Direct Antiviral Treatments for Hepatitis C Virus Have Off-Target Effects of Oncologic Relevance in Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Cell Culture and Treatments
2.2. RNA Sequencing and Bioinformatics Analysis
2.3. Patients
2.4. Statistical Analysis
3. Results
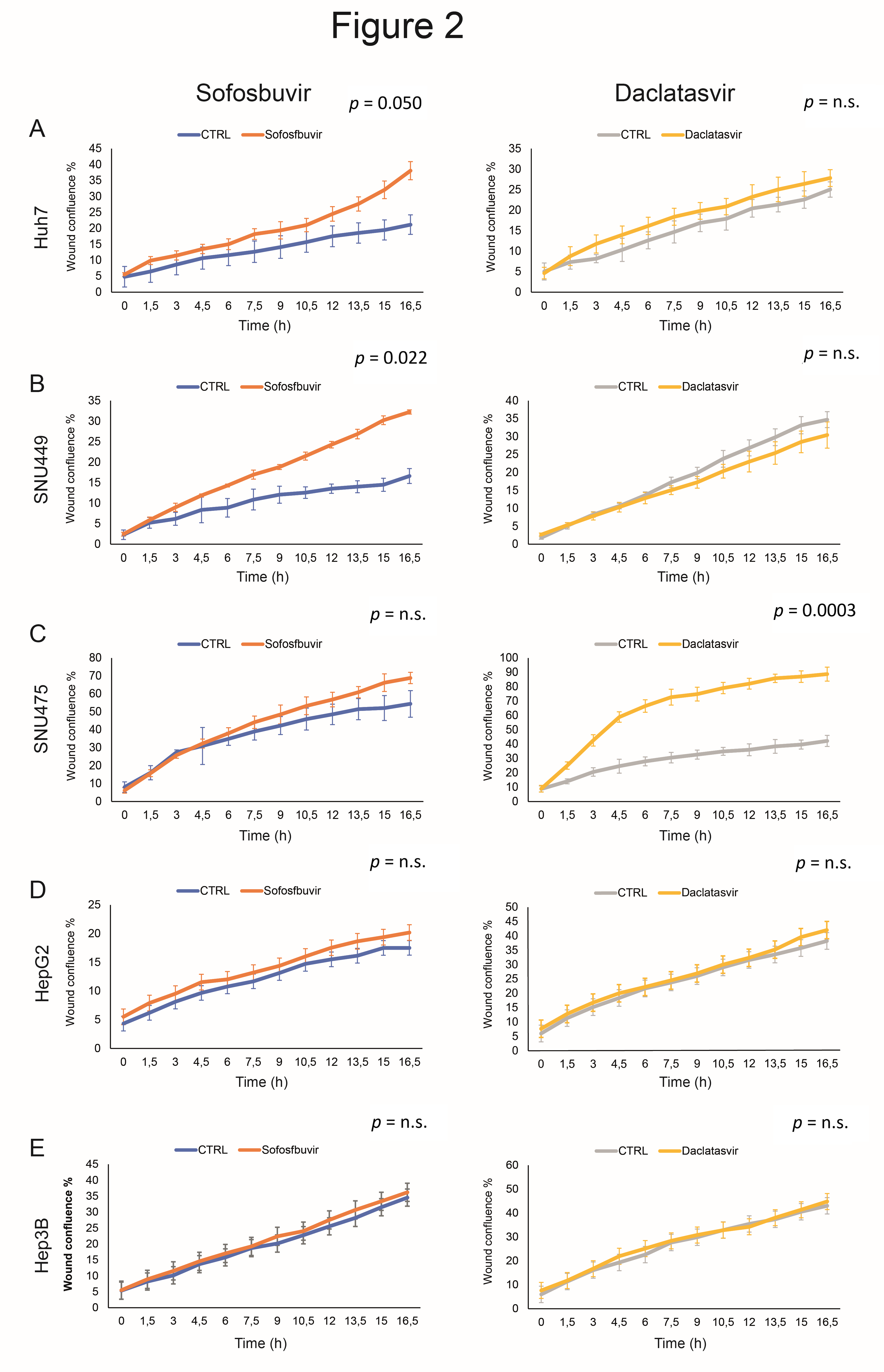
3.1. Sofosbuvir and Daclatasvir Affect Proliferation of HCC Cell Lines
3.2. Sofosbuvir and Daclatasvir Affect Migration of HCC Cells
3.3. Sofosbuvir and Daclatasvir Affect Proliferation and Migration of Non-HCC Cancer Cell Lines
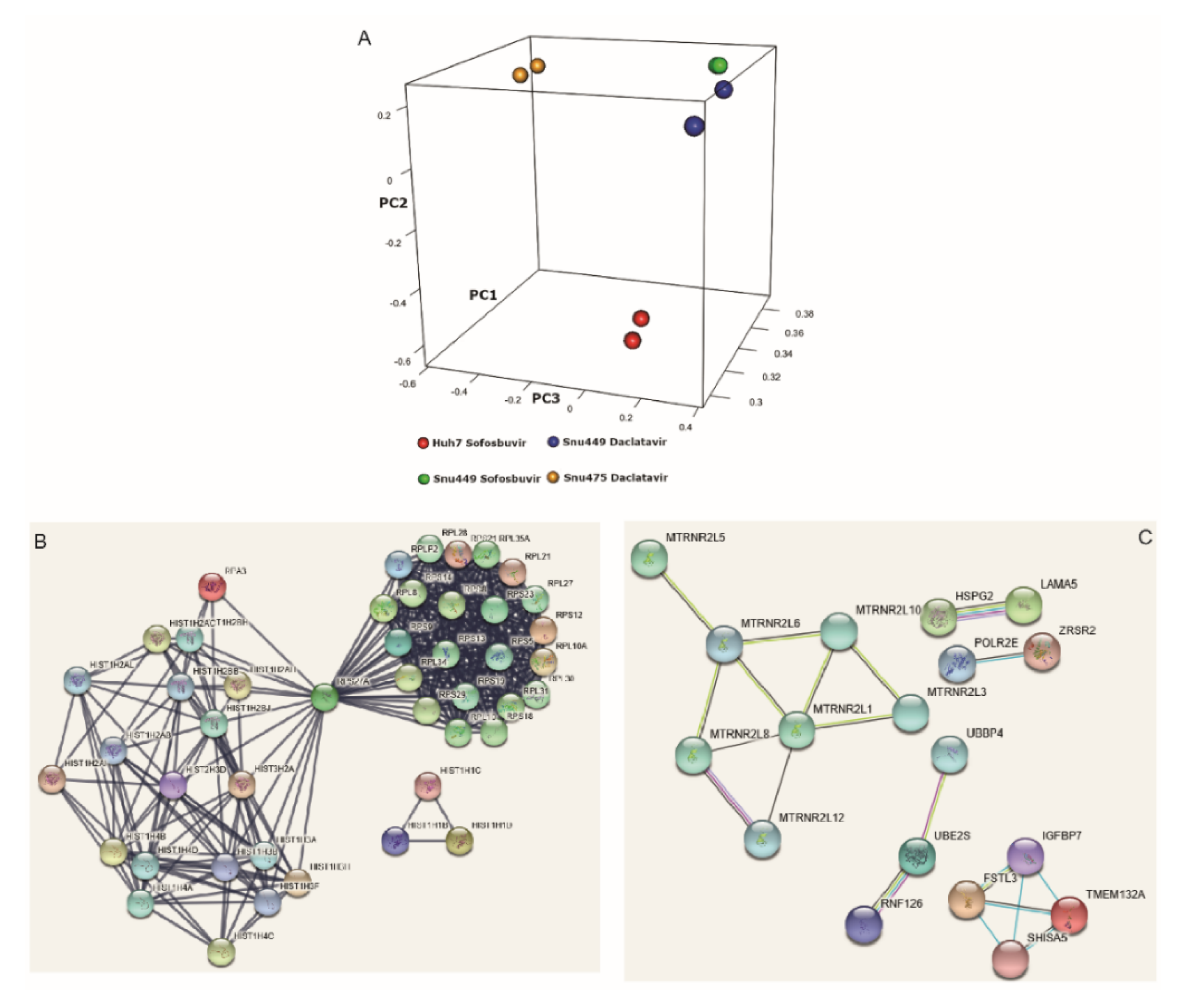
3.4. DAAs Modulate Gene Expression of HCC-Derived Cells
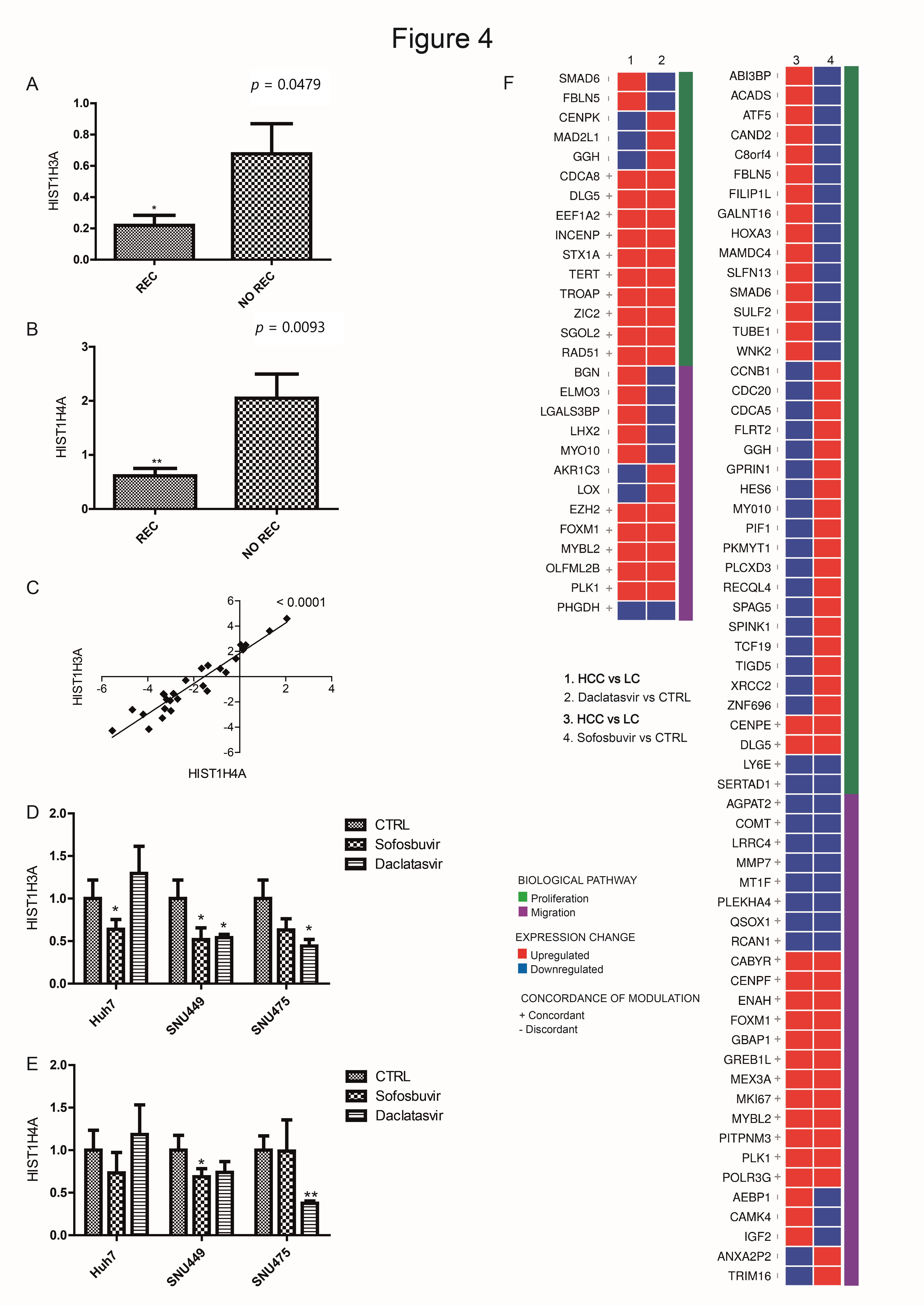
3.5. DAA Treatment Regulates HCC-Related Genes
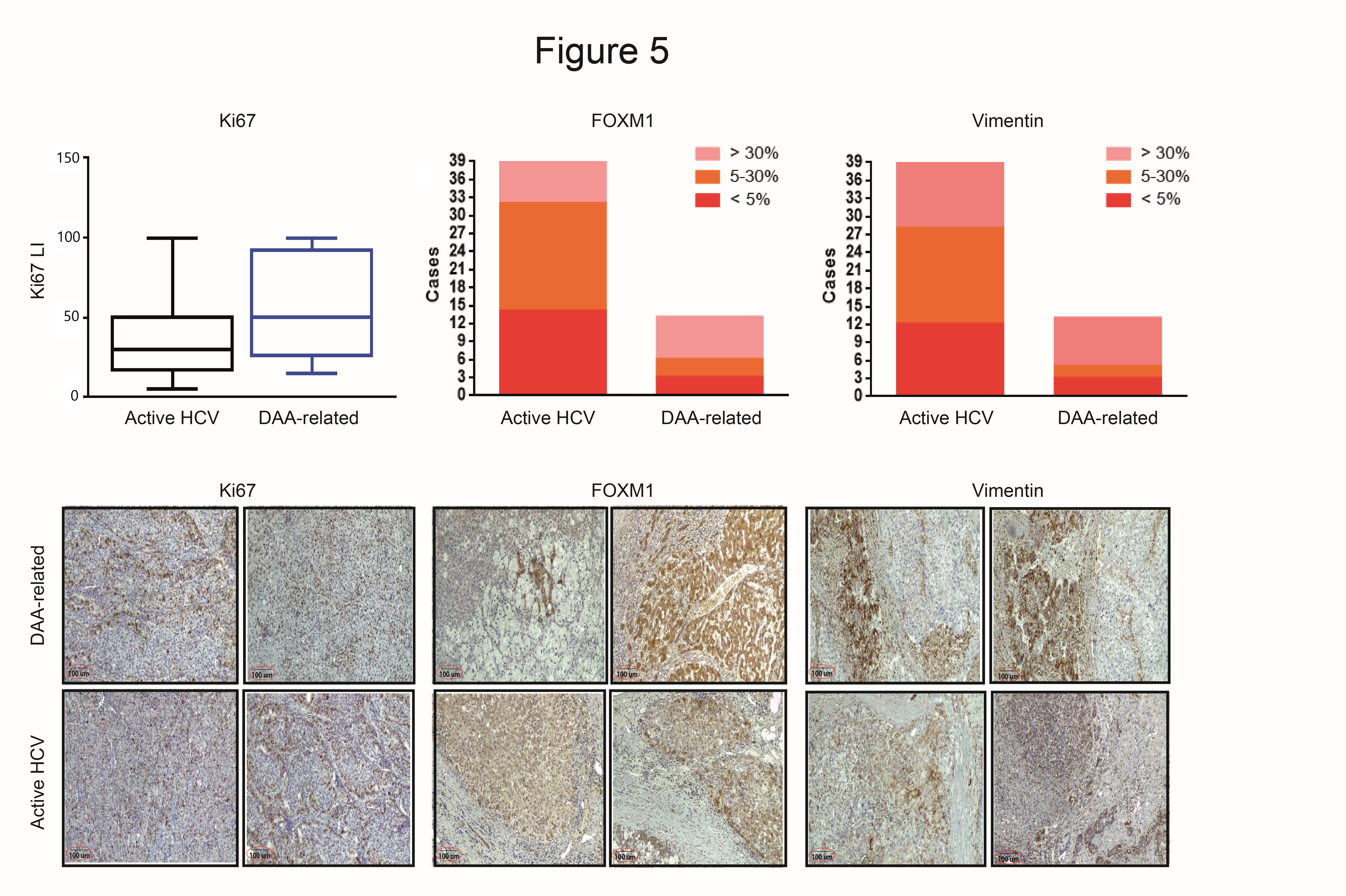
3.6. Immunohistochemistry Evaluation in HCC with and without DAAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma |
| HCV | Hepatitis C virus |
| HBV | Hepatitis B virus |
| DAAs | Direct-acting antiviral agents |
| DMSO | Dimethyl sulfoxide |
| FBS | Fetal bovine serum |
| VIM | Vimentin |
| LI | Labeling index |
| NGS | Next-generation sequencing |
| MVI | Microvascular invasion |
| TTPVI | Two-trait predictor of vascular invasion |
| NSTM | Non-smooth tumor margin |
| PTE | Peri-tumoral enhancement |
| qPCR | Quantitative polymerase chain reaction |
| IHC | Immunohistochemistry |
| CCA | Cholangiocarcinoma |
| CTRL | Control |
| EMT | Epidermal-to-mesenchymal transition |
| VEGF | Vascular endothelial growth factor |
| FFPE | Formalin-fixed, paraffin-embedded |
| PCA | Principal component analysis |
References
