Loss of Proprotein Convertase Furin in Mammary Gland Impairs proIGF1R and proIR Processing and Suppresses Tumorigenesis in Triple Negative Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Furin Is Highly Overexpressed in TNBC Tumors and Correlates with Poor Prognosis in TNBC Patients
2.2. Targeted Inactivation of Furin in the Mammary Gland via WAP-Cre Induces Normal Mammary Development
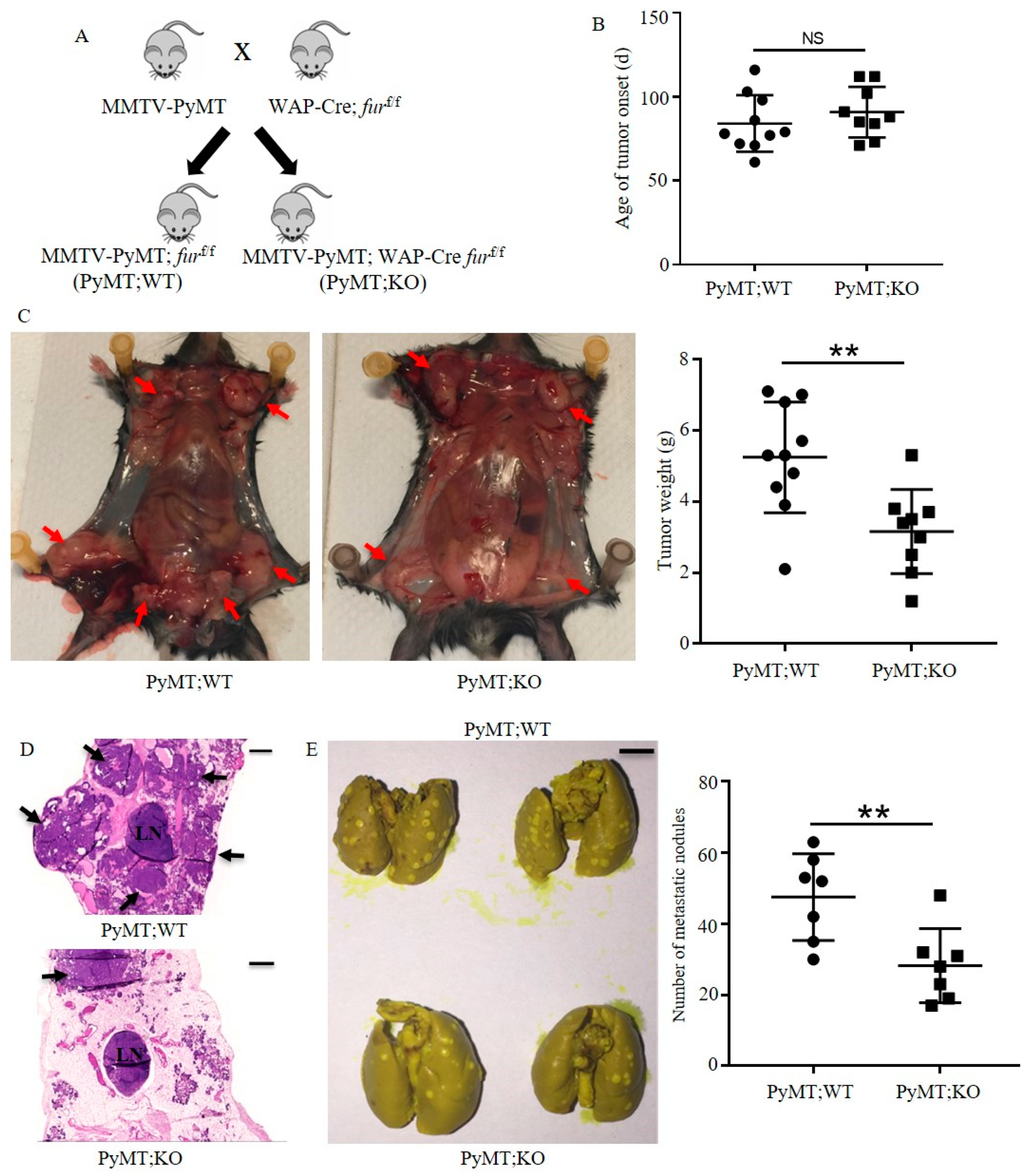
2.3. Inactivation of Furin in the Mammary Gland Inhibits Tumor Growth and Lung Metastasis in PyMT Induced TNBC Mice
2.4. Furin Inactivation in the Mammary Gland of TNBC Mice Inhibits the Proteolytic Maturation of proIGF1R and proIR and Downregulates PI3K/AKT and MAPK/ERK1/2 Expression and Activity
3. Discussion
4. Materials and Methods
4.1. Mouse Model
4.2. Animal Study
4.3. Bioinformatics Assay
4.4. Whole-Mount Staining of Mammary Glands
4.5. H&E Staining and Bouin’s Solution Staining
4.6. Quantitative RT-PCR
4.7. Isolation of Tumor Samples and Immunoblotting Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.J. Strategies for subtypes-dealing with the diversity of breast cancer: Highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, M.; Pusztai, L. Use of standard markers and incorporation of molecular markers into breast cancer therapy. Cancer 2011, 117, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Creemers, J.W.M.; Khatib, A.-M. Knock-out mouse models of proprotein convertases: Unique functions or redundancy? Front. Biosci. 2008, 13, 4960–4971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegfried, G.; Descarpentrie, J.; Evrard, S.; Khatib, A.M. Proprotein convertases: Key players in inflammation-related malignancies and metastasis. Cancer Lett. 2020, 473, 50–61. [Google Scholar] [CrossRef]
- Cheng, M.; Watson, P.H.; Paterson, J.A.; Seidah, N.; Chrétien, M.; Shiu, R.P.C. Pro-protein convertase gene expression in human breast cancer. Int. J. Cancer 1997, 71, 966–971. [Google Scholar] [CrossRef]
- Siegfried, G.; Khatib, A.M.; Benjannet, S.; Chrétien, M.; Seidah, N.G. The proteolytic processing of pro-platelet-derived growth factor-a at RRKR86 by members of the proprotein convertase family is functionally correlated to platelet-derived growth factor-A-induced functions and tumorigenicity. Cancer Res. 2003, 63, 1458–1463. [Google Scholar]
- Scamuffa, N.; Sfaxi, F.; Ma, J.; Lalou, C.; Seidah, N.; Calvo, F.; Khatib, A.M. Prodomain of the proprotein convertase subtilisin/kexin Furin (ppfurin) protects from tumor progression and metastasis. Carcinogenesis 2014, 35, 528–536. [Google Scholar] [CrossRef] [Green Version]
- Lapierre, M.; Siegfried, G.; Scamuffa, N.; Bontemps, Y.; Calvo, F.; Seidah, N.G.; Khatib, A.M. Opposing function of the proprotein convertases furin and PACE4 on breast cancer cells’ malignant phenotypes: Role of tissue inhibitors of metalloproteinase-1. Cancer Res. 2007, 67, 9030–9034. [Google Scholar] [CrossRef] [Green Version]
- Scamuffa, N.; Siegfried, G.; Bontemps, Y.; Ma, L.; Basak, A.; Cherel, G.; Calvo, F.; Seidah, N.G.; Khatib, A.-M. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J. Clin. Invest. 2008, 118, 352–363. [Google Scholar] [CrossRef] [Green Version]
