin, J.J. Expression of transporters potentially involved in the targeting of cytostatic bile acid derivatives to colon cancer and polyps. Biochem. Pharmacol. 2006, 72, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Obaidat, A.; Roth, M.; Hagenbuch, B. The expression and function of organic anion transporting polypeptides in normal tissues and in cancer. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 135–151. [Google Scholar] [CrossRef]
- Thakkar, N.; Lockhart, A.C.; Lee, W. Role of Organic Anion-Transporting Polypeptides (OATPs) in cancer therapy. AAPS J. 2015, 17, 535–545. [Google Scholar] [CrossRef]
- Nozawa, T.; Minami, H.; Sugiura, S.; Tsuji, A.; Tamai, I. Role of organic anion transporter OATP1B1 (OATP-C) in hepatic uptake of irinotecan and its active metabolite, 7-ethyl-10-hydroxycamptothecin: In vitro evidence and effect of single nucleotide polymorphisms. Drug Metab. Dispos. 2005, 33, 434–439. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, T.; Xie, C.; Liao, X.; Yu, Q.; Feng, J.; Ma, H.; Dai, J.; Li, M.; Chen, J.; et al. SLCO1B1 and SLC19A1 gene variants and irinotecan-induced rapid response and survival: A prospective multicenter pharmacogenetics study of metastatic colorectal cancer. PLoS ONE 2013, 8, e77223. [Google Scholar] [CrossRef]
- Treenert, A.; Areepium, N.; Tanasanvimon, S. Effects of ABCC2 and SLCO1B1 polymorphisms on treatment responses in thai metastatic colorectal cancer patients treated with irinotecan-based chemotherapy. Asian Pac. J. Cancer Prev. 2018, 19, 2757–2764. [Google Scholar] [CrossRef]
- Meier, Y.; Eloranta, J.J.; Darimont, J.; Ismair, M.G.; Hiller, C.; Fried, M.; Kullak-Ublick, G.A.; Vavricka, S.R. Regional distribution of solute carrier mRNA expression along the human intestinal tract. Drug Metab. Dispos. 2007, 35, 590–594. [Google Scholar] [CrossRef]
- Abe, T.; Unno, M.; Onogawa, T.; Tokui, T.; Kondo, T.N.; Nakagomi, R.; Adachi, H.; Fujiwara, K.; Okabe, M.; Suzuki, T.; et al. LST-2, a human liver-specific organic anion transporter, determines methotrexate sensitivity in gastrointestinal cancers. Gastroenterology 2001, 120, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Durmus, S.; van Hoppe, S.; Schinkel, A.H. The impact of Organic Anion-Transporting Polypeptides (OATPs) on disposition and toxicity of antitumor drugs: Insights from knockout and humanized mice. Drug Resist. Updates 2016, 27, 72–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Leake, B.F.; Kim, R.B.; Ho, R.H. Contribution of organic anion-transporting polypeptides 1A/1B to doxorubicin uptake and clearance. Mol. Pharmacol. 2017, 91, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.; Furihata, T.; Matsumoto, S.; Ishii, S.; Motohashi, S.; Yoshino, I.; Ugajin, M.; Miyajima, A.; Matsumoto, S.; Chiba, K. Identification of a new organic anion transporting polypeptide 1B3 mRNA isoform primarily expressed in human cancerous tissues and cells. Biochem. Biophys. Res. Commun. 2012, 418, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, N.; Kim, K.; Jang, E.R.; Han, S.; Kim, K.; Kim, D.; Merchant, N.; Lockhart, A.C.; Lee, W. A cancer-specific variant of the SLCO1B3 gene encodes a novel human organic anion transporting polypeptide 1B3 (OATP1B3) localized mainly in the cytoplasm of colon and pancreatic cancer cells. Mol. Pharm. 2013, 10, 406–416. [Google Scholar] [CrossRef]
- Teft, W.A.; Welch, S.; Lenehan, J.; Parfitt, J.; Choi, Y.H.; Winquist, E.; Kim, R.B. OATP1B1 and tumour OATP1B3 modulate exposure, toxicity, and survival after irinotecan-based chemotherapy. Br. J. Cancer 2015, 112, 857–865. [Google Scholar] [CrossRef]
- Yamakawa, Y.; Hamada, A.; Shuto, T.; Yuki, M.; Uchida, T.; Kai, H.; Kawaguchi, T.; Saito, H. Pharmacokinetic impact of SLCO1A2 polymorphisms on imatinib disposition in patients with chronic myeloid leukemia. Clin. Pharmacol. Ther. 2011, 90, 157–163. [Google Scholar] [CrossRef]
- Eechoute, K.; Franke, R.M.; Loos, W.J.; Scherkenbach, L.A.; Boere, I.; Verweij, J.; Gurney, H.; Kim, R.B.; Tirona, R.G.; Mathijssen, R.H.; et al. Environmental and genetic factors affecting transport of imatinib by OATP1A2. Clin. Pharmacol. Ther. 2011, 89, 816–820. [Google Scholar] [CrossRef]
- White, D.L.; Saunders, V.A.; Dang, P.; Engler, J.; Zannettino, A.C.; Cambareri, A.C.; Quinn, S.R.; Manley, P.W.; Hughes, T.P. OCT-1-mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107): Reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinib. Blood 2006, 108, 697–704. [Google Scholar] [CrossRef]
- Marin, J.J.; Romero, M.R.; Blazquez, A.G.; Herraez, E.; Keck, E.; Briz, O. Importance and limitations of chemotherapy among the available treatments for gastrointestinal tumours. Anticancer Agents Med. Chem. 2009, 9, 162–184. [Google Scholar] [CrossRef]
- Zhang, S.; Lovejoy, K.S.; Shima, J.E.; Lagpacan, L.L.; Shu, Y.; Lapuk, A.; Chen, Y.; Komori, T.; Gray, J.W.; Chen, X.; et al. Organic cation transporters are determinants of oxaliplatin cytotoxicity. Cancer Res. 2006, 66, 8847–8857. [Google Scholar] [CrossRef] [PubMed]
- Seithel, A.; Karlsson, J.; Hilgendorf, C.; Bjorquist, A.; Ungell, A.L. Variability in mRNA expression of ABC- and SLC-transporters in human intestinal cells: Comparison between human segments and Caco-2 cells. Eur. J. Pharm. Sci. 2006, 28, 291–299. [Google Scholar] [CrossRef]
- Drozdzik, M.; Busch, D.; Lapczuk, J.; Muller, J.; Ostrowski, M.; Kurzawski, M.; Oswald, S. Protein abundance of clinically relevant drug transporters in the human liver and intestine: A comparative analysis in paired tissue specimens. Clin. Pharmacol. Ther. 2019, 105, 1204–1212. [Google Scholar] [CrossRef]
- Makhtar, S.M.; Husin, A.; Baba, A.A.; Ankathil, R. Genetic variations in influx transporter gene SLC22A1 are associated with clinical responses to imatinib mesylate among Malaysian chronic myeloid leukaemia patients. J. Genet. 2018, 97, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Watkins, D.B.; Hughes, T.P.; White, D.L. OCT1 and imatinib transport in CML: Is it clinically relevant? Leukemia 2015, 29, 1960–1969. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Fukuno, S.; Yamamoto, K.; Omotani, S.; Hatsuda, Y.; Myotoku, M.; Konishi, H. Downregulation of organic cation transporter 1 and breast cancer resistance protein with the induction of Pregnane X receptor in rat kidney impaired by doxorubicin. Pharmazie 2019, 74, 744–746. [Google Scholar] [CrossRef]
- Vollmar, J.; Lautem, A.; Closs, E.; Schuppan, D.; Kim, Y.O.; Grimm, D.; Marquardt, J.U.; Fuchs, P.; Straub, B.K.; Schad, A.; et al. Loss of organic cation transporter 3 (Oct3) leads to enhanced proliferation and hepatocarcinogenesis. Oncotarget 2017, 8, 115667–115680. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guttmann, S.; Chandhok, G.; Groba, S.R.; Niemietz, C.; Sauer, V.; Gomes, A.; Ciarimboli, G.; Karst, U.; Zibert, A.; Schmidt, H.H. Organic cation transporter 3 mediates cisplatin and copper cross-resistance in hepatoma cells. Oncotarget 2018, 9, 743–754. [Google Scholar] [CrossRef]
- Le Roy, B.; Tixier, L.; Pereira, B.; Sauvanet, P.; Buc, E.; Petorin, C.; Dechelotte, P.; Pezet, D.; Balayssac, D. Assessment of the relation between the expression of oxaliplatin transporters in colorectal cancer and response to FOLFOX-4 adjuvant chemotherapy: A case control study. PLoS ONE 2016, 11, e0148739. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jong, N.N.; Nakanishi, T.; Liu, J.J.; Tamai, I.; McKeage, M.J. Oxaliplatin transport mediated by organic cation/carnitine transporters OCTN1 and OCTN2 in overexpressing human embryonic kidney 293 cells and rat dorsal root ganglion neurons. J. Pharmacol. Exp. Ther. 2011, 338, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.; Ferrara, A.M.; Giachelia, M.; Panieri, E.; Siminovitch, K.; Galeotti, T.; Larocca, L.M.; Pani, G. Association of the OCTN1/1672T variant with increased risk for colorectal cancer in young individuals and ulcerative colitis patients. Inflamm. Bowel Dis. 2012, 18, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Lancaster, C.S.; Zuo, Z.; Hu, S.; Chen, Z.; Rubnitz, J.E.; Baker, S.D.; Sparreboom, A. Inhibition of OCTN2-mediated transport of carnitine by etoposide. Mol. Cancer Ther. 2012, 11, 921–929. [Google Scholar] [CrossRef]
- Hu, S.; Franke, R.M.; Filipski, K.K.; Hu, C.; Orwick, S.J.; de Bruijn, E.A.; Burger, H.; Baker, S.D.; Sparreboom, A. Interaction of imatinib with human organic ion carriers. Clin. Cancer Res. 2008, 14, 3141–3148. [Google Scholar] [CrossRef] [PubMed]
- Angelini, S.; Pantaleo, M.A.; Ravegnini, G.; Zenesini, C.; Cavrini, G.; Nannini, M.; Fumagalli, E.; Palassini, E.; Saponara, M.; Di Battista, M.; et al. Polymorphisms in OCTN1 and OCTN2 transporters genes are associated with prolonged time to progression in unresectable gastrointestinal stromal tumours treated with imatinib therapy. Pharmacol. Res. 2013, 68, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Song, I.S.; Savaraj, N.; Siddik, Z.H.; Liu, P.; Wei, Y.; Wu, C.J.; Kuo, M.T. Role of human copper transporter Ctr1 in the transport of platinum-based antitumor agents in cisplatin-sensitive and cisplatin-resistant cells. Mol. Cancer Ther. 2004, 3, 1543–1549. [Google Scholar] [PubMed]