- Cabibbo, G.; Celsa, C.; Calvaruso, V.; Petta, S.; Cacciola, I.; Cannavo, M.R.; Madonia, S.; Rossi, M.; Magro, B.; Rini, F.; et al. Direct-acting antivirals after successful treatment of early hepatocellular carcinoma improve survival in HCV-cirrhotic patients. J. Hepatol. 2019, 71, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Conti, F.; Buonfiglioli, F.; Scuteri, A.; Crespi, C.; Bolondi, L.; Caraceni, P.; Foschi, F.G.; Lenzi, M.; Mazzella, G.; Verucchi, G.; et al. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J. Hepatol. 2016, 65, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Kozbial, K.; Moser, S.; Schwarzer, R.; Laferl, H.; Al-Zoairy, R.; Stauber, R.; Stättermayer, A.F.; Beinhardt, S.; Graziadei, I.; Freissmuth, C.; et al. Unexpected high incidence of hepatocellular carcinoma in cirrhotic patients with sustained virologic response following interferon-free direct-acting antiviral treatment. J. Hepatol. 2016, 65, 856–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reig, M.; Marino, Z.; Perello, C.; Inarrairaegui, M.; Ribeiro, A.; Lens, S.; Díaz, A.; Vilana, R.; Darnell, A.; Varela, M.; et al. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J. Hepatol. 2016, 65, 719–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ioannou, G.N.; Green, P.K.; Berry, K. HCV eradication induced by direct-acting antiviral agents reduces the risk of hepatocellular carcinoma. J. Hepatol. 2018, 68, 25–32. [Google Scholar] [CrossRef]
- Anrs Collaborative Study Group on Hepatocellular Carcinoma. Lack of evidence of an effect of direct-acting antivirals on the recurrence of hepatocellular carcinoma: Data from three ANRS cohorts. J. Hepatol. 2016, 65, 734–740. [Google Scholar] [CrossRef] [Green Version]