- Khatib, A.M.; Siegfried, G.; Prat, A.; Luis, J.; Chrétien, M.; Metrakos, P.; Seidah, N.G. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: Importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J. Biol. Chem. 2001, 276, 30686–30693. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Thorrez, L.; Siegfried, G.; Meulemans, S.; Evrard, S.; Tejpar, S.; Khatib, A.-M.; Creemers, J.W.M. The proprotein convertase furin is a pro-oncogenic driver in KRAS and BRAF driven colorectal cancer. Oncogene 2020, 39, 3571–3587. [Google Scholar] [CrossRef] [PubMed]
- Komada, M.; Hatsuzawa, K.; Shibamoto, S.; Ito, F.; Nakayama, K.; Kitamura, N. Proteolytic processing of the hepatocyte growth factor/scatter factor receptor by furin. FEBS Lett. 1993, 328, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Ma, J.; Shao, N.; Shi, Y.; Liu, R.; Li, W.; Lin, Y.; Wang, S. Co-targeting IGF-1R and autophagy enhances the effects of cell growth suppression and apoptosis induced by the IGF-1R inhibitor NVP-AEW541 in triple-negative breast cancer cells. PLoS ONE 2017, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- De Lint, K.; Poell, J.B.; Soueidan, H.; Jastrzebski, K.; Rodriguez, J.V.; Lieftink, C.; Wessels, L.F.A.; Beijersbergen, R.L. Sensitizing triple-negative breast cancer to PI3K inhibition by cotargeting IGF1R. Mol. Cancer Ther. 2016, 15, 1545–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.J.; Choi, J.S.; Seo, J.; Song, J.Y.; Eun Lee, S.; Kwon, M.J.; Kwon, M.J.; Kundu, J.; Jung, K.; Oh, E.; et al. MET is a potential target for use in combination therapy with EGFR inhibition in triple-negative/basal-like breast cancer. Int. J. Cancer 2014, 134, 2424–2436. [Google Scholar] [CrossRef] [PubMed]
- Logeat, F.; Bessia, C.; Brou, C.; LeBail, O.; Jarriault, S.; Seidah, N.G.; Israël, A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 1998, 95, 8108–8112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanchette, F.; Day, R.; Dong, W.; Laprise, M.H.; Dubois, C.M. TGFβ1 regulates gene expression of its own converting enzyme furin. J. Clin. Invest. 1997, 99, 1974–1983. [Google Scholar] [CrossRef] [Green Version]
- Bolós, V.; Mira, E.; Martínez-Poveda, B.; Luxán, G.; Cañamero, M.; Martínez-A, C.; Mañes, S.; de la Pompa, J.L. Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res. 2013, 15, R54. [Google Scholar] [CrossRef] [Green Version]
- Padua, D.; Zhang, X.H.F.; Wang, Q.; Nadal, C.; Gerald, W.L.; Gomis, R.R.; Massagué, J. TGFβ Primes Breast Tumors for Lung Metastasis Seeding through Angiopoietin-like 4. Cell 2008, 133, 66–77. [Google Scholar] [CrossRef] [Green Version]
- Wagner, K.U.; Wall, R.J.; St-Onge, L.; Gruss, P.; Wynshaw-Boris, A.; Garrett, L.; Li, M.; Furth, P.A.; Hennighausen, L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997, 25, 4323–4330. [Google Scholar] [CrossRef] [Green Version]
