High M-MDSC Percentage as a Negative Prognostic Factor in Chronic Lymphocytic Leukaemia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. The Percentage of M-MDSC Is Increased in the Peripheral Blood of CLL Patients and Correlates with The Rai Stage
2.2. M-MDSC Percentage Is Higher in Patients from High-Risk Groups
2.3. High M-MDSC Percentage Is a Negative Prognostic Marker for Time-to-Treatment and Survival Time
2.4. IDO, ARG1, NOS2, IL-10, and TGF-Β1 Are Overexpressed by CLL-Derived M-MDSC
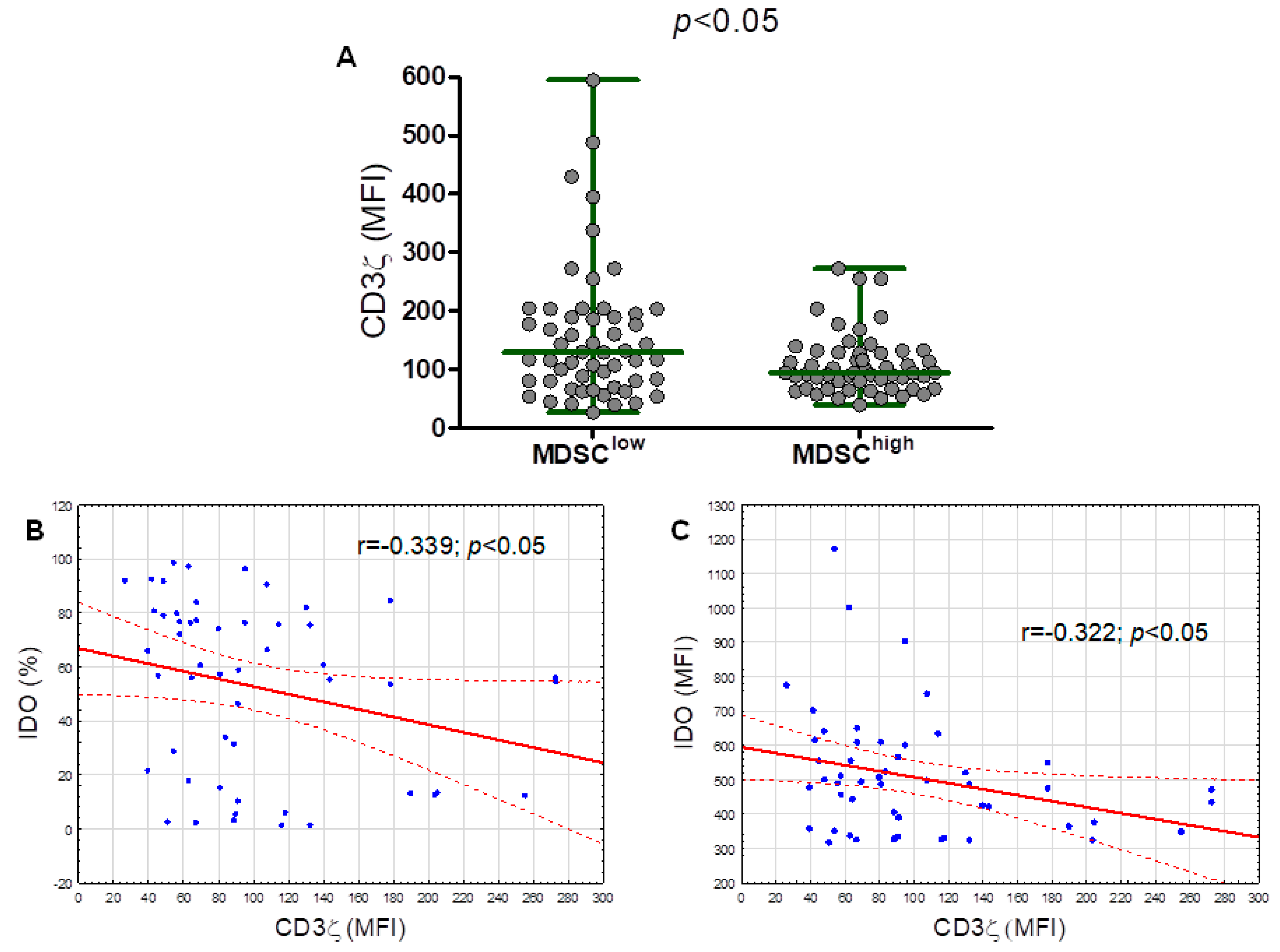
2.5. The CD3 Ζ Chain (CD247) Expression in T Lymphocytes Is Significantly Lower in the M-MDSChigh Group
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
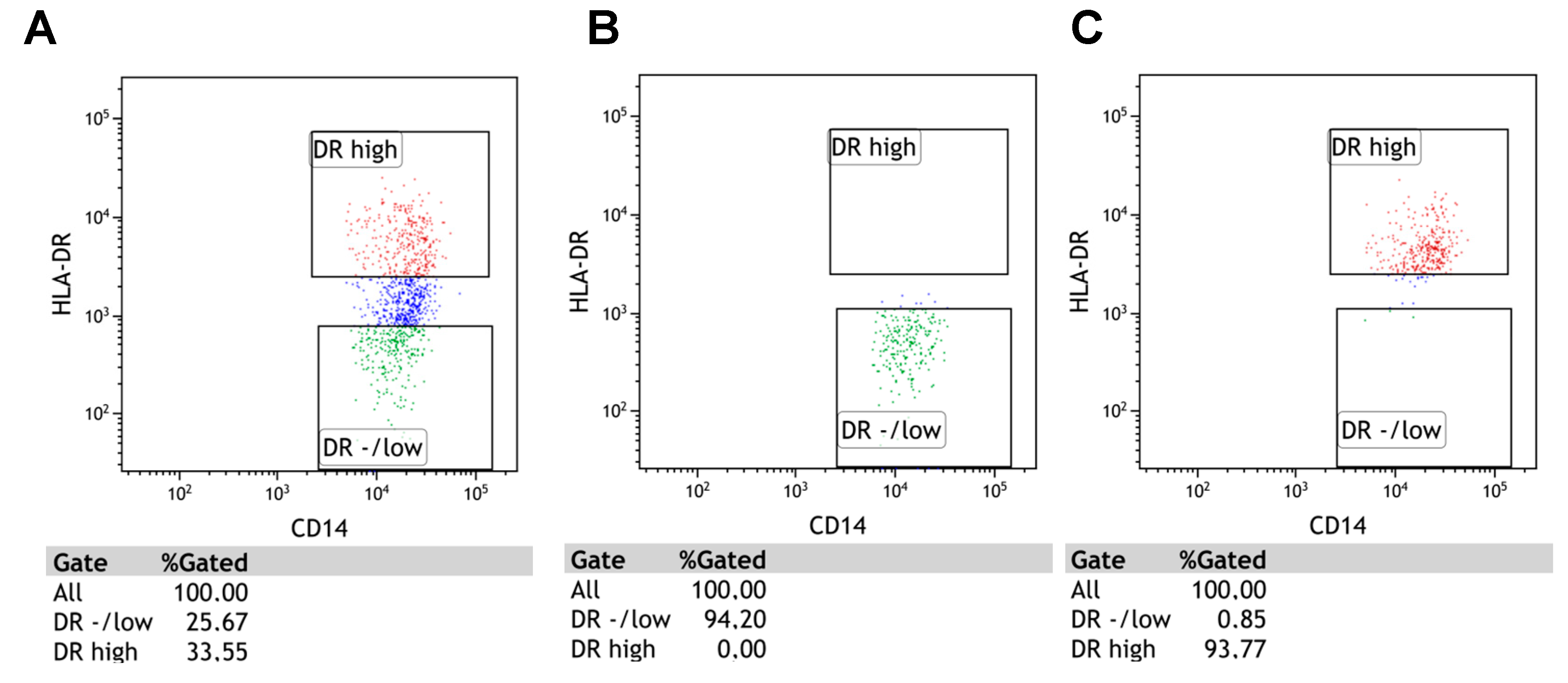
4.2. Detection of M-MDSC
4.3. Analysis of Intracellular IDO, Arg1, NOS2, IL-10, or TGF-Β1 Expressions
4.4. Flow Cytometry Analysis of M-MDSC
4.5. Flow Cytometry Sorting of M-MDSC Cells For Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-Qpcr)
4.6. RNA Preparation and RT-Qpcr for IDO, ARG1, NOS2, IL-10, and TGF-Β1
4.7. The CD3 Ζ Chain (CD247) Analysis in T Lymphocytes
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bronte, V.; Brandau, S.; Chen, S.-H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef] [Green Version]
- Vetsika, E.-K.; Koukos, A.; Kotsakis, A. Myeloid-Derived Suppressor Cells: Major Figures that Shape the Immunosuppressive and Angiogenic Network in Cancer. Cells 2019, 8, 1647. [Google Scholar] [CrossRef] [Green Version]
- Pawelec, G.; Verschoor, C.P.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells: Not Only in Tumor Immunity. Front. Immunol. 2019, 10, 1099. [Google Scholar] [CrossRef]
- Kumar, V.; Patel, S.; Tcyganov, E.; Gabrilovich, D.I. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol. 2016, 37, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Immunosenescence: The potential role of myeloid-derived suppressor cells (MDSC) in age-related immune deficiency. Cell. Mol. Life Sci. 2019, 76, 1901–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.O.; Wan, Y.Y.; Sanjabi, S.; Robertson, A.-K.L.; Flavell, R.A. Transforming growth factor-beta regulation of immune responses. Annu. Rev. Immunol. 2006, 24, 99–146. [Google Scholar] [CrossRef]