- Torres, H.A.; Vauthey, J.N.; Economides, M.P.; Mahale, P.; Kaseb, A. Hepatocellular carcinoma recurrence after treatment with direct-acting antivirals: First, do no harm by withdrawing treatment. J. Hepatol. 2016, 65, 862–864. [Google Scholar] [CrossRef]
- Reig, M.; Boix, L.; Marino, Z.; Torres, F.; Forns, X.; Bruix, J. Liver Cancer Emergence Associated with Antiviral Treatment: An Immune Surveillance Failure? Semin. Liver Dis. 2017, 37, 109–118. [Google Scholar]
- Gaglio, P.J. Extrahepatic and Intrahepatic Malignancies in Patients With HCV Who Achieve an SVR With Directly Acting Antiviral Agents: Should We be Concerned that DAA Therapy Contributed to this Phenomenon? J. Clin. Gastroenterol. 2017, 51, 657–658. [Google Scholar] [CrossRef]
- Khoury, J.; Nassar, G.; Kramsky, R.; Saadi, T. Extrahepatic Malignancies After Treatment with Direct Antiviral Agents for Chronic HCV Infection. J. Gastrointest. Cancer 2020, 51, 584–590. [Google Scholar] [CrossRef]
- Lin, R.J.; Moskovits, T.; Diefenbach, C.S.; Hymes, K.B. Development of highly aggressive mantle cell lymphoma after sofosbuvir treatment of hepatitis C. Blood Cancer J. 2016, 6, e402. [Google Scholar] [CrossRef] [Green Version]
- Vakiti, A.; Cho, M.H.; Lee, W.; Liang, J.J.; Lalos, A.T.; Fishbein, D.A. Use of direct-acting antivirals for hepatitis C viral infection and association with intrahepatic cholangiocarcinoma: Is there a linkage? J. Oncol. Pharm. Pract. 2019, 25, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Faillaci, F.; Marzi, L.; Critelli, R.; Milosa, F.; Schepis, F.; Turola, E.; Andreani, S.; Vandelli, G.; Bernabucci, V.B.; D’Ambrosio, F.; et al. Liver Angiopoietin-2 Is a Key Predictor of De Novo or Recurrent Hepatocellular Cancer After Hepatitis C Virus Direct-Acting Antivirals. Hepatology 2018, 68, 1010–1024. [Google Scholar] [CrossRef] [Green Version]