- De Vos, L.; Declercq, J.; Rosas, G.G.; Van Damme, B.; Roebroek, A.; Vermorken, F.; Ceuppens, J.; Van de Ven, W.; Creemers, J. MMTV-cre-mediated fur inactivation concomitant with PLAG1 proto-oncogene activation delays salivary gland tumorigenesis in mice. Int. J. Oncol. 2008, 32, 1073–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backman, S.A.; Ghazarian, D.; So, K.; Sanchez, O.; Wagner, K.U.; Hennighausen, L.; Suzuki, A.; Tsao, M.S.; Chapman, W.B.; Stambolic, V.; et al. Early onset of neoplasia in the prostate and skin of mice with tissue-specific deletion of Pten. Proc. Natl. Acad. Sci. USA 2004, 101, 1725–1730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantozzi, A.; Christofori, G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Gao, Y.; Welte, T.; Wang, H.; Liu, J.; Janghorban, M.; Sheng, K.; Niu, Y.; Goldstein, A.; Zhao, N.; et al. Immuno-subtyping of breast cancer reveals distinct myeloid cell profiles and immunotherapy resistance mechanisms. Nat. Cell Biol. 2019, 21, 1113–1126. [Google Scholar] [CrossRef]
- Christenson, J.L.; Butterfield, K.T.; Spoelstra, N.S.; Norris, J.D.; Josan, J.S.; Pollock, J.A.; McDonnell, D.P.; Katzenellenbogen, B.S.; Katzenellenbogen, J.A.; Richer, J.K. MMTV-PyMT and Derived Met-1 Mouse Mammary Tumor Cells as Models for Studying the Role of the Androgen Receptor in Triple-Negative Breast Cancer Progression. Horm. Cancer 2017, 8, 69–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genomics 2019, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Sfaxi, F.; Scamuffa, N.; Lalou, C.; Ma, J.; Metrakos, P.; Siegfried, G.; Ragg, H.; Bikfalvi, A.; Calvo, F.; Khatib, A.-M. Repression of liver colorectal metastasis by the serpin Spn4A a naturally occurring inhibitor of the constitutive secretory proprotein convertases. Oncotarget 2014, 5, 4195–4210. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Wang, Q.; Gao, Z.; Zhou, Z.; Peng, S.; Chang, W.L.; Lin, H.Y.; Zhang, W.; Wang, H. Proprotein Convertase Furin Regulates Apoptosis and Proliferation of Granulosa Cells in the Rat Ovary. PLoS ONE 2013, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Moses, H.; Barcellos-Hoff, M.H. TGF-β Biology in mammary development and breast cancer. Cold Spring Harb. Perspect. Biol. 2011, 3, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kleinberg, D.L.; Feldman, M.; Ruan, W. IGF-I: An essential factor in terminal end bud formation and ductal morphogenesis. J. Mammary Gland Biol. Neoplasia 2000, 5, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, G.; Basak, A.; Cromlish, J.A.; Benjannet, S.; Marcinkiewicz, J.; Chrétien, M.; Seidah, N.G.; Khatib, A. The secretory proprotein convertases furin, PC5, and PC7 activate VEGF-C to induce tumorigenesis. J. Clin. Invest. 2003, 111, 1723–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roebroek, A.J.M.; Taylor, N.A.; Louagie, E.; Pauli, I.; Smeijers, L.; Snellinx, A.; Lauwers, A.; Van De Ven, W.J.M.; Hartmann, D.; Creemers, J.W.M. Limited redundancy of the proprotein convertase furin in mouse liver. J. Biol. Chem. 2004, 279, 53442–53450. [Google Scholar] [CrossRef] [Green Version]
- Javed, A.; Lteif, A. Development of the human breast. Semin. Plast. Surg. 2013, 27, 5–12. [Google Scholar] [PubMed] [Green Version]