- Ouyang, W.; Rutz, S.; Crellin, N.K.; Valdez, P.A.; Hymowitz, S.G. Regulation and Functions of the IL-10 Family of Cytokines in Inflammation and Disease. Annu. Rev. Immunol. 2011, 29, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Sektioglu, I.M.; Carretero, R.; Bender, N.; Bogdan, C.; Garbi, N.; Umansky, V.; Umansky, L.; Urban, K.; Von Knebel-Döberitz, M.; Somasundaram, V.; et al. Macrophage-derived nitric oxide initiates T-cell diapedesis and tumor rejection. Oncoimmunology 2016, 5, e1204506. [Google Scholar] [CrossRef]
- Redd, P.S.; Ibrahim, M.L.; Klement, J.D.; Sharman, S.K.; Paschall, A.V.; Yang, D.; Nayak-Kapoor, A.; Liu, K. SETD1B Activates iNOS Expression in Myeloid-Derived Suppressor Cells. Cancer Res. 2017, 77, 2834–2843. [Google Scholar] [CrossRef] [Green Version]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Diaz-Montero, C.M.; Salem, M.L.; Nishimura, M.I.; Garrett-Mayer, E.; Cole, D.J.; Montero, A.J. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol. Immunother. 2009, 58, 49–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, V.; Hu, X.; Weber, R.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Targeting Myeloid-Derived Suppressor Cells to Bypass Tumor-Induced Immunosuppression. Front. Immunol. 2018, 9, 398. [Google Scholar] [CrossRef] [PubMed]
- Solito, S.; Marigo, I.; Pinton, L.; Damuzzo, V.; Mandruzzato, S.; Bronte, V. Myeloid-derived suppressor cell heterogeneity in human cancers. Ann. N. Y. Acad. Sci. 2014, 1319, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, N.S.; Apollonio, B.; Ramsay, A.G. Tumor microenvironment (TME)-driven immune suppression in B cell malignancy. Biochim. Biophys. Acta 2016, 1863, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Bizymi, N.; Bjelica, S.; Kittang, A.O.; Mojsilovic, S.; Velegraki, M.; Pontikoglou, C.; Roussel, M.; Ersvær, E.; Santibañez, J.F.; Lipoldová, M.; et al. Myeloid-Derived Suppressor Cells in Hematologic Diseases: Promising Biomarkers and Treatment Targets. HemaSphere 2019, 3, e168. [Google Scholar] [CrossRef]
- Lin, Y.; Gustafson, M.P.; Bulur, P.A.; Gastineau, D.A.; Witzig, T.E.; Dietz, A.B. Immunosuppressive CD14+HLA-DRlow/− monocytes in B-cell non-Hodgkin lymphoma. Blood 2011, 117, 872–881. [Google Scholar] [CrossRef] [Green Version]
- Jitschin, R.; Braun, M.; Büttner, M.; Dettmer-Wilde, K.; Bricks, J.; Berger, J.; Eckart, M.J.; Krause, S.W.; Oefner, P.J.; Le Blanc, K.; et al. CLL-cells induce IDOhi CD14+HLA-DRlo myeloid-derived suppressor cells that inhibit T-cell responses and promote TRegs. Blood 2014, 124, 750–760. [Google Scholar] [CrossRef] [Green Version]