- Freedman, H.; Winter, P.; Tuszynski, J.; Tyrrell, D.L.; Houghton, M. A computational approach for predicting off-target toxicity of antiviral ribonucleoside analogues to mitochondrial RNA polymerase. J. Biol. Chem. 2018, 293, 9696–9705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachovchin, D.A.; Koblan, L.W.; Wu, W.; Liu, Y.; Li, Y.; Zhao, P.; Woznica, I.; Shu, Y.; Lai, J.H.; Poplawski, S.E.; et al. A high-throughput, multiplexed assay for superfamily-wide profiling of enzyme activity. Nat. Chem. Biol. 2014, 10, 656–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Yazbi, A.F.; Loppnow, G.R. Probing DNA damage induced by common antiviral agents using multiple analytical techniques. J. Pharm. Biomed. Anal. 2018, 157, 226–234. [Google Scholar] [CrossRef]
- Gramantieri, L.; Baglioni, M.; Fornari, F.; Laginestra, M.A.; Ferracin, M.; Indio, V.; Ravaioli, M.; Cescon, M.; De Pace, V.; Leoni, S.; et al. LncRNAs as novel players in hepatocellular carcinoma recurrence. Oncotarget 2018, 9, 35085–35099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renzulli, M.; Buonfiglioli, F.; Conti, F.; Brocchi, S.; Serio, I.; Foschi, F.G.; Caraceni, P.; Mazzella, G.; Verucchi, G.; Golfieri, R.; et al. Imaging features of microvascular invasion in hepatocellular carcinoma developed after direct-acting antiviral therapy in HCV-related cirrhosis. Eur. Radiol. 2018, 28, 506–513. [Google Scholar] [CrossRef]
- Baghy, K.; Tatrai, P.; Regos, E.; Kovalszky, I. Proteoglycans in liver cancer. World J. Gastroenterol. 2016, 22, 379–393. [Google Scholar] [CrossRef]