- Lin, E.Y.; Jones, J.G.; Li, P.; Zhu, L.; Whitney, K.D.; Muller, W.J.; Pollard, J.W. Progression to Malignancy in the Polyoma Middle T Oncoprotein Mouse Breast Cancer Model Provides a Reliable Model for Human Diseases. Am. J. Pathol. 2003, 163, 2113–2126. [Google Scholar] [CrossRef] [Green Version]
- Baserga, R. The IGF-I receptor in cancer research. Exp. Cell Res. 1999, 253, 1–6. [Google Scholar] [CrossRef]
- Vella, V.; Milluzzo, A.; Scalisi, N.M.; Vigneri, P.; Sciacca, L. Insulin receptor isoforms in cancer. Int. J. Mol. Sci. 2018, 19, 3615. [Google Scholar] [CrossRef] [Green Version]
- Peruzzi, F.; Prisco, M.; Dews, M.; Salomoni, P.; Grassilli, E.; Romano, G.; Calabretta, B.; Baserga, R. Multiple Signaling Pathways of the Insulin-Like Growth Factor 1 Receptor in Protection from Apoptosis. Mol. Cell. Biol. 1999, 19, 7203–7215. [Google Scholar] [CrossRef] [Green Version]
- Stewart, D.P.; Marada, S.; Bodeen, W.J.; Truong, A.; Sakurada, S.M.; Pandit, T.; Pruett-Miller, S.M.; Ogden, S.K. Cleavage activates dispatched for Sonic Hedgehog ligand release. Elife 2018, 7, 1–24. [Google Scholar] [CrossRef]
- Agarwal, N.K.; Qu, C.; Kunkulla, K.; Liu, Y.; Vega, F. Transcriptional regulation of serine/threonine protein kinase (AKT) genes by glioma-associated oncogene homolog. J. Biol. Chem. 2013, 288, 15390–15401. [Google Scholar] [CrossRef] [Green Version]
- Riesterer, O.; Zingg, D.; Hummerjohann, J.; Bodis, S.; Pruschy, M. Degradation of PKB/Akt protein by inhibition of the VEGF receptor/mTOR pathway in endothelial cells. Oncogene 2004, 23, 4624–4635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duguay, S.J.; Milewski, W.M.; Young, B.D.; Nakayama, K.; Steiner, D.F. Processing of wild-type and mutant proinsulin-like growth factor-IA by subtilisin-related proprotein convertases. J. Biol. Chem. 1997, 272, 6663–6670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerratana, L.; Basile, D.; Buono, G.; De Placido, S.; Giuliano, M.; Minichillo, S.; Coinu, A.; Martorana, F.; De Santo, I.; Del Mastro, L.; et al. Androgen receptor in triple negative breast cancer: A potential target for the targetless subtype. Cancer Treat. Rev. 2018, 68, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.L.B.; Han, H.S.; Gradishar, W.J. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: A review. Breast Cancer Res. Treat. 2018, 169, 397–406. [Google Scholar] [CrossRef]
- Giltnane, J.M.; Balko, J.M. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov. Med. 2014, 17, 275–283. [Google Scholar]
- Traina, T.A.; Miller, K.; Yardley, D.A.; Eakle, J.; Schwartzberg, L.S.; O’Shaughnessy, J.; Gradishar, W.; Schmid, P.; Winer, E.; Kelly, C.; et al. Enzalutamide for the treatment of androgen receptor-expressing triple-negative breast cancer. J. Clin. Oncol. 2018, 36, 884–890. [Google Scholar] [CrossRef]
- Gucalp, A.; Tolaney, S.; Isakoff, S.J.; Ingle, J.N.; Liu, M.C.; Carey, L.A.; Blackwell, K.; Rugo, H.; Nabell, L.; Forero, A.; et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic breast cancer. Clin. Cancer Res. 2013, 19, 5505–5512. [Google Scholar] [CrossRef] [Green Version]
- Farhat, D.; Ghayad, S.E.; Icard, P.; Le Romancer, M.; Hussein, N.; Lincet, H. Lipoic acid-induced oxidative stress abrogates IGF-1R maturation by inhibiting the CREB/furin axis in breast cancer cell lines. Oncogene 2020, 39, 3604–3610. [Google Scholar] [CrossRef]