- Holzer, A.K.; Manorek, G.H.; Howell, S.B. Contribution of the major copper influx transporter CTR1 to the cellular accumulation of cisplatin, carboplatin, and oxaliplatin. Mol. Pharmacol. 2006, 70, 1390–1394. [Google Scholar] [CrossRef]
- Haslam, I.S.; Jones, K.; Coleman, T.; Simmons, N.L. Induction of P-glycoprotein expression and function in human intestinal epithelial cells (T84). Biochem. Pharmacol. 2008, 76, 850–861. [Google Scholar] [CrossRef]
- Thiebaut, F.; Tsuruo, T.; Hamada, H.; Gottesman, M.M.; Pastan, I.; Willingham, M.C. Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc. Natl. Acad. Sci. USA 1987, 84, 7735–7738. [Google Scholar] [CrossRef]
- Lagas, J.S.; van Waterschoot, R.A.; Sparidans, R.W.; Wagenaar, E.; Beijnen, J.H.; Schinkel, A.H. Breast cancer resistance protein and P-glycoprotein limit sorafenib brain accumulation. Mol. Cancer Ther. 2010, 9, 319–326. [Google Scholar] [CrossRef]
- Zheng, H.C. The molecular mechanisms of chemoresistance in cancers. Oncotarget 2017, 8, 59950–59964. [Google Scholar] [CrossRef]
- Terada, T.; Hira, D. Intestinal and hepatic drug transporters: Pharmacokinetic, pathophysiological, and pharmacogenetic roles. J. Gastroenterol. 2015, 50, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Potocnik, U.; Glavac, D.; Dean, M. Common germline MDR1/ABCB1 functional polymorphisms and haplotypes modify susceptibility to colorectal cancers with high microsatellite instability. Cancer Genet. Cytogenet. 2008, 183, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Toffoli, G.; Tumiotto, L.; Gigante, M.; Dall’Arche, M.G.; Perin, T.; Boiocchi, M. Increased chemosensitivity to doxorubicin of intrinsically multidrug-resistant human colon carcinoma cells by prolonged exposure to verapamil. Eur. J. Cancer 1993, 29A, 1776–1778. [Google Scholar] [CrossRef]
- Dahlmann, M.; Werner, R.; Kortum, B.; Kobelt, D.; Walther, W.; Stein, U. Restoring treatment response in colorectal cancer cells by targeting MACC1-dependent ABCB1 expression in combination therapy. Front. Oncol. 2020, 10, 599. [Google Scholar] [CrossRef]
- Kaiser, S.; Park, Y.K.; Franklin, J.L.; Halberg, R.B.; Yu, M.; Jessen, W.J.; Freudenberg, J.; Chen, X.; Haigis, K.; Jegga, A.G.; et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007, 8, R131. [Google Scholar] [CrossRef]
- Hinoshita, E.; Uchiumi, T.; Taguchi, K.; Kinukawa, N.; Tsuneyoshi, M.; Maehara, Y.; Sugimachi, K.; Kuwano, M. Increased expression of an ATP-binding cassette superfamily transporter, multidrug resistance protein 2, in human colorectal carcinomas. Clin. Cancer Res. 2000, 6, 2401–2407. [Google Scholar]
- Hammond, W.A.; Swaika, A.; Mody, K. Pharmacologic resistance in colorectal cancer: A review. Ther. Adv. Med. Oncol. 2016, 8, 57–84. [Google Scholar] [CrossRef]
- Vinette, V.; Placet, M.; Arguin, G.; Gendron, F.P. Multidrug resistance-associated protein 2 expression is upregulated by adenosine 5′-triphosphate in colorectal cancer cells and enhances their survival to chemotherapeutic drugs. PLoS ONE 2015, 10, e0136080. [Google Scholar] [CrossRef]
- Hlavata, I.; Mohelnikova-Duchonova, B.; Vaclavikova, R.; Liska, V.; Pitule, P.; Novak, P.; Bruha, J.; Vycital, O.; Holubec, L.; Treska, V.; et al. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis 2012, 27, 187–196. [Google Scholar] [CrossRef]
- Akerfeldt, M.C.; Tran, C.M.; Shen, C.; Hambley, T.W.; New, E.J. Interactions of cisplatin and the copper transporter CTR1 in human colon cancer cells. J. Biol. Inorg. Chem. 2017, 22, 765–774. [Google Scholar] [CrossRef]
- Grant, C.E.; Valdimarsson, G.; Hipfner, D.R.; Almquist, K.C.; Cole, S.P.; Deeley, R.G. Overexpression of multidrug resistance-associated protein (MRP) increases resistance to natural product drugs. Cancer Res. 1994, 54, 357–361. [Google Scholar] [PubMed]
- Cao, D.; Qin, S.; Mu, Y.; Zhong, M. The role of MRP1 in the multidrug resistance of colorectal cancer. Oncol. Lett. 2017, 13, 2471–2476. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Funayama, R.; Ohnuma, S.; Unno, M.; Nakayama, K. Wnt-beta-catenin signaling regulates ABCC3 (MRP3) transporter expression in colorectal cancer. Cancer Sci. 2016, 107, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Meng, F.; Wang, L.; Mao, Y.; Zhou, H.; Hua, D.; Zhang, H.; Wang, W. A polymorphism in ABCC4 is related to efficacy of 5-FU/capecitabine-based chemotherapy in colorectal cancer patients. Sci. Rep. 2017, 7, 7059. [Google Scholar] [CrossRef] [PubMed]
- Wielinga, P.; Hooijberg, J.H.; Gunnarsdottir, S.; Kathmann, I.; Reid, G.; Zelcer, N.; van der Born, K.; de Haas, M.; van der Heijden, I.; Kaspers, G.; et al. The human multidrug resistance protein MRP5 transports folates and can mediate cellular resistance against antifolates. Cancer Res. 2005, 65, 4425–4430. [Google Scholar] [CrossRef]
- Zhao, J.; Li, W.; Zhu, D.; Yu, Q.; Zhang, Z.; Sun, M.; Cai, S.; Zhang, W. Association of single nucleotide polymorphisms in MTHFR and ABCG2 with the different efficacy of first-line chemotherapy in metastatic colorectal cancer. Med. Oncol. 2014, 31, 802. [Google Scholar] [CrossRef]
- Tuy, H.D.; Shiomi, H.; Mukaisho, K.I.; Naka, S.; Shimizu, T.; Sonoda, H.; Mekata, E.; Endo, Y.; Kurumi, Y.; Sugihara, H.; et al. ABCG2 expression in colorectal adenocarcinomas may predict resistance to irinotecan. Oncol. Lett. 2016, 12, 2752–2760. [Google Scholar] [CrossRef]
- Martinez-Balibrea, E.; Martinez-Cardus, A.; Musulen, E.; Gines, A.; Manzano, J.L.; Aranda, E.; Plasencia, C.; Neamati, N.; Abad, A. Increased levels of copper efflux transporter ATP7B are associated with poor outcome in colorectal cancer patients receiving oxaliplatin-based chemotherapy. Int. J. Cancer 2009, 124, 2905–2910. [Google Scholar] [CrossRef]
- Kap, E.J.; Seibold, P.; Scherer, D.; Habermann, N.; Balavarca, Y.; Jansen, L.; Zucknick, M.; Becker, N.; Hoffmeister, M.; Ulrich, A.; et al. SNPs in transporter and metabolizing genes as predictive markers for oxaliplatin treatment in colorectal cancer patients. Int. J. Cancer 2016, 138, 2993–3001. [Google Scholar] [CrossRef]
- Kitazono, M.; Sumizawa, T.; Takebayashi, Y.; Chen, Z.S.; Furukawa, T.; Nagayama, S.; Tani, A.; Takao, S.; Aikou, T.; Akiyama, S. Multidrug resistance and the lung resistance-related protein in human colon carcinoma SW-620 cells. J. Natl. Cancer Inst. 1999, 91, 1647–1653. [Google Scholar] [CrossRef]
- Izquierdo, M.A.; Scheffer, G.L.; Flens, M.J.; Giaccone, G.; Broxterman, H.J.; Meijer, C.J.; van der Valk, P.; Scheper, R.J. Broad distribution of the multidrug resistance-related vault lung resistance protein in normal human tissues and tumors. Am. J. Pathol. 1996, 148, 877–887. [Google Scholar] [PubMed]
- Herraez, E.; Gonzalez-Sanchez, E.; Vaquero, J.; Romero, M.R.; Serrano, M.A.; Marin, J.J.; Briz, O. Cisplatin-induced chemoresistance in colon cancer cells involves FXR-dependent and FXR-independent up-regulation of ABC proteins. Mol. Pharm. 2012, 9, 2565–2576. [Google Scholar] [CrossRef] [PubMed]
- Tepsiri, N.; Chaturat, L.; Sripa, B.; Namwat, W.; Wongkham, S.; Bhudhisawasdi, V.; Tassaneeyakul, W. Drug sensitivity and drug resistance profiles of human intrahepatic cholangiocarcinoma cell lines. World J. Gastroenterol. 2005, 11, 2748–2753. [Google Scholar] [CrossRef] [PubMed]
- Rau, S.; Autschbach, F.; Riedel, H.D.; Konig, J.; Kulaksiz, H.; Stiehl, A.; Riemann, J.F.; Rost, D. Expression of the multidrug resistance proteins MRP2 and MRP3 in human cholangiocellular carcinomas. Eur. J. Clin. Investig. 2008, 38, 134–142. [Google Scholar] [CrossRef]
- Tian, Q.; Zhang, J.; Tan, T.M.; Chan, E.; Duan, W.; Chan, S.Y.; Boelsterli, U.A.; Ho, P.C.; Yang, H.; Bian, J.S.; et al. Human multidrug resistance associated protein 4 confers resistance to camptothecins. Pharm. Res. 2005, 22, 1837–1853. [Google Scholar] [CrossRef]
- Pratt, S.; Shepard, R.L.; Kandasamy, R.A.; Johnston, P.A.; Perry, W., 3rd; Dantzig, A.H. The multidrug resistance protein 5 (ABCC5) confers resistance to 5-fluorouracil and transports its monophosphorylated metabolites. Mol. Cancer Ther. 2005, 4, 855–863. [Google Scholar] [CrossRef]
- Gradilone, A.; Pulcinelli, F.M.; Lotti, L.V.; Trifiro, E.; Martino, S.; Gandini, O.; Gianni, W.; Frati, L.; Agliano, A.M.; Gazzaniga, P. Celecoxib upregulates multidrug resistance proteins in colon cancer: Lack of synergy with standard chemotherapy. Curr. Cancer Drug Targets 2008, 8, 414–420. [Google Scholar] [CrossRef]
- Lazaris, A.C.; Kavantzas, N.G.; Zorzos, H.S.; Tsavaris, N.V.; Davaris, P.S. Markers of drug resistance in relapsing colon cancer. J. Cancer Res. Clin. Oncol. 2002, 128, 114–118. [Google Scholar] [CrossRef]
- Olszewski, U.; Liedauer, R.; Ausch, C.; Thalhammer, T.; Hamilton, G. Overexpression of CYP3A4 in a COLO 205 Colon Cancer Stem Cell Model in vitro. Cancers 2011, 3, 1467–1479. [Google Scholar] [CrossRef]