- Bartolini, A.; Cardaci, S.; Lamba, S.; Oddo, D.; Marchio, C.; Cassoni, P.; Amoreo, C.A.; Corti, G.; Testori, A.; Bussolino, F.; et al. BCAM and LAMA5 Mediate the Recognition between Tumor Cells and the Endothelium in the Metastatic Spreading of KRAS-Mutant Colorectal Cancer. Clin. Cancer Res. 2016, 22, 4923–4933. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, Q.; Gu, J.; Yin, L.; Liang, S.; Wu, L.; Xu, H.; Zhao, C.; Gu, Y. Elevated mitochondrial SLC25A29 in cancer modulates metabolic status by increasing mitochondria-derived nitric oxide. Oncogene 2018, 37, 2545–2558. [Google Scholar] [CrossRef]
- Meng, F.D.; Wei, J.C.; Qu, K.; Wang, Z.X.; Wu, Q.F.; Tai, M.H.; Liu, H.-C.; Zhang, R.-Y.; Liu, C. FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 196–213. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Chen, S.; Han, J.X.; Qian, B.; Wang, X.-R.; Zhong, W.-L.; Qin, Y.; Zhang, H.; Gao, W.-F.; Lei, Y.-Y.; et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4150–4162. [Google Scholar] [CrossRef] [Green Version]
- Scatena, R.; Bottoni, P.; Botta, G.; Martorana, G.E.; Giardina, B. The role of mitochondria in pharmacotoxicology: A reevaluation of an old, newly emerging topic. Am. J. Physiol. Cell Physiol. 2007, 293, C12–C21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallace, K.B. Mitochondrial off-targets of drug therapy. Trends Pharmacol. Sci. 2008, 29, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Landry, D.M.; Hertz, M.I. Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009, 23, 2753–2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Las Heras-Rubio, A.; Perucho, L.; Paciucci, R.; Vilardell, J.; LLeonart, M.E. Ribosomal proteins as novel players in tumorigenesis. Cancer Metastasis Rev. 2014, 33, 115–141. [Google Scholar] [CrossRef]
