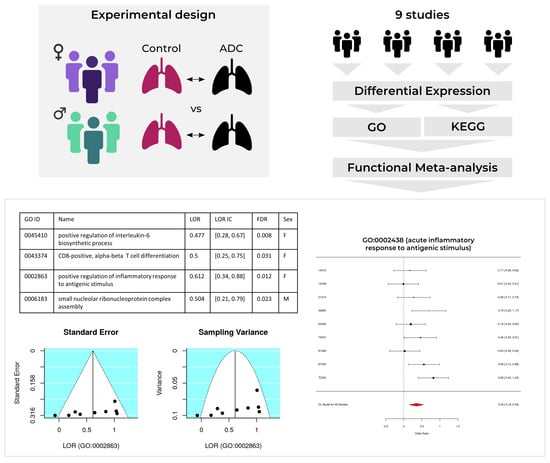
Functional Signatures in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis of Sex-Based Differences in Transcriptomic Studies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Study Search and Selection
2.2. Individual Analysis
2.3. Meta-Analysis
2.3.1. Upregulated Functions
2.3.2. Downregulated Functions
2.4. Metafun-NSCLC Web Tool
3. Discussion
4. Materials and Methods
4.1. Study Search and Selection
- Sex, disease stage, and smoking habit variables registered;
- RNA extracted directly from human lung biopsies;
- Both normal and lung adenocarcinoma samples available;
- Patients who had not undergone treatment before biopsy;
- Sample size of > 3 for case and control groups in both sexes.
4.2. Individual Transcriptomics Analysis
4.3. Functional Meta-Analysis
4.4. Metafun-NSCLC Web Tool
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Computer Code and Software
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemal, A.; Miller, K.D.; Ma, J.; Siegel, R.L.; Fedewa, S.A.; Islami, F.; Devesa, S.S.; Thun, M.J. Higher Lung Cancer Incidence in Young Women Than Young Men in the United States. N. Engl. J. Med. 2018, 378, 1999–2009. [Google Scholar] [CrossRef]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Conforti, F.; Pala, L.; Bagnardi, V.; De Pas, T.; Martinetti, M.; Viale, G.; Gelber, R.D.; Goldhirsch, A. Cancer immunotherapy efficacy and patients’ sex: A systematic review and meta-analysis. Lancet Oncol. 2018, 19, 737–746. [Google Scholar] [CrossRef]
- Barquín, M.; Calvo, V.; García-García, F.; Nuñez, B.; Sánchez-Herrero, E.; Serna-Blasco, R.; Auglytė, M.; Carcereny, E.; Rodriguez-Abreu, D.; López Castro, R.; et al. Sex is a strong prognostic factor in stage IV non-small-cell lung cancer patients and should be considered in survival rate estimation. Cancer Epidemiol. 2020, 67, 101737. [Google Scholar] [CrossRef]
- Wheatley-Price, P.; Blackhall, F.; Lee, S.-M.; Ma, C.; Ashcroft, L.; Jitlal, M.; Qian, W.; Hackshaw, A.; Rudd, R.; Booton, R.; et al. The influence of sex and histology on outcomes in non-small-cell lung cancer: A pooled analysis of five randomized trials. Ann. Oncol. 2010, 21, 2023–2028. [Google Scholar] [CrossRef]
- Fasco, M.J.; Hurteau, G.J.; Spivack, S.D. Gender-dependent expression of alpha and beta estrogen receptors in human nontumor and tumor lung tissue. Mol. Cell. Endocrinol. 2002, 188, 125–140. [Google Scholar] [CrossRef]
- Stabile, L.P.; Davis, A.L.G.; Gubish, C.T.; Hopkins, T.M.; Luketich, J.D.; Christie, N.; Finkelstein, S.; Siegfried, J.M. Human Non-Small Cell Lung Tumors and Cells Derived from Normal Lung Express Both Estrogen Receptor α and β and Show Biological Responses to Estrogen. Cancer Res. 2002, 62, 2141–2150. [Google Scholar]
- Kawai, H.; Ishii, A.; Washiya, K.; Konno, T.; Kon, H.; Yamaya, C.; Ono, I.; Minamiya, Y.; Ogawa, J. Estrogen Receptor α and β are Prognostic Factors in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2005, 11, 5084–5089. [Google Scholar] [CrossRef] [Green Version]