- Buck, E.; Sprick, M.; Gaida, M.M.; Grullich, C.; Weber, T.F.; Herpel, E.; Bruckner, T.; Koschny, R. Tumor response to irinotecan is associated with CYP3A5 expression in colorectal cancer. Oncol. Lett. 2019, 17, 3890–3898. [Google Scholar] [CrossRef]
- Untereiner, A.A.; Pavlidou, A.; Druzhyna, N.; Papapetropoulos, A.; Hellmich, M.R.; Szabo, C. Drug resistance induces the upregulation of H2S-producing enzymes in HCT116 colon cancer cells. Biochem. Pharmacol. 2018, 149, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Pavillard, V.; Agostini, C.; Richard, S.; Charasson, V.; Montaudon, D.; Robert, J. Determinants of the cytotoxicity of irinotecan in two human colorectal tumor cell lines. Cancer Chemother. Pharmacol. 2002, 49, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhang, W.; Ma, M.K.; McLeod, H.L. Human carboxylesterase 2 is commonly expressed in tumor tissue and is correlated with activation of irinotecan. Clin. Cancer Res. 2002, 8, 2605–2611. [Google Scholar] [PubMed]
- Shaojun, C.; Li, H.; Haixin, H.; Guisheng, L. Expression of Topoisomerase 1 and carboxylesterase 2 correlates with irinotecan treatment response in metastatic colorectal cancer. Cancer Biol. Ther. 2018, 19, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Ribelles, N.; Lopez-Siles, J.; Sanchez, A.; Gonzalez, E.; Sanchez, M.J.; Carabantes, F.; Sanchez-Rovira, P.; Marquez, A.; Duenas, R.; Sevilla, I.; et al. A carboxylesterase 2 gene polymorphism as predictor of capecitabine on response and time to progression. Curr. Drug Metab. 2008, 9, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, L.; Sun, Y.; Chen, Y.; Wang, X.; Xu, M.; Chi, P.; Xu, Z.; Lu, X. Overexpressed CES2 has prognostic value in CRC and knockdown CES2 reverses L-OHP-resistance in CRC cells by inhibition of the PI3K signaling pathway. Exp. Cell Res. 2020, 389, 111856. [Google Scholar] [CrossRef] [PubMed]
- Salonga, D.; Danenberg, K.D.; Johnson, M.; Metzger, R.; Groshen, S.; Tsao-Wei, D.D.; Lenz, H.J.; Leichman, C.G.; Leichman, L.; Diasio, R.B.; et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin. Cancer Res. 2000, 6, 1322–1327. [Google Scholar]
- Chai, J.; Dong, W.; Xie, C.; Wang, L.; Han, D.L.; Wang, S.; Guo, H.L.; Zhang, Z.L. MicroRNA-494 sensitizes colon cancer cells to fluorouracil through regulation of DPYD. IUBMB Life 2015, 67, 191–201. [Google Scholar] [CrossRef]
- Ahmed, F.Y.; Johnston, S.J.; Cassidy, J.; O’Kelly, T.; Binnie, N.; Murray, G.I.; van Gennip, A.H.; Abeling, N.G.; Knight, S.; McLeod, H.L. Eniluracil treatment completely inactivates dihydropyrimidine dehydrogenase in colorectal tumors. J. Clin. Oncol. 1999, 17, 2439–2445. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Luo, D.D.; Wan, S.B.; Qu, X.J. S1PR2 inhibitors potently reverse 5-FU resistance by downregulating DPD expression in colorectal cancer. Pharmacol. Res. 2020, 155, 104717. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, J.; Yao, X.; Jiang, C.; Ni, P.; Cheng, L.; Liu, J.; Ni, S.; Chen, Q.; Li, Q.; et al. Bevacizumab-enhanced antitumor effect of 5-fluorouracil via upregulation of thymidine phosphorylase through vascular endothelial growth factor A/vascular endothelial growth factor receptor 2-specificity protein 1 pathway. Cancer Sci. 2018, 109, 3294–3304. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.C.; Huang, Y.T.; Ma, C.M.; Chou, W.Y.; Lin-Chao, S. Overexpression of glutathione S-transferase and elevation of thiol pools in a multidrug-resistant human colon cancer cell line. Mol. Pharmacol. 1992, 41, 69–75. [Google Scholar] [PubMed]
- Kuo, M.T.; Bao, J.J.; Curley, S.A.; Ikeguchi, M.; Johnston, D.A.; Ishikawa, T. Frequent coordinated overexpression of the MRP/GS-X pump and gamma-glutamylcysteine synthetase genes in human colorectal cancers. Cancer Res. 1996, 56, 3642–3644. [Google Scholar] [PubMed]
- Chen, X.; Chen, X.Z.; Liu, M.; Lang, N.; Tang, Q.L.; Chen, J.; Zhao, X.F.; Bi, F. [Analysis of gene expression patterns in an irinotecan-resistance colon cancer cell by cDNA microarray]. Sichuan Da Xue Xue Bao. Yi Xue Ban = J. Sichuan Univ. Med. Sci. Ed. 2011, 42, 15–18. [Google Scholar]
- Piaggi, S.; Raggi, C.; Corti, A.; Pitzalis, E.; Mascherpa, M.C.; Saviozzi, M.; Pompella, A.; Casini, A.F. Glutathione transferase omega 1-1 (GSTO1-1) plays an anti-apoptotic role in cell resistance to cisplatin toxicity. Carcinogenesis 2010, 31, 804–811. [Google Scholar] [CrossRef]
- Cummings, J.; Boyd, G.; Ethell, B.T.; Macpherson, J.S.; Burchell, B.; Smyth, J.F.; Jodrell, D.I. Enhanced clearance of topoisomerase I inhibitors from human colon cancer cells by glucuronidation. Biochem. Pharmacol. 2002, 63, 607–613. [Google Scholar] [CrossRef]
- Cummings, J.; Ethell, B.T.; Jardine, L.; Boyd, G.; Macpherson, J.S.; Burchell, B.; Smyth, J.F.; Jodrell, D.I. Glucuronidation as a mechanism of intrinsic drug resistance in human colon cancer: Reversal of resistance by food additives. Cancer Res. 2003, 63, 8443–8450. [Google Scholar]
- Meijer, C.; Timmer, A.; De Vries, E.G.; Groten, J.P.; Knol, A.; Zwart, N.; Dam, W.A.; Sleijfer, D.T.; Mulder, N.H. Role of metallothionein in cisplatin sensitivity of germ-cell tumours. Int. J. Cancer 2000, 85, 777–781. [Google Scholar] [CrossRef]
- Hishikawa, Y.; Kohno, H.; Ueda, S.; Kimoto, T.; Dhar, D.K.; Kubota, H.; Tachibana, M.; Koji, T.; Nagasue, N. Expression of metallothionein in colorectal cancers and synchronous liver metastases. Oncology 2001, 61, 162–167. [Google Scholar] [CrossRef]
- Beckett, G.J.; Hayes, J.D. Glutathione S-transferases: Biomedical applications. Adv. Clin. Chem. 1993, 30, 281–380. [Google Scholar] [CrossRef]
- Stoehlmacher, J.; Park, D.J.; Zhang, W.; Groshen, S.; Tsao-Wei, D.D.; Yu, M.C.; Lenz, H.J. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J. Natl. Cancer Inst. 2002, 94, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Bulus, H.; Oguztuzun, S.; Guler Simsek, G.; Kilic, M.; Ada, A.O.; Gol, S.; Kocdogan, A.K.; Kaygin, P.; Bozer, B.; Iscan, M. Expression of CYP and GST in human normal and colon tumor tissues. Biotech. Histochem. 2019, 94, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, K.; Samanta, S.; Kyani, A.; Yang, S.; Tamura, S.; Ziemke, E.; Stuckey, J.A.; Li, S.; Chinnaswamy, K.; Otake, H.; et al. Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor. Nat. Commun. 2016, 7, 13084. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Yamano, T.; Igeta, M.; Imada, A.; Jihyung, S.; Babaya, A.; Hamanaka, M.; Kobayashi, M.; Tsukamoto, K.; Noda, M.; et al. UGT1A1 polymorphisms in rectal cancer associated with the efficacy and toxicity of preoperative chemoradiotherapy using irinotecan. Cancer Sci. 2018, 109, 3934–3942. [Google Scholar] [CrossRef]
- Paez, D.; Tobena, M.; Fernandez-Plana, J.; Sebio, A.; Virgili, A.C.; Cirera, L.; Barnadas, A.; Riera, P.; Sullivan, I.; Salazar, J. Pharmacogenetic clinical randomised phase II trial to evaluate the efficacy and safety of FOLFIRI with high-dose irinotecan (HD-FOLFIRI) in metastatic colorectal cancer patients according to their UGT1A 1 genotype. Br. J. Cancer 2019, 120, 190–195. [Google Scholar] [CrossRef]
- Wang, M.; Sun, D.F.; Wang, S.; Qing, Y.; Chen, S.; Wu, D.; Lin, Y.M.; Luo, J.Z.; Li, Y.Q. Polymorphic expression of UDP-glucuronosyltransferase UGTlA gene in human colorectal cancer. PLoS ONE 2013, 8, e57045. [Google Scholar] [CrossRef]
- Shimoda, R.; Achanzar, W.E.; Qu, W.; Nagamine, T.; Takagi, H.; Mori, M.; Waalkes, M.P. Metallothionein is a potential negative regulator of apoptosis. Toxicol. Sci. 2003, 73, 294–300. [Google Scholar] [CrossRef]
- Ioachim, E.E.; Goussia, A.C.; Agnantis, N.J.; Machera, M.; Tsianos, E.V.; Kappas, A.M. Prognostic evaluation of metallothionein expression in human colorectal neoplasms. J. Clin. Pathol. 1999, 52, 876–879. [Google Scholar] [CrossRef]
- Cho, Y.B.; Chung, H.J.; Lee, W.Y.; Choi, S.H.; Kim, H.C.; Yun, S.H.; Chun, H.K. Relationship between TYMS and ERCC1 mRNA expression and in vitro chemosensitivity in colorectal cancer. Anticancer Res. 2011, 31, 3843–3849. [Google Scholar]
- Carter, P.; Alifrangis, C.; Chandrasinghe, P.; Cereser, B.; Del Bel Belluz, L.; Leo, C.A.; Moderau, N.; Tabassum, N.; Warusavitarne, J.; Krell, J.; et al. The benefit of tumor molecular profiling on predicting treatments for colorectal adenocarcinomas. Oncotarget 2018, 9, 11371–11376. [Google Scholar] [CrossRef][Green Version]
- Wong, N.A.; Brett, L.; Stewart, M.; Leitch, A.; Longley, D.B.; Dunlop, M.G.; Johnston, P.G.; Lessells, A.M.; Jodrell, D.I. Nuclear thymidylate synthase expression, p53 expression and 5FU response in colorectal carcinoma. Br. J. Cancer 2001, 85, 1937–1943. [Google Scholar] [CrossRef] [PubMed]
- Johnston, P.G.; Lenz, H.J.; Leichman, C.G.; Danenberg, K.D.; Allegra, C.J.; Danenberg, P.V.; Leichman, L. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995, 55, 1407–1412. [Google Scholar] [PubMed]
- Lindebjerg, J.; Nielsen, J.N.; Hoeffding, L.D.; Jakobsen, A. Immunohistochemical expression of thymidylate synthase as predictor of response to capecitabine in patients with advanced colorectal adenocarcinoma. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2005, 113, 600–602. [Google Scholar] [CrossRef]