- Palumbo, G.A.; Parrinello, N.L.; Giallongo, C.; D’amico, E.; Zanghì, A.; Puglisi, F.; Conticello, C.; Chiarenza, A.; Tibullo, D.; Di Raimondo, F.; et al. Monocytic myeloid derived suppressor cells in hematological malignancies. Int. J. Mol. Sci. 2019, 20, 5459. [Google Scholar] [CrossRef] [Green Version]
- Argyropoulos, K.V.; Pulitzer, M.; Perez, S.; Korkolopoulou, P.; Angelopoulou, M.; Baxevanis, C.; Palomba, M.L.; Siakantaris, M. Tumor-infiltrating and circulating granulocytic myeloid-derived suppressor cells correlate with disease activity and adverse clinical outcomes in mycosis fungoides. Clin. Transl. Oncol. 2020, 22, 1059–1066. [Google Scholar] [CrossRef]
- Ten Hacken, E.; Burger, J.A. Microenvironment interactions and B-cell receptor signaling in Chronic Lymphocytic Leukemia: Implications for disease pathogenesis and treatment. Biochim. Biophys. Acta 2016, 1863, 401–413. [Google Scholar] [CrossRef]
- Hanna, B.S.; Öztürk, S.; Seiffert, M. Beyond bystanders: Myeloid cells in chronic lymphocytic leukemia. Mol. Immunol. 2019, 110, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Ten Hacken, E.; Burger, J.A. Microenvironment dependency in Chronic Lymphocytic Leukemia: The basis for new targeted therapies. Pharmacol. Ther. 2014, 144, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, M.P.; Abraham, R.S.; Lin, Y.; Wu, W.; Gastineau, D.A.; Zent, C.S.; Dietz, A.B. Association of an increased frequency of CD14+HLA-DRlo/neg monocytes with decreased time to progression in chronic lymphocytic leukaemia (CLL). Br. J. Haematol. 2012, 156, 674–676. [Google Scholar] [CrossRef] [PubMed]
- Zahran, A.M.; Moeen, S.M.; Thabet, A.F.; Rayan, A.; Abdel-Rahim, M.H.; Mohamed, W.M.Y.; Hetta, H.F. Monocytic myeloid-derived suppressor cells in chronic lymphocytic leukemia patients: A single center experience. Leuk. Lymphoma 2020. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, Y.; Huang, Q.; Qiu, L. CD14+HLA-DRlow/- expression: A novel prognostic factor in chronic lymphocytic leukemia. Oncol. Lett. 2015, 9, 1167–1172. [Google Scholar] [CrossRef] [Green Version]