- Kligerman, S.; White, C. Epidemiology of Lung Cancer in Women: Risk Factors, Survival, and Screening. Am. J. Roentgenol. 2011, 196, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Bain, C.; Feskanich, D.; Speizer, F.E.; Thun, M.; Hertzmark, E.; Rosner, B.A.; Colditz, G.A. Lung Cancer Rates in Men and Women With Comparable Histories of Smoking. JNCI J. Natl. Cancer Inst. 2004, 96, 826–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- North, C.M.; Christiani, D.C. Women and Lung Cancer: What is New? Semin. Thorac. Cardiovasc. Surg. 2013, 25, 87–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.A.; Riely, G.J.; Van Schil, P.E.; et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araujo, J.M.; Prado, A.; Cardenas, N.K.; Zaharia, M.; Dyer, R.; Doimi, F.; Bravo, L.; Pinillos, L.; Morante, Z.; Aguilar, A.; et al. Repeated observation of immune gene sets enrichment in women with non-small cell lung cancer. Oncotarget 2016, 7, 20282–20292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, K.; Li, N.; Yang, M.; Li, W. Identification of Key Genes and Pathways in Female Lung Cancer Patients Who Never Smoked by a Bioinformatics Analysis. J. Cancer 2019, 10, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Liu, L.; Chen, H.; Wang, Y.; Xu, Y.; Mao, H.; Li, J.; Mills, G.B.; Shu, Y.; Li, L.; et al. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer Cell 2016, 29, 711–722. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; He, C.-L.; Li, W.-X.; Zhang, R.-X.; Duan, Y. Transcriptome analysis reveals gender-specific differences in overall metabolic response of male and female patients in lung adenocarcinoma. PLoS ONE 2020, 15, e0230796. [Google Scholar] [CrossRef] [Green Version]
- Normand, S.-L.T. Meta-analysis: Formulating, evaluating, combining, and reporting. Stat. Med. 1999, 18, 321–359. [Google Scholar] [CrossRef]
- Higgins, J.; Thomas, J. Cochrane Handbook for Systematic Reviews of Interventions, 2nd ed.; Wiley-Blackwell: Chichester, UK, 2019. [Google Scholar]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.T.; Dracheva, T.; Rotunno, M.; Figueroa, J.D.; Liu, H.; Dasgupta, A.; Mann, F.E.; Fukuoka, J.; Hames, M.; Bergen, A.W.; et al. Gene Expression Signature of Cigarette Smoking and Its Role in Lung Adenocarcinoma Development and Survival. PLoS ONE 2008, 3, e1651. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Aerts, J.; den Hamer, B.; van IJcken, W.; den Bakker, M.; Riegman, P.; van der Leest, C.; van der Spek, P.; Foekens, J.A.; Hoogsteden, H.C.; et al. Gene Expression-Based Classification of Non-Small Cell Lung Carcinomas and Survival Prediction. PLoS ONE 2010, 5, e10312. [Google Scholar] [CrossRef] [PubMed]
- Okayama, H.; Kohno, T.; Ishii, Y.; Shimada, Y.; Shiraishi, K.; Iwakawa, R.; Furuta, K.; Tsuta, K.; Shibata, T.; Yamamoto, S.; et al. Identification of Genes Upregulated in ALK-Positive and EGFR/KRAS/ALK-Negative Lung Adenocarcinomas. Cancer Res. 2012, 72, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, M.; Yamaguchi, R.; Nakata, A.; Kohno, T.; Nagasaki, M.; Shimamura, T.; Imoto, S.; Saito, A.; Ueno, K.; Hatanaka, Y.; et al. Epidermal Growth Factor Receptor Tyrosine Kinase Defines Critical Prognostic Genes of Stage I Lung Adenocarcinoma. PLoS ONE 2012, 7, e43923. [Google Scholar] [CrossRef]
- Selamat, S.A.; Chung, B.S.; Girard, L.; Zhang, W.; Zhang, Y.; Campan, M.; Siegmund, K.D.; Koss, M.N.; Hagen, J.A.; Lam, W.L.; et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012, 22, 1197–1211. [Google Scholar] [CrossRef] [Green Version]
- Robles, A.I.; Arai, E.; Mathé, E.A.; Okayama, H.; Schetter, A.J.; Brown, D.; Petersen, D.; Bowman, E.D.; Noro, R.; Welsh, J.A.; et al. An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma Based on mRNA, microRNA, and DNA Methylation Biomarkers. J. Thorac. Oncol. 2015, 10, 1037–1048. [Google Scholar] [CrossRef] [Green Version]
- Girard, L.; Rodriguez-Canales, J.; Behrens, C.; Thompson, D.M.; Botros, I.W.; Tang, H.; Xie, Y.; Rekhtman, N.; Travis, W.D.; Wistuba, I.I.; et al. An Expression Signature as an Aid to the Histologic Classification of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2016, 22, 4880. [Google Scholar] [CrossRef] [Green Version]