- Litzenburger, B.C.; Creighton, C.J.; Tsimelzon, A.; Chan, B.T.; Hilsenbeck, S.G.; Wang, T.; Carboni, J.M.; Gottardis, M.M.; Huang, F.; Chang, J.C.; et al. High IGF-IR activity in triple-negative breast cancer cell lines and tumorgrafts correlates with Sensitivity to anti-IGF-IR therapy. Clin. Cancer Res. 2011, 17, 2314–2327. [Google Scholar] [CrossRef] [Green Version]
- Soulet, F.; Bodineau, C.; Hooks, K.B.; Descarpentrie, J.; Alves, I.D.; Dubreuil, M.; Mouchard, A.; Eugenie, M.; Hoepffner, J.-L.; Lopez, J.J.; et al. Furin-cleaved ELA/Apela precursor displays tumor suppressor function in renal cell carcinoma through mTORC1 activation. JCI Insight 2020. [Google Scholar] [CrossRef]
- Chaudhary, S.S.; Choudhary, S.; Rawat, S.; Ahir, G.; Bilgrami, A.L.; Ashraf, G.M. c-Met as a potential therapeutic target in triple negative breast cancer. In Cancer-Leading Proteases; Elsevier: Amsterdam, The Netherlands, 2020; pp. 295–326. [Google Scholar]
- Qiu, M.; Peng, Q.; Jiang, I.; Carroll, C.; Han, G.; Rymer, I.; Lippincott, J.; Zachwieja, J.; Gajiwala, K.; Kraynov, E.; et al. Specific inhibition of Notch1 signaling enhances the antitumor efficacy of chemotherapy in triple negative breast cancer through reduction of cancer stem cells. Cancer Lett. 2013, 328, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Tomé, M.; Pappalardo, A.; Soulet, F.; López, J.J.; Olaizola, J.; Leger, Y.; Dubreuil, M.; Mouchard, A.; Fessart, D.; Delom, F.; et al. Inactivation of Proprotein Convertases in T Cells Inhibits PD-1 Expression and Creates a Favorable Immune Microenvironment in Colorectal Cancer. Cancer Res. 2019, 79, 5008–5021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Khatib, A.; Creemers, J.W.M. Loss of the proprotein convertase Furin in T cells represses mammary tumorigenesis in oncogene-driven triple negative breast cancer. Cancer Lett. 2020, 484, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Khatib, A.-M.; Creemers, J.W.M. Loss of Proprotein Convertase Furin in Mammary Gland Impairs proIGF1R and proIR Processing and Suppresses Tumorigenesis in Triple Negative Breast Cancer. Cancers 2020, 12, 2686. https://doi.org/10.3390/cancers12092686
He Z, Khatib A-M, Creemers JWM. Loss of Proprotein Convertase Furin in Mammary Gland Impairs proIGF1R and proIR Processing and Suppresses Tumorigenesis in Triple Negative Breast Cancer. Cancers. 2020; 12(9):2686. https://doi.org/10.3390/cancers12092686
Chicago/Turabian StyleHe, Zongsheng, Abdel-Majid Khatib, and John W. M. Creemers. 2020. "Loss of Proprotein Convertase Furin in Mammary Gland Impairs proIGF1R and proIR Processing and Suppresses Tumorigenesis in Triple Negative Breast Cancer" Cancers 12, no. 9: 2686. https://doi.org/10.3390/cancers12092686
APA StyleHe, Z., Khatib, A.-M., & Creemers, J. W. M. (2020). Loss of Proprotein Convertase Furin in Mammary Gland Impairs proIGF1R and proIR Processing and Suppresses Tumorigenesis in Triple Negative Breast Cancer. Cancers, 12(9), 2686. https://doi.org/10.3390/cancers12092686