- Mandruzzato, S.; Brandau, S.; Britten, C.M.; Bronte, V.; Damuzzo, V.; Gouttefangeas, C.; Maurer, D.; Ottensmeier, C.; Van der Burg, S.H.; Welters, M.J.P.; et al. Toward harmonized phenotyping of human myeloid-derived suppressor cells by flow cytometry: Results from an interim study. Cancer Immunol. Immunother. 2016, 65, 161–169. [Google Scholar] [CrossRef]
- Ostrand-Rosenberg, S. Myeloid derived-suppressor cells: Their role in cancer and obesity. Curr. Opin. Immunol. 2018, 51, 68–75. [Google Scholar] [CrossRef]
- Bruns, H.; Böttcher, M.; Qorraj, M.; Fabri, M.; Jitschin, S.; Dindorf, J.; Busch, L.; Jitschin, R.; Mackensen, A.; Mougiakakos, D. CLL-cell-mediated MDSC induction by exosomal miR-155 transfer is disrupted by Vitamin D. Leukemia 2017, 31, 985–988. [Google Scholar] [CrossRef]
- Azzaoui, I.; Uhel, F.; Rossille, D.; Pangault, C.; Dulong, J.; Le Priol, J.; Lamy, T.; Houot, R.; Le Gouill, S.; Cartron, G.; et al. T-cell defect in diffuse large B-cell lymphomas involves expansion of myeloid-derived suppressor cells. Blood 2016, 128, 1081–1092. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, L.; Wang, H.; Xiong, S.; Li, Y.; Tao, Q.; Xiao, W.; Qin, H.; Wang, Y.; Zhai, Z. Tumor-induced CD14+HLA-DR−/low myeloid-derived suppressor cells correlate with tumor progression and outcome of therapy in multiple myeloma patients. Cancer Immunol. Immunother. 2015, 64, 389–399. [Google Scholar] [CrossRef]
- Law, A.M.K.; Valdes-Mora, F.; Gallego-Ortega, D. Myeloid-Derived Suppressor Cells as a Therapeutic Target for Cancer. Cells 2020, 9, 561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassetta, L.; Baekkevold, E.S.; Brandau, S.; Bujko, A.; Cassatella, M.A.; Dorhoi, A.; Krieg, C.; Lin, A.; Loré, K.; Marini, O.; et al. Deciphering myeloid-derived suppressor cells: Isolation and markers in humans, mice and non-human primates. Cancer Immunol. Immunother. 2019, 68, 687–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whiteside, T.L. Down-regulation of ?-chain expression in T cells: A biomarker of prognosis in cancer? Cancer Immunol. Immunother. 2004, 53. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.X.; Kim, T.S.; Poh, C.L. Understanding the Differentiation, Expansion, Recruitment and Suppressive Activities of Myeloid-Derived Suppressor Cells in Cancers. IJMS 2020, 21, 3599. [Google Scholar] [CrossRef]