- Mezheyeuski, A.; Bergsland, C.H.; Backman, M.; Djureinovic, D.; Sjöblom, T.; Bruun, J.; Micke, P. Multispectral imaging for quantitative and compartment-specific immune infiltrates reveals distinct immune profiles that classify lung cancer patients. J. Pathol. 2018, 244, 421–431. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, L.; Eckloff, B.W.; Deng, B.; Wang, Y.; Wampfler, J.A.; Jang, J.; Wieben, E.D.; Jen, J.; You, M.; et al. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med. Genom. 2014, 7, 486. [Google Scholar] [CrossRef] [Green Version]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, T.P. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.B.; Dawsey, S.M.; Freedman, N.D.; Inskip, P.D.; Wichner, S.M.; Quraishi, S.M.; Devesa, S.S.; McGlynn, K.A. Sex Disparities in Cancer Incidence by Period and Age. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1174–1182. [Google Scholar] [CrossRef] [Green Version]
- Lukey, M.J.; Katt, W.P.; Cerione, R.A. Targeting amino acid metabolism for cancer therapy. Drug Discov. Today 2017, 22, 796–804. [Google Scholar] [CrossRef] [Green Version]
- Poff, A.; Koutnik, A.P.; Egan, K.M.; Sahebjam, S.; D’Agostino, D.; Kumar, N.B. Targeting the Warburg effect for cancer treatment: Ketogenic diets for management of glioma. Semin. Cancer Biol. 2019, 56, 135–148. [Google Scholar] [CrossRef]
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Gaignard, P.; Savouroux, S.; Liere, P.; Pianos, A.; Thérond, P.; Schumacher, M.; Slama, A.; Guennoun, R. Effect of Sex Differences on Brain Mitochondrial Function and Its Suppression by Ovariectomy and in Aged Mice. Endocrinology 2015, 156, 2893–2904. [Google Scholar] [CrossRef]
- Mervic, L. Time Course and Pattern of Metastasis of Cutaneous Melanoma Differ between Men and Women. PLoS ONE 2012, 7, e32955. [Google Scholar] [CrossRef] [Green Version]
- Pal, S.K.; Hurria, A. Impact of Age, Sex, and Comorbidity on Cancer Therapy and Disease Progression. J. Clin. Oncol. 2010, 28, 4086–4093. [Google Scholar] [CrossRef]
- Sun, S.; Schiller, J.H.; Gazdar, A.F. Lung cancer in never smokers—A different disease. Nat. Rev. Cancer 2007, 7, 778–790. [Google Scholar] [CrossRef]
- Kim, A.M.; Tingen, C.M.; Woodruff, T.K. Sex bias in trials and treatment must end. Nature 2010, 465, 688–689. [Google Scholar] [CrossRef] [PubMed]
- Woitowich, N.C.; Beery, A.; Woodruff, T. A 10-year follow-up study of sex inclusion in the biological sciences. eLife 2020, 9, e56344. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinay, D.S.; Ryan, E.P.; Pawelec, G.; Talib, W.H.; Stagg, J.; Elkord, E.; Lichtor, T.; Decker, W.K.; Whelan, R.L.; Kumara, H.M.C.S.; et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 2015, 35, S185–S198. [Google Scholar] [CrossRef]
- Altorki, N.K.; Markowitz, G.J.; Gao, D.; Port, J.L.; Saxena, A.; Stiles, B.; McGraw, T.; Mittal, V. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat. Rev. Cancer 2019, 19, 9–31. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Jing, Y.; Li, L.; Mills, G.B.; Diao, L.; Liu, H.; Han, L. Sex-associated molecular differences for cancer immunotherapy. Nat. Commun. 2020, 11, 1779. [Google Scholar] [CrossRef] [Green Version]
- Zeng, D.-Q.; Yu, Y.-F.; Ou, Q.-Y.; Li, X.-Y.; Zhong, R.-Z.; Xie, C.-M.; Hu, Q.-G. Prognostic and predictive value of tumor-infiltrating lymphocytes for clinical therapeutic research in patients with non-small cell lung cancer. Oncotarget 2016, 7, 13765–13781. [Google Scholar] [CrossRef] [Green Version]