- Takada, H.; Kurisaki, A. Emerging roles of nucleolar and ribosomal proteins in cancer, development, and aging. Cell Mol. Life Sci. 2015, 72, 4015–4025. [Google Scholar] [CrossRef]
- Murray, J.L.; Sheng, J.; Rubin, D.H. A role for H/ACA and C/D small nucleolar RNAs in viral replication. Mol. Biotechnol. 2014, 56, 429–437. [Google Scholar] [CrossRef]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef]
- Mejlvang, J.; Feng, Y.; Alabert, C.; Neelsen, K.J.; Jasencakova, Z.; Zhao, X.; Lees, M.; Sandelin, A.; Pasero, P.; Lopes, M.; et al. New histone supply regulates replication fork speed and PCNA unloading. J. Cell Biol. 2014, 204, 29–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Clinical Feature | Post-DAA Cohort n = 13 (IHC) | Active HCV Cohort n = 39 (RNA-Seq, IHC and qPCR) | Difference |
|---|---|---|---|
| Gender (M/F) | 8/5 | 25/14 | p = n.s. |
| AFP 1 | <20 ng/mL 8 (61.5%) | 22 (56.4%) | p = n.s. |
| >20 ng/mL 5 (38.5%) | 17 (43.6%) | p = n.s. | |
| Primary/recurrence | 13/0 (100%) | 39/0 (100%) | p = n.s. |
| Maximum HCC size | <5 cm: 12 (92.3%) | <5 cm: 32 (82.1%) | p = n.s. |
| >5 cm: 1 (7.7%) | >5 cm: 7 (17.9%) | p = n.s. | |
| Unifocal | 10 (76.9%) | 34 (87.2%) | p = n.s. |
| Multifocal | 3 (23.1%) | 5 (12.8%) | p = n.s. |
| Grading G1-G2 | 6 (46.1%) | 21 (53.8%) | p = n.s. |
| Grading G3-G4 | 7 (53.8%) | 18 (46.2%) | p = n.s. |
| Etiology of CLD 2 | HCV (cleared): 13 (100%) | HCV (active): 39 (100%) |
| Pathways | p-Value | Count |
|---|---|---|
| Translation | 1.2 × 10−21 | 29 |
| Viral transcription | 1.5 × 10−18 | 20 |
| rRNA processing | 1.2 × 10−17 | 24 |
| Oxidative phosphorylation | 4.8 × 10−16 | 21 |
| Respiratory chain | 4.9 × 10−9 | 10 |
| Electron transport | 4.9 × 10−8 | 11 |
| ATP biosynthetic process | 8.5 × 10−6 | 6 |
| Histone core | 6.9 × 10−5 | 8 |