- Bruger, A.M.; Dorhoi, A.; Esendagli, G.; Barczyk-Kahlert, K.; Van der Bruggen, P.; Lipoldova, M.; Perecko, T.; Santibanez, J.; Saraiva, M.; Van Ginderachter, J.A.; et al. How to measure the immunosuppressive activity of MDSC: Assays, problems and potential solutions. Cancer Immunol. Immunother. 2019, 68, 631–644. [Google Scholar] [CrossRef]
- Tcyganov, E.; Mastio, J.; Chen, E.; Gabrilovich, D.I. Plasticity of myeloid-derived suppressor cells in cancer. Curr. Opin. Immunol. 2018, 51, 76–82. [Google Scholar] [CrossRef]
- Yazdani, Y.; Mohammadnia-Afrouzi, M.; Yousefi, M.; Anvari, E.; Ghalamfarsa, G.; Hasannia, H.; Sadreddini, S.; Jadidi-Niaragh, F. Myeloid-derived suppressor cells in B cell malignancies. Tumor Biol. 2015, 36, 7339–7353. [Google Scholar] [CrossRef]
- Özkan, B.; Lim, H.; Park, S.-G. Immunomodulatory Function of Myeloid-Derived Suppressor Cells during B Cell-Mediated Immune Responses. IJMS 2018, 19, 1468. [Google Scholar] [CrossRef] [Green Version]
- Kowalska, W.; Bojarska-Junak, A. Monocytic MDSC as a source of immunosuppressive cytokines in chronic lymphocytic leukemia (CLL) microenvironment. Folia Histochem. Cytobiol. 2020, 58, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Shimizu, K.; Shinga, J.; Hidaka, M.; Kawano, F.; Kakimi, K.; Yamasaki, S.; Asakura, M.; Fujii, S. Characterization of the myeloid-derived suppressor cell subset regulated by NK cells in malignant lymphoma. OncoImmunology 2015, 4, e995541. [Google Scholar] [CrossRef]
- Stiff, A.; Trikha, P.; Mundy-Bosse, B.; McMichael, E.; Mace, T.A.; Benner, B.; Kendra, K.; Campbell, A.; Gautam, S.; Abood, D.; et al. Nitric oxide production by myeloid-derived suppressor cells plays a role in impairing Fc receptor–mediated natural killer cell function. Clin. Cancer Res. 2018, 24, 1891–1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, Z.; Abdelaal, A.M.; Shi, L.; Liang, H.; Xiong, L.; Kidder, K.; Venkataramani, M.; Culpepper, C.; Zen, K.; Liu, Y. Arginase-1 is neither constitutively expressed in nor required for myeloid-derived suppressor cell-mediated inhibition of T-cell proliferation. Eur. J. Immunol. 2018, 48, 1046–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhyay, D.; Mukherjee, S.; Roy, S.; Dalton, J.E.; Kundu, S.; Sarkar, A.; Das, N.K.; Kaye, P.M.; Chatterjee, M. M2 Polarization of Monocytes-Macrophages is a Hallmark of Indian Post Kala-Azar Dermal Leishmaniasis. PLoS Negl. Trop. Dis. 2015, 9, e0004145. [Google Scholar] [CrossRef] [PubMed]