- Johnston, P.G.; Benson, A.B., 3rd; Catalano, P.; Rao, M.S.; O’Dwyer, P.J.; Allegra, C.J. Thymidylate synthase protein expression in primary colorectal cancer: Lack of correlation with outcome and response to fluorouracil in metastatic disease sites. J. Clin. Oncol. 2003, 21, 815–819. [Google Scholar] [CrossRef] [PubMed]
- Personeni, N.; Hendlisz, A.; Gallez, J.; Galdon, M.G.; Larsimont, D.; Van Laethem, J.L.; Nagy, N.; Barette, M.; Paesmans, M.; Cardoso, F.; et al. Correlation between the response to cetuximab alone or in combination with irinotecan and the activated/phosphorylated epidermal growth factor receptor in metastatic colorectal cancer. Semin. Oncol. 2005, 32, S59–S62. [Google Scholar] [CrossRef]
- Algars, A.; Sundstrom, J.; Lintunen, M.; Jokilehto, T.; Kytola, S.; Kaare, M.; Vainionpaa, R.; Orpana, A.; Osterlund, P.; Ristimaki, A.; et al. EGFR gene copy number predicts response to anti-EGFR treatment in RAS wild type and RAS/BRAF/PIK3CA wild type metastatic colorectal cancer. Int. J. Cancer 2017, 140, 922–929. [Google Scholar] [CrossRef]
- Siena, S.; Sartore-Bianchi, A.; Marsoni, S.; Hurwitz, H.I.; McCall, S.J.; Penault-Llorca, F.; Srock, S.; Bardelli, A.; Trusolino, L. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann. Oncol. 2018, 29, 1108–1119. [Google Scholar] [CrossRef]
- Gharib, E.; Salmanipour, R.; Nazemalhosseini Mojarad, E.; Yaghoob Taleghani, M.; Sarlak, S.; Malekzade-Moghani, M.; Nasrabadi, P.N.; Meiary, M.A.; Asadzadeh Aghdaei, H.; Zali, M.R. HER2(+) mCRC patients with exon 20 R784G substitution mutation do not respond to the cetuximab therapy. J. Cell. Physiol. 2019, 234, 13137–13144. [Google Scholar] [CrossRef]
- Hegde, P.S.; Jubb, A.M.; Chen, D.; Li, N.F.; Meng, Y.G.; Bernaards, C.; Elliott, R.; Scherer, S.J.; Chen, D.S. Predictive impact of circulating vascular endothelial growth factor in four phase III trials evaluating bevacizumab. Clin. Cancer Res. 2013, 19, 929–937. [Google Scholar] [CrossRef]
- Baba, H.; Baba, Y.; Uemoto, S.; Yoshida, K.; Saiura, A.; Watanabe, M.; Maehara, Y.; Oki, E.; Ikeda, Y.; Matsuda, H.; et al. Changes in expression levels of ERCC1, DPYD, and VEGFA mRNA after first-line chemotherapy of metastatic colorectal cancer: Results of a multicenter study. Oncotarget 2015, 6, 34004–34013. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Paccard, C.; Chiron, M.; Tabernero, J. Impact of prior bevacizumab treatment on VEGF-A and PlGF levels and outcome following second-line aflibercept treatment: Biomarker post hoc analysis of the VELOUR trial. Clin. Cancer Res. 2020, 26, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Willett, C.G.; Duda, D.G.; di Tomaso, E.; Boucher, Y.; Ancukiewicz, M.; Sahani, D.V.; Lahdenranta, J.; Chung, D.C.; Fischman, A.J.; Lauwers, G.Y.; et al. Efficacy, safety, and biomarkers of neoadjuvant bevacizumab, radiation therapy, and fluorouracil in rectal cancer: A multidisciplinary phase II study. J. Clin. Oncol. 2009, 27, 3020–3026. [Google Scholar] [CrossRef] [PubMed]
- Chiron, M.; Bagley, R.G.; Pollard, J.; Mankoo, P.K.; Henry, C.; Vincent, L.; Geslin, C.; Baltes, N.; Bergstrom, D.A. Differential antitumor activity of aflibercept and bevacizumab in patient-derived xenograft models of colorectal cancer. Mol. Cancer Ther. 2014, 13, 1636–1644. [Google Scholar] [CrossRef] [PubMed]
- Lieu, C.H.; Tran, H.; Jiang, Z.Q.; Mao, M.; Overman, M.J.; Lin, E.; Eng, C.; Morris, J.; Ellis, L.; Heymach, J.V.; et al. The association of alternate VEGF ligands with resistance to anti-VEGF therapy in metastatic colorectal cancer. PLoS ONE 2013, 8, e77117. [Google Scholar] [CrossRef]
- Lim, Y.H.; Odell, I.D.; Ko, C.J.; Choate, K.A. Somatic p.T771R KDR (VEGFR2) mutation arising in a sporadic angioma during ramucirumab therapy. JAMA Dermatol. 2015, 151, 1240–1243. [Google Scholar] [CrossRef]
- Loaiza-Bonilla, A.; Jensen, C.E.; Shroff, S.; Furth, E.; Bonilla-Reyes, P.A.; Deik, A.F.; Morrissette, J. KDR mutation as a novel predictive biomarker of exceptional response to regorafenib in metastatic colorectal cancer. Cureus 2016, 8, e478. [Google Scholar] [CrossRef]
- Callebout, E.; Ribeiro, S.M.; Laurent, S.; De Man, M.; Ferdinande, L.; Claes, K.B.M.; Van der Meulen, J.; Geboes, K.P. Long term response on Regorafenib in non-V600E BRAF mutated colon cancer: A case report. BMC Cancer 2019, 19, 567. [Google Scholar] [CrossRef]
- Li, P.; Fang, Y.J.; Li, F.; Ou, Q.J.; Chen, G.; Ma, G. ERCC1, defective mismatch repair status as predictive biomarkers of survival for stage III colon cancer patients receiving oxaliplatin-based adjuvant chemotherapy. Br. J. Cancer 2013, 108, 1238–1244. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, G.; Li, W. Elevated expression of ERCC6 confers resistance to 5-fluorouracil and is associated with poor patient survival in colorectal cancer. DNA Cell Biol. 2017, 36, 781–786. [Google Scholar] [CrossRef]
- Feng, X.; Liu, J.; Gong, Y.; Gou, K.; Yang, H.; Yuan, Y.; Xing, C. DNA repair protein XPA is differentially expressed in colorectal cancer and predicts better prognosis. Cancer Med. 2018, 7, 2339–2349. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, J.; Meng, Y.; Qu, C.; Shen, F.; Xu, L. Overexpression of xeroderma pigmentosum group C decreases the chemotherapeutic sensitivity of colorectal carcinoma cells to cisplatin. Oncol. Lett. 2018, 15, 6336–6344. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.B.; Chen, Y.; Meng, X.D.; Yu, P.; He, X.; Li, J. Nucleotide excision repair factor XPC ameliorates prognosis by increasing the susceptibility of human colorectal cancer to chemotherapy and ionizing radiation. Front. Oncol. 2018, 8, 290. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Yan, J.; Martinez-Balibrea, E.; Graziano, F.; Lenz, H.J.; Kim, H.J.; Robert, J.; Im, S.A.; Wang, W.S.; Etienne-Grimaldi, M.C.; et al. ERCC1 and ERCC2 polymorphisms predict clinical outcomes of oxaliplatin-based chemotherapies in gastric and colorectal cancer: A systemic review and meta-analysis. Clin. Cancer Res. 2011, 17, 1632–1640. [Google Scholar] [CrossRef] [PubMed]
- Pare, L.; Marcuello, E.; Altes, A.; del Rio, E.; Sedano, L.; Salazar, J.; Cortes, A.; Barnadas, A.; Baiget, M. Pharmacogenetic prediction of clinical outcome in advanced colorectal cancer patients receiving oxaliplatin/5-fluorouracil as first-line chemotherapy. Br. J. Cancer 2008, 99, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Boige, V.; Mollevi, C.; Gourgou, S.; Azria, D.; Seitz, J.F.; Vincent, M.; Bigot, L.; Juzyna, B.; Miran, I.; Gerard, J.P.; et al. Impact of single-nucleotide polymorphisms in DNA repair pathway genes on response to chemoradiotherapy in rectal cancer patients: Results from ACCORD-12/PRODIGE-2 phase III trial. Int. J. Cancer 2019, 145, 3163–3172. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, F.; Chen, K.; Wang, D.; Jiang, H.; Li, J.; Pan, F.; Chen, S.; Zhang, Y.; Ruan, Z.; et al. ERCC5 promoter polymorphisms at −763 and +25 predict the response to oxaliplatin-based chemotherapy in patients with advanced colorectal cancer. Cancer Biol. Ther. 2009, 8, 1424–1430. [Google Scholar] [CrossRef]
- Zhang, C.M.; Lv, J.F.; Gong, L.; Yu, L.Y.; Chen, X.P.; Zhou, H.H.; Fan, L. Role of deficient mismatch repair in the personalized management of colorectal cancer. Int. J. Environ. Res. Public Health 2016, 13, 892. [Google Scholar] [CrossRef]
- Jover, R.; Zapater, P.; Castells, A.; Llor, X.; Andreu, M.; Cubiella, J.; Balaguer, F.; Sempere, L.; Xicola, R.M.; Bujanda, L.; et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur. J. Cancer 2009, 45, 365–373. [Google Scholar] [CrossRef]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef]
- Alex, A.K.; Siqueira, S.; Coudry, R.; Santos, J.; Alves, M.; Hoff, P.M.; Riechelmann, R.P. Response to chemotherapy and prognosis in metastatic colorectal cancer with DNA deficient mismatch repair. Clin. Colorectal Cancer 2017, 16, 228–239. [Google Scholar] [CrossRef]
- Cercek, A.; Dos Santos Fernandes, G.; Roxburgh, C.S.; Ganesh, K.; Ng, S.; Sanchez-Vega, F.; Yaeger, R.; Segal, N.H.; Reidy-Lagunes, D.L.; Varghese, A.M.; et al. Mismatch repair-deficient rectal cancer and resistance to neoadjuvant chemotherapy. Clin. Cancer Res. 2020. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, Y.; Shen, H.; Kapesa, L.; Liu, W.; Zeng, M.; Zeng, S. Association between mismatch repair gene and irinotecan-based chemotherapy in metastatic colon cancer. Tumour Biol. 2015, 36, 9599–9609. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, X.; Zou, S.M.; Li, H.M.; Xiao, Q.; Feng, Y.R.; Huang, Y.; Feng, T.; Chen, J.N.; Lin, D.X.; et al. [Genetic variations in MLH3 and MSH2 genes are associated with the sensitivity and prognosis in locally advanced rectal cancer patients receiving preoperative chemoradiotherapy]. Zhonghua Zhong Liu Za Zhi [Chin. J. Oncol.] 2018, 40, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zeng, J.; Roychoudhury, S.; Biswas, P.; Mohapatra, B.; Ray, S.; Dowlatshahi, K.; Wang, J.; Band, V.; Talmon, G.; et al. Targeting histone chaperone FACT complex overcomes 5-fluorouracil resistance in colon cancer. Mol. Cancer Ther. 2020, 19, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Leguisamo, N.M.; Gloria, H.C.; Kalil, A.N.; Martins, T.V.; Azambuja, D.B.; Meira, L.B.; Saffi, J. Base excision repair imbalance in colorectal cancer has prognostic value and modulates response to chemotherapy. Oncotarget 2017, 8, 54199–54214. [Google Scholar] [CrossRef]