| Anaphase-promoting complex | 1.2 × 10−5 | 8 |
| NF-kappa B signaling | 4.6 × 10−5 | 7 |
| Gene Families | Genes |
|---|---|
| Mitochondrial DNA-like sequences | MTRNR2L1, MTRNR2L10, MTRNR2L12, MTRNR2L2, MTRNR2L3, MTRNR2L5, MTRNR2L6, MTRNR2L8 |
| Ribosomal protein (RP)-coding genes and pseudogenes (RPL) | RP11-329L6.1, RP11-36C20.1, RP11-75L1.2, RP11-832N8.1, RP11-889L3.1, RP3-375P9.2, RP11-1136G11.6, RP11-121L10.3, RP11-153M3.1; RPL13AP20, RPL41P2, RPL4P5, RPL7P47, RPS11P5, RPS18P9 |
| Small nuclear RNA genes (RNU) and pseudogenes (RN) | RNU4-1, RNU4-2, RNU5B-1, RNU5E-1, RNU6ATAC, U2; RN7SKP217, RN7SKP255, RN7SKP274, RN7SKP80, RN7SKP9, RN7SKP90, RN7SKP91, RN7SL616P, RN7SL704P, RNF126 |
| Ribonucleoproteins | RNY4, RNY4P1 |
| Small nucleolar RNAs | SNORA42, SCARNA13 |
| MicroRNAs | MIR3648, MIR3687 |
| Glyceraldehyde-3-phosphate dehydrogenase pseudogenes | GAPDHP1, GAPDHP38, GAPDHP60 |
| Miscellaneous | AC004453.8, AC009245.3, ACTBP2, AURKAIP1, BCL2L12, C20orf24, CHTF18, CISD3, CTD-2184D3.1, DUT, ELP5, FSTL3, FTLP3, GDF15, H1FX, HNRNPKP4, HSF1, IGFBP7, INPP5E, KIFC2, LAMA5, LDHAP3, LMF2, MAP1S, MED16, NDUFA13, NOTCH1, PCNXL3, POLR2E, PPIAP29, PTOV1, REEP4, SCAMP4, SHISA5, SLC25A29, TMEM132A, TMSB10P1, TONSL, TTYH3, TUBBP1, UBBP4, UBE2S, VKORC1 |
| Translation elongation factor pseudogenes | EEF1A1P11, EEF1A1P13, EEF1A1P25, EEF1A1P9 |
| Heat shock protein family A pseudogenes | HSP90AB2P, HSP90AB3P, HSPA8P8, HSPG2 |
| Zinc finger proteins | ZNF512B, ZNF579 |
| Gene Families | Genes |
|---|---|
| Mitochondrial DNA-like sequences: | MTRNR2L1, MTRNR2L6 |
| Ribosomal protein (RP)-coding genes and pseudogenes (RPL) | RNA5-8SP2, RPL41P2, RP11-395B7.7, RP5-857K21.11 |
| Small nuclear RNA genes (RNU) | RNU5A-1, RNU2-6P, RNU6ATAC, RNY5, RNU5A-8P |