- Ye, S.-L.; Li, X.-Y.; Zhao, K.; Feng, T. High expression of CD8 predicts favorable prognosis in patients with lung adenocarcinoma: A cohort study. Medicine 2017, 96, e6472. [Google Scholar] [CrossRef]
- Li, S.; Wang, Z.; Li, X.-J. Notch signaling pathway suppresses CD8+ T cells activity in patients with lung adenocarcinoma. Int. Immunopharmacol. 2018, 63, 129–136. [Google Scholar] [CrossRef]
- Abbas, A.K.; Trotta, E.; Simeonov, D.; Marson, A.; Bluestone, J.A. Revisiting IL-2: Biology and therapeutic prospects. Sci. Immunol. 2018, 3, eaat1482. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Lu, J.; Zeng, C.; Yang, J.; Huang, B.; Zhang, N.; Li, L.; Fu, X. IL-10 suppresses IFN-γ-mediated signaling in lung adenocarcinoma. Clin. Exp. Med. 2020, 20, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Vahl, J.M.; Friedrich, J.; Mittler, S.; Trump, S.; Heim, L.; Kachler, K.; Balabko, L.; Fuhrich, N.; Geppert, C.-I.; Trufa, D.I.; et al. Interleukin-10-regulated tumour tolerance in non-small cell lung cancer. Br. J. Cancer 2017, 117, 1644–1655. [Google Scholar] [CrossRef] [Green Version]
- Miotto, D.; Cascio, N.L.; Stendardo, M.; Querzoli, P.; Pedriali, M.; De Rosa, E.; Fabbri, L.M.; Mapp, C.E.; Boschetto, P. CD8+ T cells expressing IL-10 are associated with a favourable prognosis in lung cancer. Lung Cancer 2010, 69, 355–360. [Google Scholar] [CrossRef]
- Emmerich, J.; Mumm, J.B.; Chan, I.H.; LaFace, D.; Truong, H.; McClanahan, T.; Gorman, D.M.; Oft, M. IL-10 Directly Activates and Expands Tumor-Resident CD8+ T Cells without De Novo Infiltration from Secondary Lymphoid Organs. Cancer Res. 2012, 72, 3570. [Google Scholar] [CrossRef] [Green Version]
- Shang, G.-S.; Liu, L.; Qin, Y.-W. IL-6 and TNF-α promote metastasis of lung cancer by inducing epithelial-mesenchymal transition. Oncol. Lett. 2017, 13, 4657–4660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, E.M.; Mariano, V.S.; Pastrez, P.R.A.; Pinto, M.C.; Castro, A.G.; Syrjanen, K.J.; Longatto-Filho, A. High systemic IL-6 is associated with worse prognosis in patients with non-small cell lung cancer. PLoS ONE 2017, 12, e0181125. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.M.; Varghese, S.; Xu, H.; Alexander, H.R. Interleukin-1 and cancer progression: The emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment. J. Transl. Med. 2006, 4, 48. [Google Scholar] [CrossRef] [Green Version]
- Gottschlich, A.; Endres, S.; Kobold, S. Can we use interleukin-1β blockade for lung cancer treatment? Transl. Lung Cancer Res. 2018, 7, S160–S164. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Xu, J.; Huang, Q.; Han, J.; Duan, L.; Fan, J.; Lv, Z.; Guo, M.; Hu, G.; Chen, L.; et al. The Role of Interleukin-17 in Lung Cancer. Mediat. Inflamm. 2016, 2016, 8494079. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Qiao, W.; Jiang, Y.; Zhu, M.; Shao, J.; Ren, P.; Liu, D.; Li, W. Effect of sex on the efficacy of patients receiving immune checkpoint inhibitors in advanced non-small cell lung cancer. Cancer Med. 2019, 8, 4023–4031. [Google Scholar] [CrossRef] [Green Version]
- Conforti, F.; Pala, L.; Bagnardi, V.; Viale, G.; De Pas, T.; Pagan, E.; Pennacchioli, E.; Cocorocchio, E.; Ferrucci, P.F.; De Marinis, F.; et al. Sex-Based Heterogeneity in Response to Lung Cancer Immunotherapy: A Systematic Review and Meta-Analysis. JNCI J. Natl. Cancer Inst. 2019, 111, 772–781. [Google Scholar] [CrossRef]