- Takayama, T.; Miyanishi, K.; Hayashi, T.; Sato, Y.; Niitsu, Y. Colorectal cancer: Genetics of development and metastasis. J. Gastroenterol. 2006, 41, 185–192. [Google Scholar] [CrossRef]
- Laforest, A.; Aparicio, T.; Zaanan, A.; Silva, F.P.; Didelot, A.; Desbeaux, A.; Le Corre, D.; Benhaim, L.; Pallier, K.; Aust, D.; et al. ERBB2 gene as a potential therapeutic target in small bowel adenocarcinoma. Eur. J. Cancer 2014, 50, 1740–1746. [Google Scholar] [CrossRef]
- Schrock, A.B.; Devoe, C.E.; McWilliams, R.; Sun, J.; Aparicio, T.; Stephens, P.J.; Ross, J.S.; Wilson, R.; Miller, V.A.; Ali, S.M.; et al. Genomic profiling of small-bowel adenocarcinoma. JAMA Oncol. 2017, 3, 1546–1553. [Google Scholar] [CrossRef]
- Tominaga, T.; Iwahashi, M.; Takifuji, K.; Hotta, T.; Yokoyama, S.; Matsuda, K.; Higashiguchi, T.; Oku, Y.; Nasu, T.; Yamaue, H. Combination of p53 codon 72 polymorphism and inactive p53 mutation predicts chemosensitivity to 5-fluorouracil in colorectal cancer. Int. J. Cancer 2010, 126, 1691–1701. [Google Scholar] [CrossRef]
- Benhattar, J.; Cerottini, J.P.; Saraga, E.; Metthez, G.; Givel, J.C. p53 mutations as a possible predictor of response to chemotherapy in metastatic colorectal carcinomas. Int. J. Cancer 1996, 69, 190–192. [Google Scholar] [CrossRef]
- Toscano, F.; Parmentier, B.; Fajoui, Z.E.; Estornes, Y.; Chayvialle, J.A.; Saurin, J.C.; Abello, J. p53 dependent and independent sensitivity to oxaliplatin of colon cancer cells. Biochem. Pharmacol. 2007, 74, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Chen, G.; Qiu, Y.; Yuan, Z.; Li, H.; Yuan, X.; Sun, J.; Xu, J.; Liang, X.; Yin, P. miR-503-5p confers drug resistance by targeting PUMA in colorectal carcinoma. Oncotarget 2017, 8, 21719–21732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Deng, X.; Ren, X.; Zhang, B.; Chen, X.; Yang, J.; Ding, H.; Sui, J.; Song, X. Expression of mutant p53 and of the multidrug resistant proteins P-glycoprotein and glutathione S-transferase-pi correlated in colorectal adenocarcinoma. Scand. J. Gastroenterol. 2010, 45, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.W.; Zhao, P.; Liu, M.; Dong, X.S.; Tao, J.; Yao, X.Q.; Yin, X.H.; Li, Y.; Fu, S.B. Reversal of 5-flouroucial resistance by adenovirus-mediated transfer of wild-type p53 gene in multidrug-resistant human colon carcinoma LoVo/5-FU cells. World J. Gastroenterol. 2004, 10, 1979–1983. [Google Scholar] [CrossRef] [PubMed]
- Kandioler, D.; Mittlbock, M.; Kappel, S.; Puhalla, H.; Herbst, F.; Langner, C.; Wolf, B.; Tschmelitsch, J.; Schippinger, W.; Steger, G.; et al. TP53 mutational status and prediction of benefit from adjuvant 5-fluorouracil in stage III Colon cancer patients. EBioMedicine 2015, 2, 825–830. [Google Scholar] [CrossRef]
- Oh, H.J.; Bae, J.M.; Wen, X.; Jung, S.; Kim, Y.; Kim, K.J.; Cho, N.Y.; Kim, J.H.; Han, S.W.; Kim, T.Y.; et al. p53 expression status is associated with cancer-specific survival in stage III and high-risk stage II colorectal cancer patients treated with oxaliplatin-based adjuvant chemotherapy. Br. J. Cancer 2019, 120, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Netter, J.; Lehmann-Che, J.; Lambert, J.; Tallet, A.; Lourenco, N.; Soliman, H.; Bertheau, P.; Pariente, B.; Chirica, M.; Pocard, M.; et al. Functional TP53 mutations have no impact on response to cytotoxic agents in metastatic colon cancer. Bull. Cancer 2015, 102, 117–125. [Google Scholar] [CrossRef]
- Zhang, Y.; Geng, L.; Talmon, G.; Wang, J. MicroRNA-520g confers drug resistance by regulating p21 expression in colorectal cancer. J. Biol. Chem. 2015, 290, 6215–6225. [Google Scholar] [CrossRef]
- Wang, W.; Guo, W.; Li, L.; Fu, Z.; Liu, W.; Gao, J.; Shu, Y.; Xu, Q.; Sun, Y.; Gu, Y. Andrographolide reversed 5-FU resistance in human colorectal cancer by elevating BAX expression. Biochem. Pharmacol. 2016, 121, 8–17. [Google Scholar] [CrossRef]
- Manoochehri, M.; Karbasi, A.; Bandehpour, M.; Kazemi, B. Down-regulation of BAX gene during carcinogenesis and acquisition of resistance to 5-FU in colorectal cancer. Pathol. Oncol. Res. 2014, 20, 301–307. [Google Scholar] [CrossRef]
- Gao, C.; Wang, A.Y. Significance of increased apoptosis and Bax expression in human small intestinal adenocarcinoma. J. Histochem. Cytochem. 2009, 57, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.A.; Rego, R.L.; Foster, N.R.; Thibodeau, S.N.; Alberts, S.R.; Windschitl, H.E.; Sargent, D.J. Proapoptotic Bad and Bid protein expression predict survival in stages II and III colon cancers. Clin. Cancer Res. 2008, 14, 4128–4133. [Google Scholar] [CrossRef] [PubMed]
- Yin, A.; Jiang, Y.; Zhang, X.; Luo, H. Overexpression of FADD enhances 5-fluorouracil-induced apoptosis in colorectal adenocarcinoma cells. Med. Oncol. 2010, 27, 397–405. [Google Scholar] [CrossRef]
- Sui, G.; Qiu, Y.; Yu, H.; Kong, Q.; Zhen, B. Interleukin-17 promotes the development of cisplatin resistance in colorectal cancer. Oncol. Lett. 2019, 17, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Liang, X.; Cui, D.; Wu, Y.; Shi, W.; Liu, J. miR-1915 inhibits Bcl-2 to modulate multidrug resistance by increasing drug-sensitivity in human colorectal carcinoma cells. Mol. Carcinog. 2013, 52, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.W.; Huang, C.C.; Chang, S.W.; Chen, T.H.; Lee, H. Bcl-2 stabilization by paxillin confers 5-fluorouracil resistance in colorectal cancer. Cell Death Differ. 2015, 22, 779–789. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Liu, J.; Zhang, T. Knockdown of REG ialpha enhances the sensitivity to 5-fluorouracil of colorectal cancer cells via cyclin D1/CDK4 pathway and BAX/BCL-2 pathways. Cancer Biother. Radiopharm. 2019, 34, 362–370. [Google Scholar] [CrossRef]
- Koehler, B.C.; Jager, D.; Schulze-Bergkamen, H. Targeting cell death signaling in colorectal cancer: Current strategies and future perspectives. World J. Gastroenterol. 2014, 20, 1923–1934. [Google Scholar] [CrossRef]
- Miura, K.; Fujibuchi, W.; Ishida, K.; Naitoh, T.; Ogawa, H.; Ando, T.; Yazaki, N.; Watanabe, K.; Haneda, S.; Shibata, C.; et al. Inhibitor of apoptosis protein family as diagnostic markers and therapeutic targets of colorectal cancer. Surg. Today 2011, 41, 175–182. [Google Scholar] [CrossRef]
- Krajewska, M.; Kim, H.; Kim, C.; Kang, H.; Welsh, K.; Matsuzawa, S.; Tsukamoto, M.; Thomas, R.G.; Assa-Munt, N.; Piao, Z.; et al. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin. Cancer Res. 2005, 11, 5451–5461. [Google Scholar] [CrossRef]
- Miura, K.; Karasawa, H.; Sasaki, I. cIAP2 as a therapeutic target in colorectal cancer and other malignancies. Expert Opin. Ther. Targets 2009, 13, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Backus, H.H.; van Riel, J.M.; van Groeningen, C.J.; Vos, W.; Dukers, D.F.; Bloemena, E.; Wouters, D.; Pinedo, H.M.; Peters, G.J. Rb, mcl-1 and p53 expression correlate with clinical outcome in patients with liver metastases from colorectal cancer. Ann. Oncol. 2001, 12, 779–785. [Google Scholar] [CrossRef]
- Fang, Z.; Gong, C.; Yu, S.; Zhou, W.; Hassan, W.; Li, H.; Wang, X.; Hu, Y.; Gu, K.; Chen, X.; et al. NFYB-induced high expression of E2F1 contributes to oxaliplatin resistance in colorectal cancer via the enhancement of CHK1 signaling. Cancer Lett. 2018, 415, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hunter, T. Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 2014, 134, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Lind, D.S.; Hochwald, S.N.; Malaty, J.; Rekkas, S.; Hebig, P.; Mishra, G.; Moldawer, L.L.; Copeland, E.M., 3rd; Mackay, S. Nuclear factor-kappa B is upregulated in colorectal cancer. Surgery 2001, 130, 363–369. [Google Scholar] [CrossRef]
- Voboril, R.; Hochwald, S.N.; Li, J.; Brank, A.; Weberova, J.; Wessels, F.; Moldawer, L.L.; Camp, E.R.; MacKay, S.L. Inhibition of NF-kappa B augments sensitivity to 5-fluorouracil/folinic acid in colon cancer. J. Surg. Res. 2004, 120, 178–188. [Google Scholar] [CrossRef]
- Korber, M.I.; Staribacher, A.; Ratzenbock, I.; Steger, G.; Mader, R.M. NFkappaB-associated pathways in progression of chemoresistance to 5-fluorouracil in an in vitro model of colonic carcinoma. Anticancer Res. 2016, 36, 1631–1639. [Google Scholar]
- Liu, B.; Xu, T.; Xu, X.; Cui, Y.; Xing, X. Biglycan promotes the chemotherapy resistance of colon cancer by activating NF-kappaB signal transduction. Mol. Cell. Biochem. 2018, 449, 285–294. [Google Scholar] [CrossRef]
- Yuan, Z.; Liang, X.; Zhan, Y.; Wang, Z.; Xu, J.; Qiu, Y.; Wang, J.; Cao, Y.; Le, V.M.; Ly, H.T.; et al. Targeting CD133 reverses drug-resistance via the AKT/NF-kappaB/MDR1 pathway in colorectal cancer. Br. J. Cancer 2020, 122, 1342–1353. [Google Scholar] [CrossRef]