| Small nucleolar RNAs and pseudogenes | SNORA54, SNORA26, SCARNA6, SNORA71D, Y, U2; RN7SKP9 |
| MicroRNAs | MIR3648 |
| Histones | HIST1H3A, HIST1H4D, HIST1H4A, HIST1H2AB, HIST1H2AI |
| Miscellaneous | LAMA5, HSPG2, ERVK13-1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giovannini, C.; Fornari, F.; Indio, V.; Trerè, D.; Renzulli, M.; Vasuri, F.; Cescon, M.; Ravaioli, M.; Perrucci, A.; Astolfi, A.; et al. Direct Antiviral Treatments for Hepatitis C Virus Have Off-Target Effects of Oncologic Relevance in Hepatocellular Carcinoma. Cancers 2020, 12, 2674. https://doi.org/10.3390/cancers12092674
Giovannini C, Fornari F, Indio V, Trerè D, Renzulli M, Vasuri F, Cescon M, Ravaioli M, Perrucci A, Astolfi A, et al. Direct Antiviral Treatments for Hepatitis C Virus Have Off-Target Effects of Oncologic Relevance in Hepatocellular Carcinoma. Cancers. 2020; 12(9):2674. https://doi.org/10.3390/cancers12092674
Chicago/Turabian StyleGiovannini, Catia, Francesca Fornari, Valentina Indio, Davide Trerè, Matteo Renzulli, Francesco Vasuri, Matteo Cescon, Matteo Ravaioli, Alessia Perrucci, Annalisa Astolfi, and et al. 2020. "Direct Antiviral Treatments for Hepatitis C Virus Have Off-Target Effects of Oncologic Relevance in Hepatocellular Carcinoma" Cancers 12, no. 9: 2674. https://doi.org/10.3390/cancers12092674
APA StyleGiovannini, C., Fornari, F., Indio, V., Trerè, D., Renzulli, M., Vasuri, F., Cescon, M., Ravaioli, M., Perrucci, A., Astolfi, A., Piscaglia, F., & Gramantieri, L. (2020). Direct Antiviral Treatments for Hepatitis C Virus Have Off-Target Effects of Oncologic Relevance in Hepatocellular Carcinoma. Cancers, 12(9), 2674. https://doi.org/10.3390/cancers12092674