- Marvel, D.; Gabrilovich, D.I. Myeloid-derived suppressor cells in the tumor microenvironment: Expect the unexpected. J. Clin. Investig. 2015, 125, 3356–3364. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.J.; Montserrat, E.; Rai, K.R.; et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008, 111, 5446–5456. [Google Scholar] [CrossRef] [Green Version]
- Rai, K.R.; Sawitsky, A.; Cronkite, E.P.; Chanana, A.D.; Levy, R.N.; Pasternack, B.S. Clinical staging of chronic lymphocytic leukemia. Blood 1975, 46, 219–234. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Bojarska-Junak, A.; Waldowska, M.; Woś, J.; Chocholska, S.; Hus, I.; Tomczak, W.; Dzik, M.; Hus, M.; Roliński, J. Intracellular IL-4 and IFN-γ expression in iNKT cells from patients with chronic lymphocytic leukemia. Oncol. Lett. 2018, 15, 1580–1590. [Google Scholar] [CrossRef] [Green Version]
- Hus, I.; Podhorecka, M.; Bojarska-Junak, A.; Roliński, J.; Schmitt, M.; Sieklucka, M.; Wąsik-Szczepanek, E.; Dmoszyńska, A. The clinical significance of ZAP-70 and CD38 expression in B-cell chronic lymphocytic leukaemia. Ann. Oncol. 2006, 17, 683–690. [Google Scholar] [CrossRef]






| Variable | All Patients | M-MDSClow | M-MDSChigh |
|---|---|---|---|
| No. of patients (%) | 110 | 56 (50.1) | 54 (49.9) |
| Sex | |||
| Female (%) | 60 (51.8) | 26 (46.4) | 34 (63.0) |
| Male (%) | 50 (48.2) | 30 (53.6) | 20 (37.0) |
| Rai Stage | |||
| 0 (%) | 56 (50.9) | 33 (58.9) | 23 (42.6) |
| I (%) | 25 (22.7) | 15 (26.8) | 10 (18.5) |
| II (%) | 17 (15.5) | 6 (10.7) | 11 (20.4) |
| III (%) | 7 (6.4) | 1 (1.8) | 6 (11.1) |
| IV (%) | 5 (4.5) | 1 (1.8) | 4 (7.4) |
| ZAP-70 (cut-off 20%) | |||
| Positive (%) | 38 (34.5) | 15 (26.8) | 23 (42.6) |
| Negative (%) | 72 (65.5) | 41 (73.2) | 31 (57.4) |
| CD38 (cut-off 30%) | |||
| Positive (%) | 37 (33.6) | 14 (55.3) | 23(24.6) |
| Negative (%) | 73 (66.4) | 42 (44.7) | 31 (75.4) |
| IGHV gene mutation status | |||
| Mutated (%) | 26 (23.6) | 17 (30.4) | 9 (16.7) |
| Unmutated (%) | 19 (17.3) | 6 (10.7) | 13 (24.1) |
| Not available (%) | 65 (59.1) | 33 (58.9) | 32 (59.2) |
| Cytogenetic abnormalities | |||
| del(17p13.1) (%) | 5 (4.5) | 4 (7.1) | 1 (1.9) |
| del(11q22.3) (%) | 14 (12.7) | 5 (8.9) | 9 (16.6) |
| del(17p13.1) and del(11q22.3) (%) | 2 (1.8) | 1 (1.8) | 1 (1.9) |
| Without del(17p13.1) and del(11q22.3) (%) | 89 (81.0) | 46 (82.2) | 43 (79.6) |