- Chen, S.P.; Wu, C.C.; Lin, S.Z.; Kang, J.C.; Su, C.C.; Chen, Y.L.; Lin, P.C.; Chiu, S.C.; Pang, C.Y.; Harn, H.J. Prognostic significance of interaction between somatic APC mutations and 5-fluorouracil adjuvant chemotherapy in Taiwanese colorectal cancer subjects. Am. J. Clin. Oncol. 2009, 32, 122–126. [Google Scholar] [CrossRef]
- Zheng, G.; Tseng, L.H.; Chen, G.; Haley, L.; Illei, P.; Gocke, C.D.; Eshleman, J.R.; Lin, M.T. Clinical detection and categorization of uncommon and concomitant mutations involving BRAF. BMC Cancer 2015, 15, 779. [Google Scholar] [CrossRef] [PubMed]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2018, 19, 603–615. [Google Scholar] [CrossRef]
- Douillard, J.Y.; Oliner, K.S.; Siena, S.; Tabernero, J.; Burkes, R.; Barugel, M.; Humblet, Y.; Bodoky, G.; Cunningham, D.; Jassem, J.; et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N. Engl. J. Med. 2013, 369, 1023–1034. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Lenz, H.J.; Kohne, C.H.; Heinemann, V.; Tejpar, S.; Melezinek, I.; Beier, F.; Stroh, C.; Rougier, P.; van Krieken, J.H.; et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J. Clin. Oncol. 2015, 33, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Selvarajan, K.; Hasan, M.R.; Chan, A.P.; Jin, C.; Kim, J.; Chan, S.K.; Le, N.D.; Kim, Y.B.; Tai, I.T. Inhibition of COX-2 in colon cancer modulates tumor growth and MDR-1 expression to enhance tumor regression in therapy-refractory cancers in vivo. Neoplasia 2012, 14, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Saikawa, Y.; Sugiura, T.; Toriumi, F.; Kubota, T.; Suganuma, K.; Isshiki, S.; Otani, Y.; Kumai, K.; Kitajima, M. Cyclooxygenase-2 gene induction causes CDDP resistance in colon cancer cell line, HCT-15. Anticancer Res. 2004, 24, 2723–2728. [Google Scholar]
- Adam, L.; San Lucas, F.A.; Fowler, R.; Yu, Y.; Wu, W.; Liu, Y.; Wang, H.; Menter, D.; Tetzlaff, M.T.; Ensor, J., Jr.; et al. DNA sequencing of small bowel adenocarcinomas identifies targetable recurrent mutations in the ERBB2 signaling pathway. Clin. Cancer Res. 2019, 25, 641–651. [Google Scholar] [CrossRef]
- He, L.; Zhu, H.; Zhou, S.; Wu, T.; Wu, H.; Yang, H.; Mao, H.; SekharKathera, C.; Janardhan, A.; Edick, A.M.; et al. Wnt pathway is involved in 5-FU drug resistance of colorectal cancer cells. Exp. Mol. Med. 2018, 50, 101. [Google Scholar] [CrossRef]
- Aghabozorgi, A.S.; Bahreyni, A.; Soleimani, A.; Bahrami, A.; Khazaei, M.; Ferns, G.A.; Avan, A.; Hassanian, S.M. Role of adenomatous polyposis coli (APC) gene mutations in the pathogenesis of colorectal cancer; current status and perspectives. Biochimie 2019, 157, 64–71. [Google Scholar] [CrossRef]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology 2015, 149, 1204–1225.e12. [Google Scholar] [CrossRef]
- Ozawa, T.; Kazama, S.; Akiyoshi, T.; Murono, K.; Yoneyama, S.; Tanaka, T.; Tanaka, J.; Kiyomatsu, T.; Kawai, K.; Nozawa, H.; et al. Nuclear Notch3 expression is associated with tumor recurrence in patients with stage II and III colorectal cancer. Ann. Surg. Oncol. 2014, 21, 2650–2658. [Google Scholar] [CrossRef]
- Fre, S.; Hannezo, E.; Sale, S.; Huyghe, M.; Lafkas, D.; Kissel, H.; Louvi, A.; Greve, J.; Louvard, D.; Artavanis-Tsakonas, S. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE 2011, 6, e25785. [Google Scholar] [CrossRef]
- Jun, S.Y.; Kim, M.; Jin Gu, M.; Kyung Bae, Y.; Chang, H.K.; Sun Jung, E.; Jang, K.T.; Kim, J.; Yu, E.; Woon Eom, D.; et al. Clinicopathologic and prognostic associations of KRAS and BRAF mutations in small intestinal adenocarcinoma. Mod. Pathol. 2016, 29, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kopetz, S.; Lothe, R.A. Biomarker-guided therapy for colorectal cancer: Strength in complexity. Nat. Rev. Clin. Oncol. 2020, 17, 11–32. [Google Scholar] [CrossRef] [PubMed]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, L.; Esposito, F.; Thomson, T.M.; Maurel, J. The tumor microenvironment in colorectal cancer therapy. Cancers 2019, 11, 1172. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef]
- Chen, J.; Ding, Z.; Peng, Y.; Pan, F.; Li, J.; Zou, L.; Zhang, Y.; Liang, H. HIF-1alpha inhibition reverses multidrug resistance in colon cancer cells via downregulation of MDR1/P-glycoprotein. PLoS ONE 2014, 9, e98882. [Google Scholar] [CrossRef]
- Selvakumaran, M.; Yao, K.S.; Feldman, M.D.; O’Dwyer, P.J. Antitumor effect of the angiogenesis inhibitor bevacizumab is dependent on susceptibility of tumors to hypoxia-induced apoptosis. Biochem. Pharmacol. 2008, 75, 627–638. [Google Scholar] [CrossRef]
- Tang, Y.A.; Chen, Y.F.; Bao, Y.; Mahara, S.; Yatim, S.; Oguz, G.; Lee, P.L.; Feng, M.; Cai, Y.; Tan, E.Y.; et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1alpha and TGF-beta2 to promote chemoresistance in colorectal cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E5990–E5999. [Google Scholar] [CrossRef] [PubMed]
- Lotti, F.; Jarrar, A.M.; Pai, R.K.; Hitomi, M.; Lathia, J.; Mace, A.; Gantt, G.A., Jr.; Sukhdeo, K.; DeVecchio, J.; Vasanji, A.; et al. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. J. Exp. Med. 2013, 210, 2851–2872. [Google Scholar] [CrossRef]
- Malesci, A.; Bianchi, P.; Celesti, G.; Basso, G.; Marchesi, F.; Grizzi, F.; Di Caro, G.; Cavalleri, T.; Rimassa, L.; Palmqvist, R.; et al. Tumor-associated macrophages and response to 5-fluorouracil adjuvant therapy in stage III colorectal cancer. Oncoimmunology 2017, 6, e1342918. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yao, S.; Hu, Y.; Feng, Y.; Li, M.; Bian, Z.; Zhang, J.; Qin, Y.; Qi, X.; Zhou, L.; et al. The immune-microenvironment confers chemoresistance of colorectal cancer through macrophage-derived IL6. Clin. Cancer Res. 2017, 23, 7375–7387. [Google Scholar] [CrossRef] [PubMed]
- Correale, P.; Rotundo, M.S.; Botta, C.; Del Vecchio, M.T.; Ginanneschi, C.; Licchetta, A.; Conca, R.; Apollinari, S.; De Luca, F.; Tassone, P.; et al. Tumor infiltration by T lymphocytes expressing chemokine receptor 7 (CCR7) is predictive of favorable outcome in patients with advanced colorectal carcinoma. Clin. Cancer Res. 2012, 18, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Galluzzi, L.; Viaud, S.; Vetizou, M.; Daillere, R.; Merad, M.; Kroemer, G. Cancer and the gut microbiota: An unexpected link. Sci. Transl. Med. 2015, 7, 271ps1. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Yang, Y.N.; Zhang, R.; Du, J.W.; Yuan, H.H.; Li, Y.J.; Wei, X.L.; Du, X.X.; Jiang, S.L.; Han, Y. Predictive role of UCA1-containing exosomes in cetuximab-resistant colorectal cancer. Cancer Cell Int. 2018, 18, 164. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Zhou, Y.; Tozzi, F.; Chen, J.; Fan, F.; Xia, L.; Wang, J.; Gao, G.; Zhang, A.; Xia, X.; Brasher, H.; et al. Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 2012, 72, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Giallombardo, M.; Taverna, S.; Alessandro, R.; Hong, D.; Rolfo, C. Exosome-mediated drug resistance in cancer: The near future is here. Ther. Adv. Med. Oncol. 2016, 8, 320–322. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yan, C.; Mu, L.; Huang, K.; Li, X.; Tao, D.; Wu, Y.; Qin, J. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS ONE 2015, 10, e0125625. [Google Scholar] [CrossRef]
- Kikuchi, K.; Hoshino, D. Sensitization of HT29 colorectal cancer cells to vemurafenib in three-dimensional collagen cultures. Cell Biol. Int. 2020, 44, 621–629. [Google Scholar] [CrossRef]
- Li, C.; Singh, B.; Graves-Deal, R.; Ma, H.; Starchenko, A.; Fry, W.H.; Lu, Y.; Wang, Y.; Bogatcheva, G.; Khan, M.P.; et al. Three-dimensional culture system identifies a new mode of cetuximab resistance and disease-relevant genes in colorectal cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E2852–E2861. [Google Scholar] [CrossRef] [PubMed]
- Szarynska, M.; Olejniczak, A.; Kobiela, J.; Laski, D.; Sledzinski, Z.; Kmiec, Z. Cancer stem cells as targets for DC-based immunotherapy of colorectal cancer. Sci. Rep. 2018, 8, 12042. [Google Scholar] [CrossRef]
- Cao, H.; Xu, E.; Liu, H.; Wan, L.; Lai, M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol. Res. Pract. 2015, 211, 557–569. [Google Scholar] [CrossRef]
- Druzhkova, I.; Ignatova, N.; Prodanets, N.; Kiselev, N.; Zhukov, I.; Shirmanova, M.; Zagainov, V.; Zagaynova, E. E-cadherin in colorectal cancer: Relation to chemosensitivity. Clin. Colorectal Cancer 2019, 18, e74–e86. [Google Scholar] [CrossRef]
- Lee, M.R.; Ji, S.Y.; Mia-Jan, K.; Cho, M.Y. Chemoresistance of CD133(+) colon cancer may be related with increased survivin expression. Biochem. Biophys. Res. Commun. 2015, 463, 229–234. [Google Scholar] [CrossRef]
- Pothuraju, R.; Rachagani, S.; Krishn, S.R.; Chaudhary, S.; Nimmakayala, R.K.; Siddiqui, J.A.; Ganguly, K.; Lakshmanan, I.; Cox, J.L.; Mallya, K.; et al. Molecular implications of MUC5AC-CD44 axis in colorectal cancer progression and chemoresistance. Mol. Cancer 2020, 19, 37. [Google Scholar] [CrossRef]