- Vijayan, D.; Smyth, M.J.; Teng, M.W.L. Purinergic Receptors: Novel Targets for Cancer Immunotherapy. In Oncoimmunology: A Practical Guide for Cancer Immunotherapy; Zitvogel, L., Kroemer, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 115–141. [Google Scholar]
- Crain, J.M.; Nikodemova, M.; Watters, J.J. Expression of P2 nucleotide receptors varies with age and sex in murine brain microglia. J. Neuroinflamm. 2009, 6, 24. [Google Scholar] [CrossRef] [Green Version]
- Savio, L.E.B.; de Andrade Mello, P.; da Silva, C.G.; Coutinho-Silva, R. The P2X7 Receptor in Inflammatory Diseases: Angel or Demon? Front. Pharmacol. 2018, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.-P.; Zhang, X.-X.; Jiang, S.-H.; Tao, L.-Y.; Li, Q.; Zhu, L.-L.; Yang, M.-W.; Huo, Y.-M.; Jiang, Y.-S.; Tian, G.-A.; et al. Targeting Purinergic Receptor P2Y2 Prevents the Growth of Pancreatic Ductal Adenocarcinoma by Inhibiting Cancer Cell Glycolysis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 1318–1330. [Google Scholar] [CrossRef] [Green Version]
- Tak, E.; Jun, D.Y.; Kim, S.-H.; Park, G.-C.; Lee, J.; Hwang, S.; Song, G.-W.; Lee, S.-G. Upregulation of P2Y2 nucleotide receptor in human hepatocellular carcinoma cells. J. Int. Med. Res. 2016, 44, 1234–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asif, A.; Khalid, M.; Manzoor, S.; Ahmad, H.; Rehman, A.U. Role of purinergic receptors in hepatobiliary carcinoma in Pakistani population: An approach towards proinflammatory role of P2X4 and P2X7 receptors. Purinergic Signal. 2019, 15, 367–374. [Google Scholar] [CrossRef]
- Xu, M.; Almasi, S.; Yang, Y.; Yan, C.; Sterea, A.M.; Rizvi Syeda, A.K.; Shen, B.; Richard Derek, C.; Huang, P.; Gujar, S.; et al. The lysosomal TRPML1 channel regulates triple negative breast cancer development by promoting mTORC1 and purinergic signaling pathways. Cell Calcium 2019, 79, 80–88. [Google Scholar] [CrossRef]
- Zhu, Y.; Shao, X.; Wang, X.; Liu, L.; Liang, H. Sex disparities in cancer. Cancer Lett. 2019, 466, 35–38. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Senescence and immortalization: Role of telomeres and telomerase. Carcinogenesis 2005, 26, 867–874. [Google Scholar] [CrossRef]
- Datta, A.; Brosh, R.M. New Insights Into DNA Helicases as Druggable Targets for Cancer Therapy. Front. Mol. Biosci. 2018, 5, 59. [Google Scholar] [CrossRef]
- Luo, J.; Dai, X.; Hu, H.; Chen, J.; Zhao, L.; Yang, C.; Sun, J.; Zhang, L.; Wang, Q.; Xu, S.; et al. Fluzoparib increases radiation sensitivity of non-small cell lung cancer (NSCLC) cells without BRCA1/2 mutation, a novel PARP1 inhibitor undergoing clinical trials. J. Cancer Res. Clin. Oncol. 2020, 146, 721–737. [Google Scholar] [CrossRef]
- Abbotts, R.; Topper, M.J.; Biondi, C.; Fontaine, D.; Goswami, R.; Stojanovic, L.; Choi, E.Y.; McLaughlin, L.; Kogan, A.A.; Xia, L.; et al. DNA methyltransferase inhibitors induce a BRCAness phenotype that sensitizes NSCLC to PARP inhibitor and ionizing radiation. Proc. Natl. Acad. Sci. USA 2019, 116, 22609. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Athar, A.; Füllgrabe, A.; George, N.; Iqbal, H.; Huerta, L.; Ali, A.; Snow, C.; Fonseca, N.A.; Petryszak, R.; Papatheodorou, I.; et al. ArrayExpress update-from bulk to single-cell expression data. Nucleic Acids Res. 2019, 47, D711–D715. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maglott, D.; Ostell, J.; Pruitt, K.D.; Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005, 33, D54–D58. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Montaner, D.; Dopazo, J. Multidimensional Gene Set Analysis of Genomic Data. PLoS ONE 2010, 5, e10348. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef]