| Patients requiring therapy (%) | 26 (23.6) | 7 (12.5) | 19 (35.2) |
| Untreated patients (%) | 84 (76.4) | 49 (87.5) | 35 (34.8) |
| No. of Deaths (%) | 8 (7.3) | 1 (1.8) | 7 (12.9) |
| Age at diagnosis (years) * | 65 (47–87) | 66 (48–87) | 64 (47–85) |
| WBC count (G/L) * | 23.9 (10.1–298.4) | 20.79 (10.72–26.8) a | 27.11 (10.1–298.4) a |
| Lymphocyte count (G/L) * | 17.86 (5.2–284.9) | 16.39 (5.2–53.5) b | 22.7 (5.5–284.9) b |
| LDH (IU/L) * | 366 (178–618) | 366 (246–618) | 368 (178–579) |
| Haemoglobin (g/dL) * | 13.9 (8.1–17.2) | 14.0 (11.4–17.2) | 13.8 (8.1–16.2) |
| Platelets (G/L) * | 191 (49–414) | 198 (90–414) | 182 (142–394) |
| β2M (mg/dL) * | 2.4 (1.3–5.4) | 2.26 (1.4–4.2) | 2.59 (1.3–5.4) |
| % CD19+/CD5+/ZAP-70+ cells * | 13.2 (0.2–50.3) | 11.97 (0.37–44.3) | 14.5 (0.2–50.3) |
| % CD19+/CD5+/CD38+ cells * | 6.9 (0.2–88.7) | 4.99 (0.2–88.7) | 9.38 (0.2–80.9) |
| Variations | Univariate # | Multivariate | |||
|---|---|---|---|---|---|
| Median TTT (months) | HR (95% CI) | p | HR (95% CI) | p | |
| Age | |||||
| ≥65 years | 29 | 0.77 (0.35–1.70) | 0.525 | ||
| <65 years | 34 | ||||
| ZAP-70 | |||||
| ≥20% | 21 | 5.76 (2.48–13.38) | <0.0001 | 3.88 (1.43–10.47) | <0.01 |
| <20% | 35 | ||||
| CD38 | |||||
| ≥30% | 20 | 4.53 (1.99–10.30) | <0.01 | 1.64 (0.60–4.50) | 0.333 |
| <30% | 34 | ||||
| Β2M | |||||
| ≥3.5 mg/dL | 15 | 4.96 (2.24–10.97) | <0.0001 | 4.29 (2.01–11.16) | 0.085 |
| <3.5 mg/dL | 36 | ||||
| del(17p13.1) or del(11q22.3) | |||||
| Positive | 20 | 3.22 (1.44–7.19) | <0.01 | 2.59 (1.12–6.02) | <0.05 |
| Negative | 32 | ||||
| LDH | |||||
| ≥ULN * | 29 | 2.29 (1.10–4.76) | 0.155 | ||
| <ULN | 34 | ||||
| IGHV mutation status# | |||||
| Unmutated | 40 | 4.04 (1.27–12.80) | 0.056 | ||
| Mutated | 44 | ||||
| M-MDSC | |||||
| ≥9.35% | 26 | 0.31 (0.13–0.72) | <0.01 | 0.39 (0.16–0.96) | <0.05 |
| <9.35% | 40 | ||||
| Variations | Univariate # | Multivariate | |||
|---|---|---|---|---|---|
| Median OS (months) | HR (95% CI) | p | HR (95% CI) | p | |
| ≥65 years | 36 | 1.46 (0.34–6.13) | 0.603 | ||
| <65 years | 40 | ||||
| ZAP-70 | |||||
| ≥20% | 35 | 1.21 (0.28–5.05) | <0.01 | 0.39 (0.08–1.83) | 0.238 |
| <20% | 41 | ||||
| CD38 | |||||
| ≥30% | 37 | 2.00 (0.50–8.01) | 0.326 | ||
| <30% | 38 | ||||
| Β2M | |||||
| ≥3.5 mg/dL | 31 | 10.26 (2.06–50.89) | <0.01 | 10.36 (1.85–58.20) | <0.01 |