- Meng, Q.; Wu, W.; Pei, T.; Li, L.; Tang, X.; Sun, H. Novel markers for circulating tumor stem cells in colorectal carcinoma. Am. J. Transl. Res. 2016, 8, 4233–4241. [Google Scholar] [PubMed]
- Wang, Q.; Shi, Y.L.; Zhou, K.; Wang, L.L.; Yan, Z.X.; Liu, Y.L.; Xu, L.L.; Zhao, S.W.; Chu, H.L.; Shi, T.T.; et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 2018, 9, 739. [Google Scholar] [CrossRef] [PubMed]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [PubMed]
- Pohl, A.; El-Khoueiry, A.; Yang, D.; Zhang, W.; Lurje, G.; Ning, Y.; Winder, T.; Hu-Lieskoven, S.; Iqbal, S.; Danenberg, K.D.; et al. Pharmacogenetic profiling of CD133 is associated with response rate (RR) and progression-free survival (PFS) in patients with metastatic colorectal cancer (mCRC), treated with bevacizumab-based chemotherapy. Pharm. J. 2013, 13, 173–180. [Google Scholar] [CrossRef]
- Fujiwara-Tani, R.; Sasaki, T.; Ohmori, H.; Luo, Y.; Goto, K.; Nishiguchi, Y.; Mori, S.; Nakashima, C.; Mori, T.; Miyagawa, Y.; et al. Concurrent expression of CD47 and CD44 in colorectal cancer promotes malignancy. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2019, 86, 182–189. [Google Scholar] [CrossRef]
- Munro, M.J.; Wickremesekera, S.K.; Peng, L.; Tan, S.T.; Itinteang, T. Cancer stem cells in colorectal cancer: A review. J. Clin. Pathol. 2018, 71, 110–116. [Google Scholar] [CrossRef]
- Paquet-Fifield, S.; Koh, S.L.; Cheng, L.; Beyit, L.M.; Shembrey, C.; Molck, C.; Behrenbruch, C.; Papin, M.; Gironella, M.; Guelfi, S.; et al. Tight junction protein Claudin-2 Promotes self-renewal of human colorectal cancer stem-like cells. Cancer Res. 2018, 78, 2925–2938. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.J.; Chen, X.W.; Zhang, W.J.; Wang, J.Z.; Ouyang, M.Z.; Zhong, Q.; Liu, C.C. Twist1 is a potential prognostic marker for colorectal cancer and associated with chemoresistance. Am. J. Cancer Res. 2015, 5, 2000–2011. [Google Scholar]
- Bamodu, O.A.; Yang, C.K.; Cheng, W.H.; Tzeng, D.T.W.; Kuo, K.T.; Huang, C.C.; Deng, L.; Hsiao, M.; Lee, W.H.; Yeh, C.T. 4-Acetyl-antroquinonol B suppresses SOD2-enhanced cancer stem cell-like phenotypes and chemoresistance of colorectal cancer cells by inducing hsa-miR-324 re-expression. Cancers 2018, 10, 269. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, X.; Du, L.; Wang, Y.; Liu, X.; Tian, H.; Wang, L.; Li, P.; Zhao, Y.; Duan, W.; et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol. Cancer 2019, 18, 43. [Google Scholar] [CrossRef]
- Kjersem, J.B.; Ikdahl, T.; Lingjaerde, O.C.; Guren, T.; Tveit, K.M.; Kure, E.H. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol. Oncol. 2014, 8, 59–67. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.J.; Carter, J.V.; Burton, J.F.; Oxford, B.G.; Schmidt, M.N.; Hallion, J.C.; Galandiuk, S. The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: A systematic review. Int. J. Cancer 2018, 142, 2501–2511. [Google Scholar] [CrossRef]
- Eyking, A.; Reis, H.; Frank, M.; Gerken, G.; Schmid, K.W.; Cario, E. MiR-205 and MiR-373 are associated with aggressive human mucinous colorectal cancer. PLoS ONE 2016, 11, e0156871. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol. Cancer Ther. 2017, 16, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Z.; Wang, H.; Tang, Z.; Liu, Y.; Liang, Z.; Deng, X.; Zhao, M.; Fu, Q.; Li, L.; et al. VPS33B negatively modulated by nicotine functions as a tumor suppressor in colorectal cancer. Int. J. Cancer 2020, 146, 496–509. [Google Scholar] [CrossRef]
- Sun, L.; Ke, J.; He, Z.; Chen, Z.; Huang, Q.; Ai, W.; Wang, G.; Wei, Y.; Zou, X.; Zhang, S.; et al. HES1 promotes colorectal cancer cell resistance to 5-Fu by inducing of EMT and ABC transporter proteins. J. Cancer 2017, 8, 2802–2808. [Google Scholar] [CrossRef]
- Kim, H.B.; Lim, H.J.; Lee, H.J.; Park, J.H.; Park, S.G. Evaluation and clinical significance of jagged-1-activated notch signaling by APEX1 in colorectal cancer. Anticancer Res. 2019, 39, 6097–6105. [Google Scholar] [CrossRef]
- Aguilera, O.; Gonzalez-Sancho, J.M.; Zazo, S.; Rincon, R.; Fernandez, A.F.; Tapia, O.; Canals, F.; Morte, B.; Calvanese, V.; Orgaz, J.L.; et al. Nuclear DICKKOPF-1 as a biomarker of chemoresistance and poor clinical outcome in colorectal cancer. Oncotarget 2015, 6, 5903–5917. [Google Scholar] [CrossRef]
- Enyu, L.; Zhengchuan, N.; Jiayong, W.; Benjia, L.; Qi, S.; Ruixi, Q.; Cheng, P.; Khan, A.Q.; Wei, S.; Jun, N. Integrin beta6 can be translationally regulated by eukaryotic initiation factor 4E: Contributing to colonic tumor malignancy. Tumour Biol. 2015, 36, 6541–6550. [Google Scholar] [CrossRef]
- Bao, Y.; Lu, Y.; Wang, X.; Feng, W.; Sun, X.; Guo, H.; Tang, C.; Zhang, X.; Shi, Q.; Yu, H. Eukaryotic translation initiation factor 5A2 (eIF5A2) regulates chemoresistance in colorectal cancer through epithelial mesenchymal transition. Cancer Cell Int. 2015, 15, 109. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chan, K.; Qi, Y.; Lu, L.; Ning, F.; Wu, M.; Wang, H.; Wang, Y.; Cai, S.; Du, J. Participation of CCL1 in Snail-Positive Fibroblasts in Colorectal Cancer Contribute to 5-Fluorouracil/Paclitaxel Chemoresistance. Cancer Res. Treat. 2018, 50, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Mizushima, T.; Yokoyama, Y.; Hirose, H.; Wu, X.; Qian, Y.; Ikehata, K.; Miyoshi, N.; Takahashi, H.; Haraguchi, N.; et al. Sox2 is associated with cancer stem-like properties in colorectal cancer. Sci. Rep. 2018, 8, 17639. [Google Scholar] [CrossRef] [PubMed]
- Francescangeli, F.; Contavalli, P.; De Angelis, M.L.; Careccia, S.; Signore, M.; Haas, T.L.; Salaris, F.; Baiocchi, M.; Boe, A.; Giuliani, A.; et al. A pre-existing population of ZEB2(+) quiescent cells with stemness and mesenchymal features dictate chemoresistance in colorectal cancer. J. Exp. Clin. Cancer Res. CR 2020, 39, 2. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Song, C.; Gu, X.; Wang, M.; Miao, D.; Lv, J.; Liu, Y. Ubiquitin-specific peptidase 22 contributes to colorectal cancer stemness and chemoresistance via Wnt/beta-catenin pathway. Cell. Physiol. Biochem. 2018, 46, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Zhao, C.H.; Ding, H.W.; Wu, Q.; Ren, T.S.; Wang, J.; Chen, C.Q.; Zhao, Q.C. A novel inhibitor of ADAM17 sensitizes colorectal cancer cells to 5-Fluorouracil by reversing Notch and epithelial-mesenchymal transition in vitro and in vivo. Cell Prolif. 2018, 51, e12480. [Google Scholar] [CrossRef]
- Garcia-Heredia, J.M.; Verdugo Sivianes, E.M.; Lucena-Cacace, A.; Molina-Pinelo, S.; Carnero, A. Numb-like (NumbL) downregulation increases tumorigenicity, cancer stem cell-like properties and resistance to chemotherapy. Oncotarget 2016, 7, 63611–63628. [Google Scholar] [CrossRef]
- Usui, T.; Sakurai, M.; Umata, K.; Elbadawy, M.; Ohama, T.; Yamawaki, H.; Hazama, S.; Takenouchi, H.; Nakajima, M.; Tsunedomi, R.; et al. Hedgehog signals mediate anti-cancer drug resistance in three-dimensional primary colorectal cancer organoid culture. Int. J. Mol. Sci. 2018, 19, 1098. [Google Scholar] [CrossRef]
- Qian, Y.; Wu, X.; Yokoyama, Y.; Okuzaki, D.; Taguchi, M.; Hirose, H.; Wang, J.; Hata, T.; Inoue, A.; Hiraki, M.; et al. E-cadherin-Fc chimera protein matrix enhances cancer stem-like properties and induces mesenchymal features in colon cancer cells. Cancer Sci. 2019, 110, 3520–3532. [Google Scholar] [CrossRef]
| Protein | Change | Drugs Affected | Consequences | References |
|---|---|---|---|---|
| Uptake Transporters (MOC-1a) | ||||
| OATP1B1 | GV (OATP1B1*15 haplotype) | Irinotecan, Methotrexate | Lower response in vitro and in patients | [11,15,16,18,19] |
| OATP1B3 | GV (Cancer-type) | Irinotecan | Reduced PFS | [21,22,23] |
| OATP1A2 | Downregulation | Imatinib, Methotrexate | Reduced drug uptake | [16,27] |
| OCT1 | Downregulation | Imatinib, Doxorubicin | Lower sensitivity in vitro; lower clinical response | [32,33,34,35] |
| OCT3 | Impaired expression | Irinotecan, Imatinib, Cisplatin, 5-FU, FOLFOX | Lower clinical response | [38,39] |
| OCTN2 | GV (rs2631367, rs2631372) | Imatinib, Etoposide | Lower sensitivity in vitro | [38,42,43,44] |
| CTR1 | Downregulation | Cisplatin | Lower sensitivity in vitro | [60] |
| Efflux transporters (MOC-1b) | ||||
| MDR1 | Upregulation | Doxorubicin, Etoposide, Irinotecan | Lower sensitivity in vitro | [47] |
| MRP1 | Upregulation | Doxorubicin, Etoposide, 5-FU, Oxaliplatin | Lower sensitivity in vitro | [61,62] |
| MRP2 | Upregulation | Cisplatin | Lower sensitivity in vitro | [56] |
| MRP3 | Upregulation | Doxorubicin, Etoposide | Lower sensitivity in vitro | [63] |
| MRP4 | GV (rs3742106) | 5-FU, Capecitabine | Lower clinical response | [64] |
| MRP5 | Upregulation | 5-FU, Methotrexate | Lower sensitivity in vitro | [65] |
| BCRP | GV (rs2231137, rs2231142) | Irinotecan | Lower sensitivity in vitro; Lower clinical response | [66,67] |
| ATP7B | Upregulation | Oxaliplatin | Poor clinical outcome | [68] |
| ABCA9 | GV | Oxaliplatin | Reduced OS and response | [69] |
| LRP | Upregulation | Doxorubicin, Etoposide | Lower sensitivity in vitro | [70,71] |
| Protein | Change | Drugs Affected | Consequences | References |
|---|---|---|---|---|