- García-García, F. Methods of Functional Enrichment Analysis in Genomic Studies; Universitat de València: València, Spain, 2016. [Google Scholar]
- Viechtbauer, W. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw. 2010, 36, 104408. [Google Scholar] [CrossRef] [Green Version]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control. Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Egger, M. Funnel plots for detecting bias in meta-analysis: Guidelines on choice of axis. J. Clin. Epidemiol. 2001, 54, 1046–1055. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]





| Study | Platform | Publication |
|---|---|---|
| GSE10072 | Affymetrix Human Genome U133A Array | [23] |
| GSE19188 | Affymetrix Human Genome U133 Plus 2.0 Array | [24] |
| GSE31210 | Affymetrix Human Genome U133 Plus 2.0 Array | [25,26] |
| GSE32863 | Illumina HumanWG-6 v3.0 Expression BeadChip | [27] |
| GSE63459 | Illumina HumanRef-8 v3.0 Expression BeadChip | [28] |
| GSE75037 | Illumina HumanWG-6 v3.0 Expression BeadChip | [29] |
| GSE81089 | Illumina HiSeq 2500 | [30] |
| GSE87340 | Illumina HiSeq 2000 | [31] |
| TCGA | Illumina HiSeq 2000 | [22] |
| Study | Significant GO BP Terms | Significant GO MF Terms | Significant GO CC Terms | Significant KEGG Pathways | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Total | Up | Down | Total | Up | Down | Total | Up | Down | Total | |
| GSE10072 | 26 | 153 | 179 | 29 | 8 | 37 | 16 | 40 | 56 | 1 | 5 | 6 |
| GSE19188 | 7 | 12 | 19 | 0 | 3 | 3 | 11 | 20 | 31 | 1 | 4 | 5 |
| GSE31210 | 21 | 2 | 23 | 0 | 0 | 0 | 4 | 0 | 4 | 8 | 2 | 10 |
| GSE32863 | 428 | 51 | 479 | 28 | 5 | 33 | 40 | 41 | 81 | 27 | 6 | 33 |
| GSE63459 | 0 | 26 | 26 | 0 | 3 | 3 | 8 | 27 | 35 | 1 | 4 | 5 |
| GSE75037 | 245 | 35 | 280 | 14 | 4 | 18 | 14 | 21 | 35 | 22 | 5 | 27 |
| GSE81089 | 2 | 1 | 3 | 7 | 1 | 8 | 7 | 4 | 11 | 1 | 1 | 2 |
| GSE87340 | 178 | 62 | 240 | 3 | 0 | 3 | 48 | 13 | 61 | 26 | 1 | 27 |
| TCGA | 294 | 228 | 522 | 28 | 30 | 58 | 21 | 70 | 91 | 30 | 17 | 47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Díez, I.; Hidalgo, M.R.; Malmierca-Merlo, P.; Andreu, Z.; Romera-Giner, S.; Farràs, R.; de la Iglesia-Vayá, M.; Provencio, M.; Romero, A.; García-García, F. Functional Signatures in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis of Sex-Based Differences in Transcriptomic Studies. Cancers 2021, 13, 143. https://doi.org/10.3390/cancers13010143
Pérez-Díez I, Hidalgo MR, Malmierca-Merlo P, Andreu Z, Romera-Giner S, Farràs R, de la Iglesia-Vayá M, Provencio M, Romero A, García-García F. Functional Signatures in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis of Sex-Based Differences in Transcriptomic Studies. Cancers. 2021; 13(1):143. https://doi.org/10.3390/cancers13010143
Chicago/Turabian StylePérez-Díez, Irene, Marta R. Hidalgo, Pablo Malmierca-Merlo, Zoraida Andreu, Sergio Romera-Giner, Rosa Farràs, María de la Iglesia-Vayá, Mariano Provencio, Atocha Romero, and Francisco García-García. 2021. "Functional Signatures in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis of Sex-Based Differences in Transcriptomic Studies" Cancers 13, no. 1: 143. https://doi.org/10.3390/cancers13010143
APA StylePérez-Díez, I., Hidalgo, M. R., Malmierca-Merlo, P., Andreu, Z., Romera-Giner, S., Farràs, R., de la Iglesia-Vayá, M., Provencio, M., Romero, A., & García-García, F. (2021). Functional Signatures in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis of Sex-Based Differences in Transcriptomic Studies. Cancers, 13(1), 143. https://doi.org/10.3390/cancers13010143