| <3.5 mg/dL | 40 | ||||
| del(17p13.1) or del(11q22.3) | |||||
| Positive | 33 | 4.15 (1.04–16.63) | <0.01 | 3.35 (0.82–13.80) | 0.092 |
| Negative | 43 | ||||
| LDH | |||||
| ≥ULN * | 32 | 0.41 (0.09–1.73) | 0.22 | ||
| <ULN | 41 | ||||
| M-MDSC | |||||
| ≥9.35% | 33 | 0.22 (0.05–0.86) | <0.05 | 0.18 (0.02–1.44) | 0.107 |
| <9.35% | 45 | ||||
| Variable | Unit | HV (n = 20) Median (IQR) | CLL (n = 60) Median (IQR) | p |
|---|---|---|---|---|
| IDO | % | 2.32 (1.40–4.42) | 60.51 (19.77–79.46) | <0.0001 |
| ΔMFI | 214.6 (167.9–329.6) | 350.0 (295.6–475.3) | <0.05 | |
| Arg1 | % | 51.69 (16.36–65.33) | 86.19 (70.64-99.43) | <0.0001 |
| ΔMFI | 359.8 (317.4–597.9) | 608.5 (390.7–770.6) | <0.05 | |
| NOS2 | % | 21.64 (12.23–42.30) | 70.77 (47.04–92.08) | <0.0001 |
| ΔMFI | 255.7 (199.1–319.7) | 468.0 (215.8–655.7) | <0.01 | |
| IL-10 | % | 0.66 (0.12–1.64) | 27.34 (16.3–71.03) | <0.0001 |
| ΔMFI | 275.3 (195.2–323.6) | 390.0 (328.1–477.5) | <0.01 | |
| TGF-β | % | 1.44 (0.70–4.64) | 49.93 (22.17–81.29) | <0.0001 |
| ΔMFI | 326.2 (229.0–397.7) | 430.3 (332.3–553.5) | <0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarobkiewicz, M.; Kowalska, W.; Chocholska, S.; Tomczak, W.; Szymańska, A.; Morawska, I.; Wojciechowska, A.; Bojarska-Junak, A. High M-MDSC Percentage as a Negative Prognostic Factor in Chronic Lymphocytic Leukaemia. Cancers 2020, 12, 2614. https://doi.org/10.3390/cancers12092614
Zarobkiewicz M, Kowalska W, Chocholska S, Tomczak W, Szymańska A, Morawska I, Wojciechowska A, Bojarska-Junak A. High M-MDSC Percentage as a Negative Prognostic Factor in Chronic Lymphocytic Leukaemia. Cancers. 2020; 12(9):2614. https://doi.org/10.3390/cancers12092614
Chicago/Turabian StyleZarobkiewicz, Michał, Wioleta Kowalska, Sylwia Chocholska, Waldemar Tomczak, Agata Szymańska, Izabela Morawska, Agnieszka Wojciechowska, and Agnieszka Bojarska-Junak. 2020. "High M-MDSC Percentage as a Negative Prognostic Factor in Chronic Lymphocytic Leukaemia" Cancers 12, no. 9: 2614. https://doi.org/10.3390/cancers12092614
APA StyleZarobkiewicz, M., Kowalska, W., Chocholska, S., Tomczak, W., Szymańska, A., Morawska, I., Wojciechowska, A., & Bojarska-Junak, A. (2020). High M-MDSC Percentage as a Negative Prognostic Factor in Chronic Lymphocytic Leukaemia. Cancers, 12(9), 2614. https://doi.org/10.3390/cancers12092614