| CYP3A5, CYP3A4 | Upregulation | Irinotecan (SN-38) | Enhanced drug inactivation | [79,80] |
| CYP1A2, CYP2A6 | Upregulation | 5-FU | Enhanced drug inactivation | [81] |
| CES2 | Downregulation | Irinotecan | Reduced drug activation | [82,84] |
| DPYP | Upregulation | 5-FU | Reduced clinical response | [87,88] |
| TYMP | Downregulation | 5-FU | Reduced drug activation | [87] |
| γ-GCS | Upregulation | Cisplatin, Doxorubicin | Enhanced drug inactivation | [92,93] |
| GSTA1 | Upregulation | Irinotecan (SN-38) | Enhanced drug inactivation | [94] |
| GSTO1 | Upregulation | Cisplatin | Enhanced drug inactivation | [95] |
| GSTP1 | Upregulation | Anthracyclines | Enhanced drug inactivation | [92] |
| UGTs | Upregulation | Irinotecan (SN-38) | Enhanced drug inactivation | [96,97] |
| MT | Upregulation | Cisplatin | Reduced sensitivity in vitro and poor clinical prognosis * | [98,99] |
| Protein | Change | Drugs Affected | Consequences | References |
|---|---|---|---|---|
| EGFR | Low gene copy number | Cetuximab Panitumumab | Reduced response in patients with wild-type KRAS | [116] |
| pEGFR | Low levels | Cetuximab | Reduced clinical response | [115] |
| ERBB2 | Upregulation and R784G mutation | Cetuximab | Reduced clinical response | [118] |
| PlGF | High serum levels | Bevacizumab | Reduced clinical response | [121] |
| TYMS | Downregulation | 5-FU | Worse outcome * | [109,110] |
| VEGF-A | High serum levels | Bevacizumab | Reduced clinical response | [121] |
| VEGFR-1 | High serum levels | Bevacizumab | Reduced clinical response | [122] |
| VEGFR-2 | T771R mutation | Ramucirumab | Reduced clinical response | [125] |
| Protein | Change | Drug Affected | Consequences | References |
|---|---|---|---|---|
| Nucleotide Excision Repair (NER) | ||||
| ERCC1 | High expression | Oxaliplatin | Reduced efficacy | [128] |
| ERCC1 | GV (rs11615, rs10412761) | Oxaliplatin, 5-FU, Capecitabine | Reduced efficacy | [133,134,135] |
| ERCC2 | GV (rs13181, rs1799787) | Oxaliplatin, 5-FU, Capecitabine | Reduced efficacy | [133,134,135] |
| ERCC6 | High expression | 5-FU | Reduced efficacy | [129] |
| XPC | High expression | Cisplatin | Drug resistance * | [131,132] |
| Mismatch Repair (MMR) | ||||
| Several | Defective MMR | 5-FU, Oxaliplatin | Reduced efficacy | [138,139,140] |
| Protein | Change | Drugs Affected | Consequences | References |
|---|---|---|---|---|
| Pro-Apoptotic Factors (MOC-5a) | ||||
| BAD | Downregulation | 5-FU | Apoptosis inhibition | [162] |
| BAX | Downregulation and inactivating mutations | 5-FU | Apoptosis inhibition | [159,160] |
| BID | Downregulation | 5-FU | Apoptosis inhibition | [162] |
| FADD | Downregulation | 5-FU | Apoptosis inhibition | [163] |
| miR-520g | Upregulation | 5-FU, Oxaliplatin | No cell cycle arrest; apoptosis inhibition; p21 downregulation | [158] |
| p53 | Inactivating mutations | 5-FU, FOLFOX | No cell cycle arrest; apoptosis inhibition | [149,150,155,156] |
| Oxaliplatin | miR-503-5p upregulation; PUMA downregulation; apoptosis inhibition | [151,152] | ||
| 5-FU | Associated with enhanced MDR1 and GSTP expression | [153,154] | ||
| Survival Pathways (MOC-5b) | ||||
| APC | Inactivating mutations | 5-FU | Stimulation of Wnt/β-catenin | [180] |
| BCL-2 | Upregulation | 5-FU | Apoptosis inhibition | [166,167] |
| Biglycan | Upregulation | 5-FU | Increased activity of the NFκB pathway | [178] |
| BRAF | Inactivating mutations | Vemurafenib, Dabrafenib, Encorafenib | Increased proliferation | [181,182] |
| CD133 | Upregulation | Doxorubicin | Increased activity of the NFκB pathway; MDR1 upregulation | [179] |
| CHK1 | Upregulation | 5-FU, Oxaliplatin | No cell cycle arrest; apoptosis inhibition | [173,174] |
| IAP2 | Modulation of caspase 3/7 activity | 5-FU | Apoptosis inhibition | [171] |
| IL-17 | Upregulation of p-AKT, mTOR and BCL-2; Suppression of BAX | Cisplatin | Apoptosis inhibition | [164] |
| KRAS | Activating mutations | Cetuximab, Panitumumab, others | Increased proliferation | [148,183,184] |
| MCL-1 | Perinuclear expression | 5-FU | No cell cycle arrest; apoptosis inhibition | [172] |
| NFκB | Increased activity | 5-FU, Gemcitabine | Upregulation of anti-apoptotic factors | [175,176,177] |
| Notch | Increased activity | 5-FU, Cisplatin | Upregulation of COX2; MDR1 and MRP1 upregulation | [185,186] |
| RNF43 | Inactivating mutations | Dacomitinib | Stimulation of Wnt/β-catenin | [187] |
| Wnt/β-catenin | Increased activity | 5-FU | Stimulation of cell proliferation | [188] |
| ZNRF3 | Inactivating mutations | Dacomitinib | Stimulation of Wnt/β-catenin | [187] |
| Factor | Change | Drugs Affected | Consequences | Reference |
|---|---|---|---|---|
| HIF-1α | Upregulation | 5-FU | MDR1 upregulation; lower response to treatment | [199] |
| Upregulation | Bevacizumab | Lower apoptosis in resistant cells in vitro | [200] | |
| HIF-1α, TGF-β | High expression | 5-FU, Oxaliplatin | Increased GLI2 expression; lower drug effect in vitro | [201] |
| IL-17A | Increased production | 5-FU, Oxaliplatin | Reduced drug effect on CSCs | [202] |
| Gut microbiota | Fusobacterium nucleatum | Oxaliplatin, Capecitabine | Lower response to treatment | [207] |
| Gammaproteobacteria | Gemcitabine | Drug inactivation; reduced efficacy in vivo | [208] | |
| UCA1 | Upregulation | Cetuximab | Reduced drug efficacy in vitro and in patients | [209] |
| Factor | Change | Drugs Affected | Consequences | References |
|---|---|---|---|---|
| Cell Adhesion Proteins | ||||
| CD133 | Downregulation | Bevacizumab | Increased DPR | [224] |
| CD133 | Upregulation | 5-FU | Reduced sensitivity in vitro | [219] |
| CD262 | Upregulation | 5-FU, Cisplatin | Reduced sensitivity in vitro | [221] |
| CD44 | Upregulation | 5-FU, Oxaliplatin | Reduced sensitivity in vitro and in vivo | [220] |
| CD44, CD47 | Upregulation | Nivolumab | Reduced DFS | [225] |
| E-cadherin | Downregulation | 5-FU, Irinotecan, Oxaliplatin | Higher sensitivity in vitro | [217] |
| LGR5 | Upregulation | 5-FU, Capecitabine, Oxaliplatin | Reduced DFS and OS | [222] |
| Enzymes | ||||
| ALDH | Upregulation | 5-FU | Reduced RFS | [227] |
| Non-Coding RNAs | ||||
| miR-324-5p | Downregulation | 5-FU, Oxaliplatin | Reduced clinical response | [229] |
| miR-128-3p | Downregulation | Oxaliplatin | Reduced PFS | [230] |
| miR-148a, miR-27b | Upregulation | 5-FU, Oxaliplatin | Reduced PFS | [231] |
| miR-200a, miR-200c, miR-429 | Downregulation | Adjuvant chemotherapy | Reduced OS | [232] |
| miR-205, miR-373 | Upregulation | 5-FU, Oxaliplatin | Increased cancer progression | [233] |
| miR-92a-3p | Upregulation | 5-FU, Oxaliplatin | Reduced clinical response | [234] |
| MALAT1 | Upregulation | 5-FU, Oxaliplatin | Reduced OS and PFS | [235] |
| Survival Pathways | ||||
| EGFR | Overactivation | Oxaliplatin | Reduced sensitivity in vitro and in vivo | [236] |
| Notch | Overactivation | 5-FU, Oxaliplatin, Irinotecan | Reduced DFS and OS | [237,238] |
| Wnt/β-catenin | Overactivation | 5-FU, Oxaliplatin, Irinotecan | Reduced OS | [239] |
| Transcription Factors | ||||
| eIF4E | Upregulation | 5-FU | Reduced sensitivity in vitro | [240] |
| eIF5A2 | Upregulation | Doxorubicin | Reduced sensitivity in vitro | [241] |
| SNAI1 | Upregulation | 5-FU, Paclitaxel | Reduced sensitivity in vitro and in vivo | [242] |
| SOX2, OCT4, NANOG | Upregulation | 5-FU, Oxaliplatin | Reduced OS and RFS | [243] |
| TWIST1 | Upregulation | 5-FU, Oxaliplatin | Reduced OS | [228] |
| ZEB2 | Upregulation | 5-FU, Oxaliplatin | Reduced RFS | [244] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marin, J.J.G.; Macias, R.I.R.; Monte, M.J.; Herraez, E.; Peleteiro-Vigil, A.; Blas, B.S.d.; Sanchon-Sanchez, P.; Temprano, A.G.; Espinosa-Escudero, R.A.; Lozano, E.; et al. Cellular Mechanisms Accounting for the Refractoriness of Colorectal Carcinoma to Pharmacological Treatment. Cancers 2020, 12, 2605. https://doi.org/10.3390/cancers12092605
Marin JJG, Macias RIR, Monte MJ, Herraez E, Peleteiro-Vigil A, Blas BSd, Sanchon-Sanchez P, Temprano AG, Espinosa-Escudero RA, Lozano E, et al. Cellular Mechanisms Accounting for the Refractoriness of Colorectal Carcinoma to Pharmacological Treatment. Cancers. 2020; 12(9):2605. https://doi.org/10.3390/cancers12092605
Chicago/Turabian StyleMarin, Jose J.G., Rocio I.R. Macias, Maria J. Monte, Elisa Herraez, Ana Peleteiro-Vigil, Beatriz Sanchez de Blas, Paula Sanchon-Sanchez, Alvaro G. Temprano, Ricardo A. Espinosa-Escudero, Elisa Lozano, and et al. 2020. "Cellular Mechanisms Accounting for the Refractoriness of Colorectal Carcinoma to Pharmacological Treatment" Cancers 12, no. 9: 2605. https://doi.org/10.3390/cancers12092605
APA StyleMarin, J. J. G., Macias, R. I. R., Monte, M. J., Herraez, E., Peleteiro-Vigil, A., Blas, B. S. d., Sanchon-Sanchez, P., Temprano, A. G., Espinosa-Escudero, R. A., Lozano, E., Briz, O., & Romero, M. R. (2020). Cellular Mechanisms Accounting for the Refractoriness of Colorectal Carcinoma to Pharmacological Treatment. Cancers, 12(9), 2605. https://doi.org/10.3390/cancers12092605








